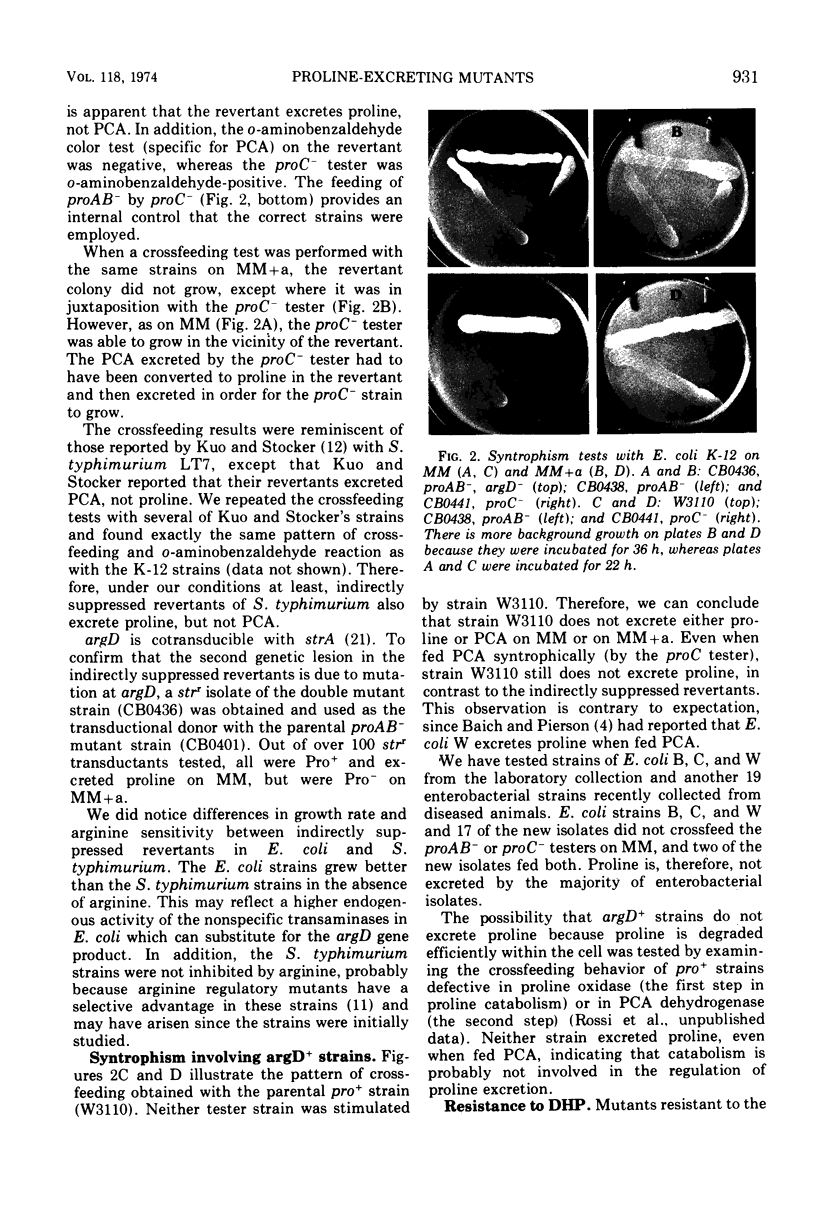
Abstract
The last step in proline biosynthesis in Escherichia coli K-12, Salmonella typhimurium LT7, and a number of other enterobacterial isolates is regulated so that no proline is excreted, even if excess Δ1-pyrroline-5-carboxylate, the immediate precursor of proline, is added to a culture. In proline auxotrophs blocked at an early step in proline biosynthesis (proA or proB), reversion to prototrophy is often due to a mutation in the arginine pathway which diverts N-acetyl glutamate γ-semialdehyde to proline synthesis, thus bypassing the proA or proB block. In such double mutants (proAB, argD), the last step in proline synthesis appears to be unregulated, since proline is excreted. Feedback inhibition and repression of the arginine pathway overcomes indirect suppression (restoring the Pro− phenotype), but proline regulation is not restored; double mutants still excrete proline when fed Δ1-pyrroline-5-carboxylate exogeneously. A new class of proline analogue-resistant mutant, due to mutation at argD, is also described.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baich A., Pierson D. J. Control of proline synthesis in Escherichia coli. Biochim Biophys Acta. 1965 Jul 8;104(2):397–404. doi: 10.1016/0304-4165(65)90345-4. [DOI] [PubMed] [Google Scholar]
- Baich A. Proline synthesis in Escherichia coli. A proline-inhibitable glutamic acid kinase. Biochim Biophys Acta. 1969 Dec 30;192(3):462–467. doi: 10.1016/0304-4165(69)90395-x. [DOI] [PubMed] [Google Scholar]
- Baich A. The biosynthesis of proline in Escherichia coli: phosphate-dependent glutamate -semialdehyde dehydrogenase (NADP), the second enzyme in the pathway. Biochim Biophys Acta. 1971 Jul 20;244(1):129–134. doi: 10.1016/0304-4165(71)90129-2. [DOI] [PubMed] [Google Scholar]
- Condamine H. Sur la régulation de la production de proline chez E. Coli K 12. Ann Inst Pasteur (Paris) 1971 Feb;120(2):126–143. [PubMed] [Google Scholar]
- Curtiss R., 3rd, Charamella L. J., Berg C. M., Harris P. E. Kinetic and genetic analyses of D-cycloserine inhibition and resistance in Escherichia coli. J Bacteriol. 1965 Nov;90(5):1238–1250. doi: 10.1128/jb.90.5.1238-1250.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORINI L., GUNDERSEN W., BURGER M. Genetics of regulation of enzyme synthesis in the arginine biosynthetic pathway of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1961;26:173–182. doi: 10.1101/sqb.1961.026.01.022. [DOI] [PubMed] [Google Scholar]
- Gorini L., Beckwith J. R. Suppression. Annu Rev Microbiol. 1966;20:401–422. doi: 10.1146/annurev.mi.20.100166.002153. [DOI] [PubMed] [Google Scholar]
- Hirota Y., Inuzuka M., Tomoeda M. Elective selection of proline-requiring mutants. J Bacteriol. 1966 Jun;91(6):2392–2392. doi: 10.1128/jb.91.6.2392-.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itikawa H., Baumberg S., Vogel H. J. Enzymic basis for a genetic suppression: accumulation and deacylation of N-acetylglutamic gamma-semialdehyde in enterobacterial mutants. Biochim Biophys Acta. 1968 Jul 9;159(3):547–550. doi: 10.1016/0005-2744(68)90142-3. [DOI] [PubMed] [Google Scholar]
- Kuo T. T., Stocker B. A. Suppression of proline requirement of proA and proAB deletion mutants in Salmonella typhimurium by mutation to arginine requirement. J Bacteriol. 1969 May;98(2):593–598. doi: 10.1128/jb.98.2.593-598.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAAS W. K. Studies on repression of arginine biosynthesis in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1961;26:183–191. doi: 10.1101/sqb.1961.026.01.023. [DOI] [PubMed] [Google Scholar]
- Rossi J. J., Berg C. M. Differential recovery of auxotrophs after penicillin enrichment in Escherichia coli. J Bacteriol. 1971 May;106(2):297–300. doi: 10.1128/jb.106.2.297-300.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STRECKER H. J., MELA P. The interconversion of glutamic acid and proline. Biochim Biophys Acta. 1955 Aug;17(4):580–581. doi: 10.1016/0006-3002(55)90422-4. [DOI] [PubMed] [Google Scholar]
- Smith-Keary P. F. Changes in the pattern of reversion of a proline-requiring mutant of Salmonella typhimurium. Mutat Res. 1971 Mar;11(3):279–292. [PubMed] [Google Scholar]
- Smith-Keary P. F. Partial reversions of a proline auxotroph of Salmonella typhimurium. Cross-feeding associated with cell-to-cell contact. Mutat Res. 1972 Jul;15(3):259–272. doi: 10.1016/0027-5107(72)90073-5. [DOI] [PubMed] [Google Scholar]
- Taylor A. L. Current linkage map of Escherichia coli. Bacteriol Rev. 1970 Jun;34(2):155–175. doi: 10.1128/br.34.2.155-175.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristram H., Thurston C. F. Control of proline biosynthesis by proline and proline analogues. Nature. 1966 Oct 1;212(5057):74–75. doi: 10.1038/212074a0. [DOI] [PubMed] [Google Scholar]




