Abstract
Relaxation of imprinting at the insulin-like growth factor II (IFG-II)/H19 locus is a major mechanism involved in the onset of sporadic Wilms tumor and several other embryonal tumors. The high prevalence of histologically abnormal foci in kidney adjacent to Wilms tumors suggests that tumor-predisposing genetic/epigenetic lesion might also be found at high frequency in Wilms tumor-bearing kidneys. Focusing on Wilms tumors with relaxation of IFG-II imprinting, we determined the frequency of epigenetic change at the IFG-II/H19 locus in adjacent kidney. In all kidneys adjacent to these Wilms tumors, we detected substantial mosaicism for a population of cells with relaxation of IFG-II imprinting and biallelic H19 methylation, regardless of whether the patient had a tumor-predisposing syndrome or not. The high proportion of epigenetically modified cells among “normal” tissue indicates that the epigenetic error occurred very early in development, before the onset of Wilms tumor. Not only does this suggest that the major Wilms tumor-predisposing event occurs within the first few days of development, but it also suggests that sporadic Wilms tumor may represent one end of a spectrum of overgrowth disorders characterized by mosaic epigenetic change at the IFG-II/H19 locus.
Keywords: genomic imprinting, DNA methylation, Wilms tumor
Several lines of evidence have implicated insulin-like growth factor II (IFG-II) in the onset of Wilms tumor. IFG-II is transcribed at high levels both in Wilms tumors and the embryonal renal blastema from which the tumors arise (1, 2). The IFG-II gene is located within a region of frequent loss of heterozygosity (LOH), which leads to duplication of the active paternal copy (3–5). Furthermore, the imprinting of IFG-II is relaxed in approximately one-third of Wilms tumors, resulting in transcription of IFG-II from both alleles (6, 7). Activation of the maternal IFG-II allele is accompanied by DNA methylation and transcriptional silencing of the adjacent H19 maternal allele (8–10). The coordinate and opposite expression patterns of H19 and IFG-II suggest that these genes constitute a single epigenetic locus (11).
Epigenetic changes at IFG-II/H19 are implicated as one of the earliest events leading to Wilms tumor onset. In support of this are observations that in predisposed individuals with the Beckwith–Wiedemann syndrome (BWS) or somatic overgrowth, IFG-II is expressed biallelically and H19 is methylated on both alleles in some cases (12–15). Immature structures composed of blastemal rests or immature glomeruli, are also often found in the kidney adjacent to Wilms tumors and the “normal” renal tissue of individuals with BWS (16). In view of these findings, it seems likely that the normal kidney tissue of sporadic Wilms tumor patients may carry genetic or epigenetic abnormalities that predispose it to tumorigenesis.
Relaxation (loss) of IFG-II/H19 imprinting (LOI) is a good candidate for one of the main initial events occurring in the developing kidney of individuals with sporadic Wilms tumor. We have investigated this possibility by examining the normal kidney tissues adjacent to Wilms tumors with relaxed IFG-II/H19 imprinting. This involved estimating the extent of biallelic H19 methylation in normal kidney tissues and correlating this with the amount of biallelic IFG-II expression. These experiments showed that in every kidney tissue examined, there was significant mosaicism for cells with relaxed IFG-II/H19 imprinting.
MATERIALS AND METHODS
Tissues.
Twenty-three unilateral Wilms tumors were classified according to whether they had biallelic IFG-II expression or normal IFG-II expression as described (7, 10). LOH from 11p15 was measured by comparing the genotype of normal DNA (peripheral blood or kidney) with tumor DNA using an ApaI restriction fragment length polymorphism in IFG-II exon 9 (17) or the tyrosine hydroxylase microsatellite polymorphism (18). Of the eight kidneys adjacent to Wilms tumors with IFG-II LOI, one was from a child with generalized somatic overgrowth (13) (case 2 in Table 1), another was from a child with lower limb hemihyperplasia (case 3 in Table 1), and the remainder had no syndromal features.
Table 1.
Results of mosaicism by H19 methylation Southern blots and IFG-II SNuPE assay in normal kidney and blood samples of Wilms tumour patients
| Sources | Ratio of H19 methylation,* mean ± SD | % mosaicism† (H19 Southern blots) | % mosaicism‡ (IFG-II SNuPE) |
|---|---|---|---|
| Adult normal kidney | 1.01 ± 0.06 [10] | ||
| NonLOI, nonLOH kidney | 0.97 ± 0.05 [8] | 0 | 0 [6] |
| LOI kidney | |||
| Case 1 | 3.59 ± 0.37 (3) | 56 | 76 |
| Case 2 | 5.40 ± 0.25 (3) | 69 | 85 |
| Case 3 | 3.36 ± 0.32 (3) | 54 | 66 |
| Case 4 | 2.34 ± 0.24 (3) | 40 | NI |
| Case 5 | 2.20 ± 0.15 (2) | 38 | 23 |
| Case 6 | 1.45 ± 0.18 (3) | 18 | 26 |
| Case 7 | 1.15 ± 0.07 (3) | 7 | 6 |
| Case 8 | 1.29 ± 0.10 (2) | 13 | 9 |
| LOH kidney | |||
| Case 9 | 3.52 ± 0.13 (3) | 56 | NA |
| Cases 10–15 | 1.04 ± 0.07 (2) | <5 | NA |
| Normal blood | 0.99 ± 0.06 [4] | 0 [3] | |
| Blood from nonLOI, nonLOH | 0.90 ± 0.09 [6] | 0 | § |
| Blood from LOI | |||
| Case 1 | 0.92 ± 0.08 (2) | 0 | 0 |
| Case 2 | 2.43 ± 0.21 (3) | 42 | 32 |
| Case 3 | 0.98 ± 0.07 (2) | 0 | 0 |
| Case 6 | 0.89 ± 0.07 (2) | 0 | § |
| Blood from LOH | |||
| Case 9 | 0.95 ± 0.10 (2) | 0 | NA |
| Cases 10–13 | 0.88 ± 0.14 (2) | 0 | NA |
The number of cases examined is shown in square brackets. The number of independent experiments analyzed for each case is shown in parentheses. NI, not informative at the exon 9 ApaI polymorphism; NA, not applicable. The ratios of intensities between methylated and unmethylated alleles are summarized. The average of the ratio of normal individual cases was standardized to 1.00 in each blot.
Proportion of cells with H19 methylated biallelically.
Proportion of cells with biallelic IFG-II expression. Cases which are polymorphic at the exon 9 ApaI site were used. Each sample was analyzed by two or more independent SNuPE assays. The mean value is shown for each case.
Samples were not available.
H19 Promoter Methylation.
DNA samples (5 μg) were digested with both RsaI and HpaII, electrophoresed in 1.2% agarose, and transferred to Hybond N+ (Amersham). Filters were hybridized at 65°C with a 32P-labeled 383-bp DNA fragment from the H19 promoter region (see Fig. 1). Band intensities were quantitated using a Fuji BAS-1500 Bio-imaging analyzer.
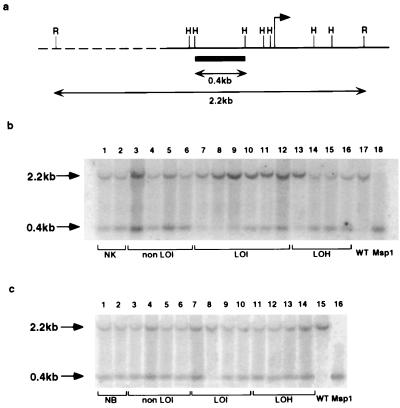
Figure 1.
(a) HpaII methylation-sensitive restriction sites in the H19 promoter region. The arrow indicates the transcription start and the probe is represented by solid box. R, RsaI site; H, HpaII site. (b) H19 Southern blot. DNA samples from the following tissues were analyzed: normal kidney from renal cell carcinomas (lanes 1 and 2); kidney from Wilms tumors with normal IFG-II imprinting (lanes 3–6); kidney from Wilms tumors with relaxed IFG-II/H19 imprinting (lanes 7–12, corresponding to cases 1–6); kidney from Wilms tumors with 11p15 LOH (lane 13, case 9, and lanes 14–16, cases 10–12, respectively); and a representative Wilms tumor with relaxed IFG-II/H19 imprinting (lanes 17 and 18). DNA samples in lanes 1–17 were digested with RsaI/HpaII, and the sample in lane 18, with RsaI/MspI. The average ratio between the upper (methylated) and lower (unmethylated) bands in normal kidney was standardized to 1.00 using 10 DNA samples from normal kidney tissues adjacent to adult renal cell carcinomas (data not shown). (c) H19 Southern blots of peripheral blood DNA. The following samples were analyzed: normal individuals (lanes 1 and 2); patients with tumors showing normal IFG-II/H19 imprinting (lanes 3–6); patients with tumors showing relaxed IFG-II/H19 imprinting (lanes 7–10, cases 1, 2, 3, and 6, respectively); and patients with tumors showing 11p15 LOH (lanes 11–14, cases 9–12, respectively). A Wilms tumor with loss of IFG-II imprinting was used as a control (lanes 15 and 16). Samples 1–15 were digested with RsaI and HpaII. Lane 16 was digested with MspI. All DNA samples had a similar unmethylated/methylated H19 allele ratio except for case 8 (see Table 1).
Reverse Transcriptase–PCR.
Reverse transcriptase–PCR of IFG-II exons 8 and 9 was done as described (7). PWO polymerase and primers P1 and P3 were used to amplify a region of exons 8 and 9 encompassing a polymorphic ApaI site (17). Forty cycles of amplification were done using 10 sec at 95°C, 10 sec at 56°C, and 1 min at 72°C. The cDNA-PCR products were reamplified in some cases with low expression. Amplified cDNA products were purified by gel electrophoresis.
Single Nucleotide Primer Extension (SNuPE) Assay.
A schematic of the reaction is shown in Fig. 2. This assay is based on a single nucleotide difference between allelic RNAs as described (19, 20). The sequence difference was located within an ApaI polymorphism in IFG-II exon 9 (17). The SNuPE reaction used 10 ng of purified cDNA-PCR fragment and consisted of one cycle of 95°C for 30 sec, 42°C for 30 sec, and 72°C for 1 min. Sample were electrophoresed on a 15% denaturing polyacrylamide gel, and bands were quantified by phosphoimager analysis.
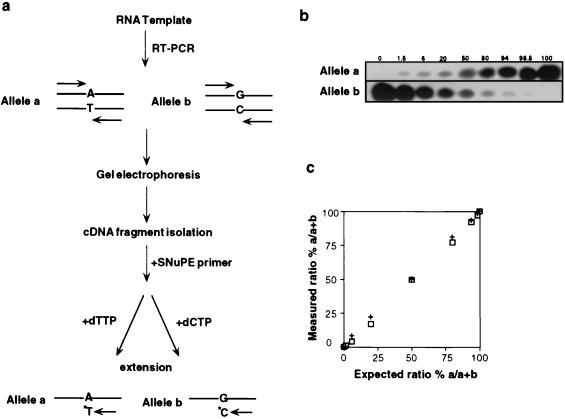
Figure 2.
(a) Outline of Reverse transcriptase–PCR SNuPE assay. (b) Evaluation of SNuPE assay. Linearity was examined by combining varying amounts of DNA-PCR products amplified from either an a/a homozygote or b/b homozygote. PCR products were mixed keeping the total DNA constant at 20 ng. SNuPE assay was then done as described in Materials and Methods, and alleles a and b intensities quantified by phosphoimager analysis. (c) Assessment of SNuPE assay linearity. The expected ratio % a/a + b was plotted as a function of the measured ratio % a/a + b calculated from Fig. 1b. The measured ratio was normalized to 1:1 using the allele a/b 50:50 ratio in Fig. 1b. This experiment was done twice (+, □).
Quantification of IFG-II Biallelic Expression and H19 Methylation.
The percentage of cells with H19 biallelic methylation was calculated using 100(x − 1)/(x + 1), where x is the methylated H19/unmethylated H19 allele ratio determined from Fig. 1. The percentage of cells with biallelic expression of IFG-II, assuming equal expression levels from active alleles, was extrapolated from the SNuPE analyses using the following expressions: 100(a/b) or 100(b/a), where a and b are the intensities determined from phosphoimager analysis after subtraction of background intensities, and the standardization of a/b heterozygote DNA to a 1:1 ratio.
RESULTS
H19 Methylation Is Mosaic in Kidney and Peripheral Blood of Wilms Tumor Patients.
The methylation status of part of the H19 promoter was examined in the unaffected adjacent kidney and peripheral blood of Wilms tumor patients to determine whether aberrant methylation of H19 was present in normal tissues.
A DNA probe flanked by HpaII/MspI sites was used such that after digestion with HpaII and RsaI, the methylated and unmethylated alleles of H19 could be distinguished (Fig. 1a). In DNA samples from kidneys adjacent to Wilms tumors with normal IFG-II imprinting, the ratios between the methylated allele and unmethylated alleles were similar and standardized to unity (Fig. 1b, lanes 3–6). Ten additional kidney samples associated with renal cell carcinoma were also analyzed as a control tissue, and a similar 1:1 ratio was obtained (Table 1). In contrast, in all eight samples derived from kidneys adjacent to tumors with biallelic IFG-II expression (and loss of H19 expression), there was a significantly increased proportion of the methylated H19 allele (six DNA samples are shown in Fig. 1a, lanes 7–12). In these kidney DNA samples, 7–69% of cells was estimated to be methylated on both H19 alleles (Table 1). These data suggest that the maternal H19 allele was methylated early in kidney development and led to a mosaic population of cells in which H19 was either monoallelically or biallelically methylated. In comparison, mosaicism was not detectable in kidney tissues adjacent to 10 renal cell carcinomas or eight Wilms tumors with normal IFG-II/H19 imprinting.
One kidney sample adjacent to a tumor with 11p15 LOH showed increased methylation on one H19 allele (Fig. 1a, lane 13; see Table 1 for detailed results). This finding is consistent with a previous report in which the DNA from normal tissues from 4 of 67 Wilms tumor patients was shown to have partial 11p15 LOH (21).
To determine whether the mosaicism was restricted to kidney tissues, we measured the H19 methylation status of peripheral blood DNA from Wilms tumor patients. Four cases of age-matched blood from individuals without Wilms tumor were used as controls. Increased H19 methylation was detected in one of four blood samples from patients having tumors with IFG-II LOI (case 2, Table 1). This individual was previously reported to have gigantism and constitutional relaxation of IFG-II imprinting (13, 15).
Loss of IFG-II Imprinting Is Mosaic in Normal Tissues of Wilms Tumor Patients.
To measure the extent of biallelic IFG-II expression, we developed a quantitative SNuPE assay that exploited the single nucleotide difference at the polymorphic ApaI site in exon 9 (17) (Fig. 2a). The sensitivity and linearity of the assay was first determined by mixing DNA samples containing varying ratios of the ApaI a and b alleles. As shown in Fig. 2b, primer extension with the relevant 32P-nucleotide resulted in specific labeling of either the a or the b IFG-II alleles. This assay was linear with a lower detection limit of less than 1.5% in a mixed allele population (Fig. 2c).
The IFG-II SNuPE assay was used to measure the extent of IFG-II biallelic expression in normal tissues of Wilms tumor patients. In the kidney adjacent to Wilms tumors with normal IFG-II imprinting, IFG-II was expressed exclusively from either the a or the b alleles, with no detectable expression from the opposite allele (Fig. 3a, lanes 5–9). However, in all kidney tissues adjacent to Wilms tumors with biallelic IFG-II expression, IFG-II was expressed to varying degrees from both alleles (Fig. 3a, lanes 10–14). Biallelic IFG-II expression was found in all seven kidneys examined, with the proportion of mosaicism was ranged from 6% to 85% (see Table 1 for detailed results). In four cases where parental DNA was available and informative, the more strongly expressed allele was of paternal origin (data not shown).
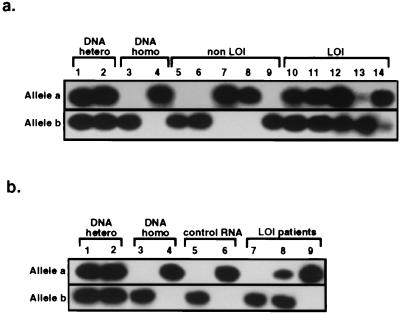
Figure 3.
Quantification of IFG-II allelic expression in normal tissues of patients with Wilms tumors by SNuPE assay. (a) Kidney samples. Samples analyzed were as follows: a/b heterozygote DNA (lanes 1 and 2); b/b and a/a homozygote DNA (lanes 3 and 4); RNA from normal kidney adjacent to Wilms tumors with normal IFG-II/H19 imprinting (lanes 5–9); and RNA from normal kidney adjacent to Wilms tumors with relaxed IFG-II/H19 imprinting (lanes 10–14, cases 1, 2, 3, 7, and 8, respectively). (b) Peripheral blood samples. The following samples were analyzed: a/b heterozygote DNA controls (lanes 1 and 2); b/b and a/a homozygote DNA (lanes 3 and 4, respectively); leukocyte RNA from unaffected individuals (lanes 5 and 6); and leukocyte RNA from patients with Wilms tumors with relaxed IFG-II/H19 imprinting (lanes 7–9, cases 1–3, respectively).
The pattern of IFG-II allelic expression was also studied in the peripheral blood leukocyte RNA from individuals with Wilms tumor. In one of three available informative blood samples, IFG-II was expressed biallelically, with the proportion of mosaicism estimated at 32% (case 2 in Fig. 3b, lane 8, and Table 1). In this same Wilms tumor patient, 42% of blood leukocytes was methylated on both H19 alleles (see above). In contrast, monoallelic RNA expression was detected in two other RNA samples (Fig. 3b, lanes 7 and 9) and in samples from unaffected individuals (Fig. 3b, lanes 5 and 6).
Correlation Between H19 Biallelic Methylation and IFG-II Biallelic Expression in Tumor-Bearing Kidney.
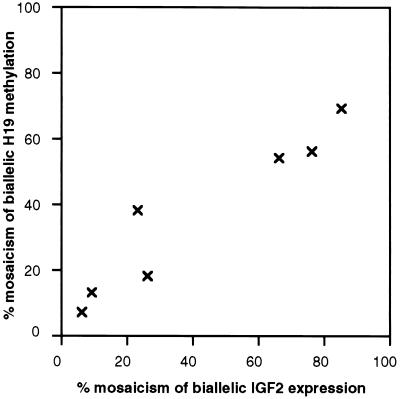
As described above, mosaic biallelic IFG-II expression and H19 methylation were detected in all kidney tissues adjacent to Wilms tumors with relaxed IFG-II/H19 imprinting. To determine the mechanism of the epigenetic changes in these normal tissues, whether coordinated or random, a comparison was made between the extent of H19 methylation and IFG-II biallelic expression. The data from Table 1 were plotted, and as shown in Fig. 4, there was a linear correlation between H19 methylation and biallelic IFG-II expression. These data are consistent with the presence of a population of cells within the normal kidney in which the relaxation of IFG-II and H19 imprinting was mechanistically coordinated. Furthermore, because the mosaicism was substantial, the event leading to mosaicism must have occurred during development of the embryo.
Figure 4.
Correlation between (i) percentage of cells mosaic for H19 biallelic methylation and (ii) estimated percentage of cells with biallelic IFG-II expression in kidney tissues adjacent to Wilms tumors with relaxed IFG-II/H19 imprinting. Individual values for the percent mosaicism were obtained from the H19 Southern blot (Fig. 1b) and the IFG-II SNuPE assay from Fig. 3a.
DISCUSSION
The major finding of this paper is that in every Wilms tumor in which imprinting of the IFG-II/H19 locus was relaxed (eight of eight), a substantial proportion of the cells in the adjacent normal kidney showed comparable epigenetic changes, i.e., biallelic IFG-II expression and biallelic H19 methylation. The high proportion of epigenetically modified cells in the normal kidney indicates that an epigenetic error must have occurred early in development, preceding the onset of Wilms tumor.
Approximately one-third of Wilms tumors have been documented to show relaxation of imprinting of IFG-II (LOI) with concomitant biallelic methylation of H19 (6–10). Previous reports have suggested that comparable epigenetic change at the IFG-II/H19 locus can occur in the somatic tissues of children with tumor-predisposing overgrowth syndrome (12, 14, 15). Although abnormal H19 methylation has previously been reported in a minority of Wilms tumor-bearing kidneys (9), our results indicate that epigenetic change affecting both H19 and IFG-II is invariably present in “normal” kidney adjacent to tumors with IFG-II LOI, regardless of whether the patient had a tumor-predisposing syndrome or not.
Given that Wilms tumors may also acquire a paternal epigenotype by maternal 11p15 LOH, it is of interest that Chao et al. have shown that normal kidney tissue adjacent to some Wilms tumors with 11p LOH were also mosaic for a population of cells with paternal 11p15 isoallelism (localized LOH) (21). Both 11p15 LOH with its paternal duplication of 11p15 and relaxation of IFG-II/H19 imprinting may therefore be regarded as functionally equivalent, very early events in tumorigenesis, both of which lead to enhanced production of IFG-II mRNA and the loss of H19 expression. It is plausible that enhanced IFG-II expression predisposes the developing kidney to malignant transformation by altering the proliferation/differentiation balance. The fundamental role of IFG-II imprinting in tumorigenesis has been highlighted by the observation that simian virus 40 T antigen mice display early focal activation of the silent IFG-II allele in the pancreas and subsequently develop pancreatic hyperplasia and pancreatoblastoma (22). Although it remains possible that the tumor-predisposing event is not the loss of IFG-II imprinting itself, the accumulated data indicate that this event must, at least, involve a gene in the imprinted 11p15 locus which contains H19 and IFG-II.
Nephrogenic rests in the adjacent kidney of many Wilms tumors are believed to represent precursor lesions. Rests have also been found in the kidneys of young infants, although at a much lower frequency (16), suggesting that the kidneys of Wilms tumor patients are predisposed to the development of this early lesion. Because the proportion of nephrogenic rests in the kidney tissues was very small (data not shown) compared with the proportion of cells with biallelic H19 methylation, the epigenetic modification must affect a much greater proportion of kidney cells than that constituted by rests. It is probable that the evolution of rests requires a second event, for example, mutations in WT1 as has previously been documented (23).
Relaxation of IFG-II imprinting and mosaic uniparental disomy of chromosome 11p has previously been found in the BWS 12, 14, 24–26), and in children with non-syndromic somatic overgrowth (15). BWS has a number of variable clinical features, which include somatic overgrowth and a predisposition to embryonal malignancies. The extent of and location of mosaicism of genetic/epigenetic changes may be responsible for the variable nature of diagnostic features of BWS. Mosaicism has previously been proposed within the context of a two-hit tumor suppressor gene model (27). We now provide evidence that relaxation of IFG-II/H19 imprinting can occur in somatic tissues without obvious growth abnormalities. Similarly, mosaic paternal uniparental disomy of 11p15 has been detected in a variety of tissues in two children with Wilms tumor who showed normal somatic growth and differentiation (21). Our findings suggest that all apparently sporadic Wilms tumors with relaxation of IFG-II imprinting are a manifestation of an early somatic epigenetic error and thus form one end of a spectrum of disease which, at the other extreme, is recognized as BWS. While it is formally possible that the observed mosaicism could be due to two populations of cells that express IFG-II from either the paternal or maternal allele, this is unlikely given that both H19 alleles were extensively methylated in some kidney samples.
Germ-line inactivation of the maternal H19 allele in mice has been shown to result in biallelic IFG-II expression and proportional overgrowth akin to features of the BWS (28). The mechanism by which the maternally inherited H19/IFG-II locus acquires a paternal epigenotype in Wilms tumor and BWS is, however, not yet understood. One possibility involves an “imprint transfer” mechanism from the methylated to unmethylated H19 allele (29). Alternatively, the function of a distant imprinting control element may be involved. In this regard, it has recently been shown in some BWS patients that a cluster of translocations involving the maternally inherited 11p15 region occur within 100 kb of the IFG-II gene (30). Two transcripts within this region correspond to untranslated RNAs (31) and may play a role in regulating the imprinting of this chromosomal region in a manner similar to transcripts of the SNRPN gene which function as a chromosome 15 imprinting switch (32).
Acknowledgments
We thank David Becroft and Michael Eccles for helpful comments. This work was supported by the Cancer Society of New Zealand, the Health Research Council (New Zealand), and the New Zealand Lotteries Grants Board.
ABBREVIATIONS
- BWS
Beckwith–Wiedemann syndrome
- IFG-II
insulin-like growth factor II
- LOH
loss of heterozygosity
- LOI
relaxation of IFG-II/H19 imprinting
- SNuPE
single nucleotide primer extension
References
- 1.Reeve A E, Eccles M R, Wilkins R J W, Bell G I, Millow L J. Nature (London) 1985;317:258–260. doi: 10.1038/317258a0. [DOI] [PubMed] [Google Scholar]
- 2.Scott J, Cowell J, Robertson M E, Priestley L M, Wadey R, Hopkins B, Pritchard J, Bell G I, Rall L B, Graham C F, Knott T J. Nature (London) 1985;317:261–262. doi: 10.1038/317260a0. [DOI] [PubMed] [Google Scholar]
- 3.Fearon E R, Vogelstein B, Feinberg A P. Nature (London) 1984;309:176–178. doi: 10.1038/309176a0. [DOI] [PubMed] [Google Scholar]
- 4.Reeve A E, Sih S A, Raizis A M, Feinberg A P. Mol Cell Biol. 1989;9:1799–1803. doi: 10.1128/mcb.9.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastie N D. Annu Rev Genet. 1994;28:523–528. doi: 10.1146/annurev.ge.28.120194.002515. [DOI] [PubMed] [Google Scholar]
- 6.Rainier S, Johnson L A, Dobry C J, Ping A J, Grundy P E, Feinberg A P. Nature (London) 1993;362:747–749. doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa O, Eccles M R, Szeto J, McNoe L A, Yun K, Maw M A, Smith P J, Reeve A E. Nature (London) 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- 8.Steenman M J C, Rainier S, Dobry C J, Grundy P, Horon I L, Feinberg A P. Nat Genet. 1994;7:433–439. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 9.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, Kraus W, Gerald W, Tycko B. Nat Genet. 1994;7:440–447. doi: 10.1038/ng0794-440. [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi T, Sullivan M J, Ogawa O, Reeve A E. Proc Natl Acad Sci USA. 1995;92:2159–2163. doi: 10.1073/pnas.92.6.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zemel S, Bartolomei M S, Tilghman S M. Nat Genet. 1992;2:61–65. doi: 10.1038/ng0992-61. [DOI] [PubMed] [Google Scholar]
- 12.Weksberg R, Shem D R, Song Q L, Squire J. Nat Genet. 1993;5:143–149. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa O, Becroft D M, Morison I M, Eccles M R, Skeen J S, Mauger D C, Reeve A E. Nat Genet. 1993;5:408–412. doi: 10.1038/ng1293-408. [DOI] [PubMed] [Google Scholar]
- 14.Brown K W, Villar A J, Bickmore W, Clayton-Smith J, Catchpoole D, Maher E R, Reik W. Hum Mol Genet. 1996;5:2027–2032. doi: 10.1093/hmg/5.12.2027. [DOI] [PubMed] [Google Scholar]
- 15.Morison I M, Becroft D M, Taniguchi T, Woods C G, Reeve A E. Nat Med. 1996;2:311–316. doi: 10.1038/nm0396-311. [DOI] [PubMed] [Google Scholar]
- 16.Beckwith J B. Med Pediatr Oncol. 1993;21:158–168. doi: 10.1002/mpo.2950210303. [DOI] [PubMed] [Google Scholar]
- 17.Tadokoro K, Fujii H, Inoue T, Yamada M. Nucleic Acids Res. 1991;19:6967. doi: 10.1093/nar/19.24.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards A, Civitello A, Hammond H A, Caskey C T. Am J Hum Genet. 1991;49:746–756. [PMC free article] [PubMed] [Google Scholar]
- 19.Singer-Sam J, Chapman V, LeBon J M, Riggs A D. Proc Natl Acad Sci USA. 1992;89:10469–10473. doi: 10.1073/pnas.89.21.10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo P E, Mann J R. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- 21.Chao L-Y, Huff V, Tomlinson G, Riccardi V M, Strong L C, Saunders G F. Nat Genet. 1993;3:127–131. doi: 10.1038/ng0293-127. [DOI] [PubMed] [Google Scholar]
- 22.Christofori G, Naik P, Hanahan D. Nat Genet. 1995;10:196–201. doi: 10.1038/ng0695-196. [DOI] [PubMed] [Google Scholar]
- 23.Park S, Bernard A, Bove K E, Sens D A, Hazen-Martin D J, Garvin A J, Haber D A. Nat Genet. 1993;5:363–367. doi: 10.1038/ng1293-363. [DOI] [PubMed] [Google Scholar]
- 24.Henry I, Bonaiti-Pellie C, Chehensse V, Beldjord C, Schwartz C, Utermann G, Junien C. Nature (London) 1991;351:665–667. doi: 10.1038/351665a0. [DOI] [PubMed] [Google Scholar]
- 25.Grundy P, Telzerow P, Paterson M C, Haber D, Berman B, Li F, Garber J. Lancet. 1991;338:1079–1080. doi: 10.1016/0140-6736(91)91937-p. [DOI] [PubMed] [Google Scholar]
- 26.Reik W, Brown K W, Slatter R E, Sartori P, Elliott M, Maher E R. Hum Mol Genet. 1994;3:1297–1301. doi: 10.1093/hmg/3.8.1297. [DOI] [PubMed] [Google Scholar]
- 27.Sapienza C. Mol Carcinog. 1990;3:118–121. doi: 10.1002/mc.2940030303. [DOI] [PubMed] [Google Scholar]
- 28.Leighton P A, Ingram R S, Eggenschwiler J, Efstratiadis A, Tilghman S M. Nature (London) 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 29.Bestor T H, Tycko B. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 30.Hoovers J M N, Kalikin L M, Johnson L A, Alders M, Redeker B, Law D J, Bliek J, Steenman M, Benedict M, Wiegant J, Lengauer C, Taillon-Miller P, Schlessinger D, Edwards M C, Elledge S J, Ivens A, Westerveld A, Little P, Mannens M, Feinberg A P. Proc Natl Acad Sci USA. 1995;92:12346–12460. doi: 10.1073/pnas.92.26.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M P, Hu R-J, Johnson L A, Feinberg A P. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 32.Dittrich B, Buiting K, Korn B, Rickard S, Buxton J, Saitoh S, Nicholls R D, Poustka A, Winterpacht A, Zabel B, Horsthemke B. Nat Genet. 1996;14:163–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]