Abstract
The 4,188-kb circular genome of Bacillus subtilis 168 was artificially dissected into two stable circular chromosomes in vivo, one being the 3,878-kb main genome and the other the 310-kb subgenome that was recovered as covalently closed circular DNA in CsCl-ethidium bromide ultracentrifugation. The minimal requirements to physically separate the 310-kb DNA segment out of the genome were two interrepeat homologous sequences and an origin of DNA replication between them. The subgenome originated from the 1,255–1,551-kb region of the B. subtilis genome was essential for the cell to survive because the subgenome was not lost from the cell. The finding that the B. subtilis genome has a potential to be divided and the resulting two replicons stably maintained may shed light on origins and formation mechanisms of giant plasmids or second chromosomes present in many bacteria. Similar excision or its reversal process, i.e., integration of large sized covalently closed circular DNA pieces into the main genome, implies significant roles of subgenomes in the exchange of genetic information and size variation of bacterial genomes in bacterial evolution.
Keywords: recombination, neomycin resistance, main genome, covalently closed circular DNA, replication origin
The bacterial chromosome structure has been thought to be stably maintained under constant environments (1), although some plasticity tolerating DNA rearrangements such as inversion, deletion, and duplication has been reported (2–4). There are several bacteria that have a second chromosome (chromosome II) (5–7) or megabase-sized plasmids (7–11), although the origins and dynamics of their formation mechanisms are not clear, and the discrimination of plasmids from second chromosomes has not been well defined. Meanwhile there are numerous examples that DNA pieces shuttle between an autonomously replicating state and an integrated form in the host genome. Well known examples are, the genome of Escherichia coli phage lambda that exists in two states, one the lysogenic state, and the other the state showing lytic cycle (12) and the F-factor that shuttles between the Hfr and episome forms (13).
B. subtilis 168, an endospore forming gram-positive soil bacterium, has a single 4,188-kb circular genome whose structure is considered to be stably maintained (1, 14). This bacterium carries no indigenous plasmid although there are a number of foreign plasmids capable of replication in this host (15, 16). In this study we show evidence that a 310-kb genomic DNA segment of this bacterium can be excised and replicate as an independent replicon from the genome. The established covalently closed circular form of DNA was found in the cell and its presence was essential for the cell to grow. We propose to term the autonomously replicating entity a subgenome of bacteria.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
B. subtilis 168 trpC2 was obtained from the Bacillus Genetic Stock Center (Columbus, OH). All the BEST strains in this report are derived from this by integration of antibiotic resistance markers. The SfiI-NotI physical map (14) of the B. subtilis 168 genome and the I-CeuI map, equivalent to the ribosomal RNA gene operon (rrn) map (17) are described in Fig. 1. Preparation and transformation of E. coli JA221 (F− hsdR hsdM+ trp leu lacY recA1, ref. 14) competent cells were done by the method of Mandel and Higa (19). Protocols for preparation and transformation of competent B. subtilis cells were described (14). Luria–Bertani broth (20) was used for growth of E. coli, and Luria–Bertani and antibiotic medium 3 (Difco) for B. subtilis. Nutritional requirements for B. subtilis were tested using Spizizen plates with appropriate amino acids (21). Bacteria were grown at 37°C in all media.
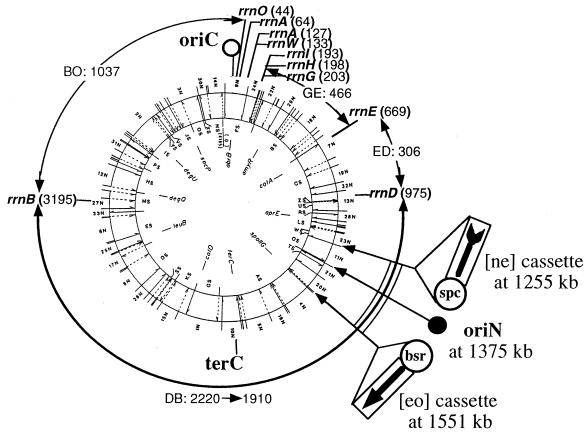
Figure 1.
SfiI-NotI-I-CeuI map of the B. subtilis 168 genome. The physical map of the 4,188-kb B. subtilis genome for NotI (inner circle), SfiI (outer circle), and I-CeuI map (equivalent of the rrn map) is shown. The sizes of the I-CeuI fragments >100 kb are shown by arcs with two arrowheads. The loci of oriC and terC have been determined (14, 18). Strain BEST4173 has the [ne] cassette at 1,255 kb, the [eo] cassette at 1,551 kb, and the oriN (closed circle) at 1,375 kb of the physical map as described in the materials and methods section. The spectinomycin resistance gene (spc) included in the [ne] cassette and the blasticidin S resistance gene (bsr) in the [eo] cassette are shown. The 300-kb DNA segment (1,255–1,551 kb) is shown by double arcs.
pNEXT33 and pNEXT41 (the NotI-linking clones carrying NotI site at 1,255 and 3,791 kb, respectively) were described previously (14, 22). In this study we used two truncated neomycin resistance gene alleles; one carries a deletion at the C terminus (ne), while the other carries a deletion at N terminus (eo). Construction of pBEST518 that carries the [ne] cassette and pBEST524B that carries the [eo] cassette was described previously (23).
The [ne] and [eo] gene cassettes used in this study were prepared from pBEST518 and pBEST524B. Briefly, the 2.4-kb [ne] cassette composed of a (ne) segment (1.1 kb) and the spectinomycin resistance gene (spc, 1.3 kb), the 1.4-kb [eo] cassette composed of a (eo) segment (0.79 kb) and the blasticidin S resistance gene (bsr, 0.6 kb) were excised with NotI from pBEST518 and pBEST524B, respectively. The [ne] cassette and the [eo] cassette were inserted into the NotI sites of pNEXT33 and pNEXT41, respectively, resulting in pNEXT33FA and pNEXT41GB.
The oriN sequence is the replication origin of a 70-kb low-copy number plasmid pLS32 obtained from a B. natto strain (24). Plasmid pBET131 (14.3 kb) is composed of an oriN-carrying 7-kb BamHI segment from pLS32, 5.4-kb pUH101 (a pBR322 derivative plasmid carrying the chloramphenicol resistance gene), and a 1.9-kb fragment carrying the tetracycline resistance gene (T.T., Y. Ohshiro, and M. Ogura, unpublished work). To integrate pBET131 in the BEST5018 genome, a pBR322 sequence was first created at 1,375 kb of the BEST5018 genome. The pBR322 sequence marked with erythromycin resistance gene of BEST2124 (25) was transferred to the 1,375-kb site of BEST5018 by transformation, resulted in BEST4170 (SpcR, BSR, pBR322::EmR at 1,375 kb). BEST4173 (Fig. 1) was obtained from BEST4170 by replacement of the erythromycin marker with the cat-oriN-tet array of pBET131. BEST4173, selected by chloramphenicol and tetracycline, showed erythromycin sensitive.
Plasmid transformants of E. coli were selected by tetracycline, 15 μg/ml; chloramphenicol, 5 μg/ml; spectinomycin, 100 μg/ml; and ampicillin, 100 μg/ml. Transformants of B. subtilis were selected by chloramphenicol, 5 μg/ml; blasticidin S, 500 μg/ml; tetracycline, 15 μg/ml, and spectinomycin, 50 μg/ml.
In Vitro DNA Manipulations.
Endonucleases and T4 DNA ligase were obtained from Toyobo (Osaka), except for SfiI and I-CeuI from New England Biolabs, NotI from Takara Shuzo (Kyoto), and I-SceI from Boehringer Mannheim.
Preparation of B. subtilis Chromosomal DNA and Southern Hybridization.
Intact unsheared chromosomal DNA for contour-clamped homogeneous electric field gel electrophoretic analysis was prepared in agarose plugs as described previously (1, 14, 18). Running conditions are described in the Fig. 3. Chromosomal DNA for analysis by conventional gel electrophoresis was prepared by a liquid isolation method (26). Agarose gel (1.0% wt/vol) in TBE solution (50 mM Tris⋅borate, pH 8.0/1.0 mM EDTA) was used for conventional gel electrophoresis at room temperature. After electrophoresis the DNA was stained in ethidium bromide solution (600 ng/ml) for 15 min and photographed. Southern hybridization experiments using a nonradioactive labeling nucleotide, digoxigenin-11-dUTP (Boehringer Mannheim), were done according to the supplier’s manual. Isolation of the 310-kb subgenome was performed by lysis of the host cell, followed by centrifugation of the total lysate in a CsCl gradient in the presence of ethidium bromide (Fig. 3a).
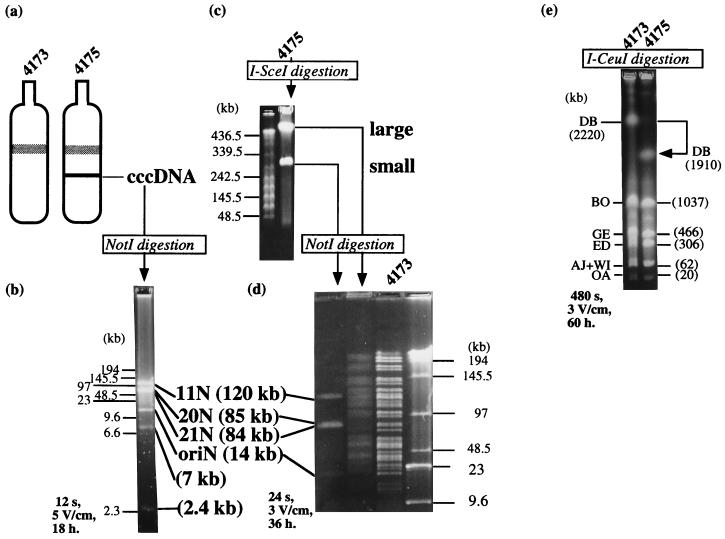
Figure 3.
Isolation and characterization of the 310-kb subgenome. (a) A total DNA lysate gently prepared from 10-ml cultures of BEST4173 and BEST4175 were centrifuged in a CsCl-ethidium bromide gradient at 140,000 × g for 18 hr in the vertical rotor using Beckman L-80 ultracentrifuge. Only the BEST4175 sample gave an extra band that was subsequently recovered, desalted and destained by extensive dialysis against TE solution (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA). (b) The cccDNA sample was digested with NotI and resolved by pulsed-field gel electrophoresis. The cccDNA gave rise to six NotI segments that are predicted in Fig. 2. (c) I-SceI digestion of BEST4175 DNA gave two linear DNA fragments in pulsed-field gel electrophoresis. The large and small DNAs were isolated and subjected to NotI digestion. Concatemeric lambda DNA (48.5 × n kb) was used as size markers. (d) The small I-SceI fragment gave NotI fragments identical to those of the cccDNA. The 7- and 2.4-kb NotI fragments were verified for their presence in a separate experiment (data not shown). A NotI digest of the large I-SceI fragment was resolved in pulsed-field gel electrophoresis and found to be composed of the genomic NotI fragments lacking the subgenome fragments, as shown by the comparison with the NotI fragments of BEST4173 DNA in the next lane. Concatemeric lambda DNA (48.5 × n kb) and HindIII digested lambda DNA are size markers. (e) Total DNAs from BEST4173 and BEST4175 were digested with I-CeuI. The DB fragment derived from the I-CeuI sites in the rrnD and rrnB operons shown in Fig. 1. In BEST4175 only the 2,220-kb DB fragment was shortened to the 1,910-kb band, which verifies that the main genome carried a deletion of a DNA segment with the size equivalent to the subgenome. The subgenome remained as circular DNA and did not enter the gel. Specific running conditions for pulsed-field gel electrophoresis, pulse time, voltages, and running time are shown in each figure.
RESULTS
Isolation of the B. subtilis Mutant Carrying a Subgenome.
We have developed a method that allows isolation of B. subtilis mutants bearing a rearrangement of a specific DNA segment of the genome (23, 27). The method employs intrachromosomal homologous recombination between two truncated neomycin resistance gene alleles (indicated as [ne] and [eo] in Fig. 1) integrated at the ends of the DNA region to be rearranged. Integration of the [ne] cassette at 1,255 kb was done by transformation of linearized pNEXT33FA, selecting for spectinomycin resistance transformant BEST2161. Integration of the [eo] cassette at 1,551 kb of the BEST2161 genome was similarly done using linearized pNEXT41GB. BEST5018 was obtained as blasticidin S resistant transformant of BEST2161. Presence and orientation of the [ne] and [eo] cassettes in the BEST5018 genome was verified by Southern hybridization using both plasmids as probes (data not shown). Strain BEST5018 did not produce a spontaneous neomycin-resistant recombinants.
To test if about 300-kb genomic segment (1,255–1,551 kb) can be established as an autonomous replicon, strain BEST4173 was constructed from BEST5018 as described in Materials and Methods. Presence of the [ne], [eo], and oriN of BEST4173 shown in Fig. 1 was verified by Southern hybridization experiments using pNEXT33FA, pNEXT41GB, and pBET131 as probes (data not shown). It was expected that a recombinational event occurring between the common 590-bp homologous region of the integrated [ne] and [eo] sequence (shown by × in Fig. 2) excises the 300-kb segment out of the genome and gives rise to the functional neomycin resistance gene [neo] that renders the cell neomycin resistant. A number of neomycin-resistant colonies were obtained from a BEST4173 culture. We investigated one neomycin-resistant strain, BEST4175, to determine if it bore two circular DNAs as expected; one main genome composed of 3,878 kb and the other 310-kb subgenome.
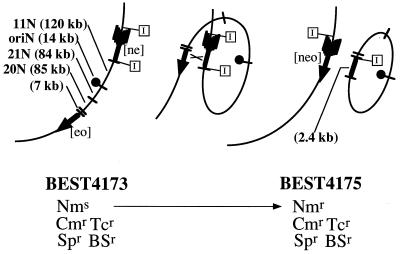
Figure 2.
How to excise a 300-kb genomic DNA of the B. subtilis and make it replicate autonomously. Enlargement of the 1255–1551-kb region of the BEST4173 genome (Left), a hypothetical intermediate of the intrachromosomal recombination (md Middle), and BEST4175 (Right) genome. The intact neomycin resistance gene [neo] was formed by reciprocal homologous recombination shown by × between a 590-bp common region in the [ne] and [eo]. BEST4173 was neomycin sensitive and the strain that established the subgenome (BEST4175) were obtained by resistance to neomycin (5 μg/ml). The spectinomycin resistance gene (spc) included in the [ne] cassette and the blasticidin S resistance gene (bsr) in the [eo] cassette that render BEST4173 and BEST4175 spectinomycin resistance (Spr) and blasticidin S resistance (BSr) are omitted in the figure. The chloramphenicol resistance (Cmr) and tetracycline resistance (Tcr) are linked to the oriN sequence. See the details in Materials and Methods. The [ne] fragment has two I-SceI sites at its both ends shown by “I” in rectangles. Upon recombination one I-SceI site is allotted to the subgenome and the other remains in the main genome, so that I-SceI linearizes both the main and subgenomes as seen in Fig. 3c. The 310-kb DNA contains five NotI fragments, 120 kb (11N), 85 kb (20N), 84 kb (21N), 14 kb (oriN), and 7 kb. The subgenome has an additional 2.4-kb NotI fragment generated by the recombination. The rate of generation of neomycin-resistant cells in the BEST4173 culture was estimated to be ≈10−7 per cell per cell division at 37°C.
Purification and Characterization of the Subgenome.
The subgenome was verified to have the expected structure by the following two observations: (i) The cccDNA obtained from strain BEST4175 by CsCl-ethidium bromide ultracentrifugation (Fig. 3a) was recovered gently to avoid physical shearing and subjected to NotI digestion. It gave six NotI fragments (shown in Fig. 3b) with sizes expected to be included in the subgenome as indicated in Fig. 2. (ii) The two unique I-SceI recognition sites at both ends of the [ne] allele (Fig. 2) should be allotted to the two genomes if the subgenome is formed in the predicted manner shown in Fig. 2, and therefore I-SceI endonuclease should cleave the BEST4175 genome into two linear forms. As shown in Fig. 3c, two I-SceI fragments, one with a size of ≈310 kb and the other beyond the size markers, were obtained from BEST4175 DNA. Both I-SceI fragments were isolated from the gel and digested with NotI. The NotI digestion profiles presented in Fig. 3d show that the 310-kb DNA was identical to the cccDNA and the large fragment had the structure of the main genome lacking the six NotI fragments. These results indicated that the 310-kb fragment autonomously replicates as a circular form in BEST4175.
The Main Genome Structure.
Structure of the main genome was further studied by using I-CeuI endonuclease. According to the I-CeuI map of the B. subtilis genome, the DB fragment includes the 310-kb region (see Fig. 1). Theoretically, excision of the 310-kb segment renders the 2,220-kb DB fragment shorter by 310 kb. Digestion of the BEST4173 and BEST4175 DNAs with I-CeuI produced the same banding patterns except the DB fragment as shown in Fig. 3e. As expected, the DB fragment of the BEST4175 genome showed a net 310 kb decrease, an equivalent to the size of the subgenome that remained as cccDNA and was not visible in the gel of Fig. 3e.
Genetic Stability of the Subgenome.
Digestion of the total DNA of BEST4175 with NotI or SfiI gave the similar banding pattern and intensity of DNA fragments as those of the BEST4173 DNA (data not shown). There was no gross difference in growth rates of BEST4173 and 4175 when cultured in Luria–Bertani or synthetic media at 37°C, or observed by colony size on the plate with or without antibiotics, indicating few segregational event, if any. These observations suggested the presence of the two genomes at 1:1 ratio. For the genetic stability of the subgenome, six hundred BEST4175 colonies formed after >25 generations without any antibiotic selection were tested for the three markers, one (neomycin resistance) for the main genome, and the other two (chloramphenicol and spectinomycin resistance) for the subgenome. No colony had lost these markers. This indicates that subgenome-free-cell did not appear and is consistent with the observation that no neomycin-resistant cell was produced from strain BEST5018 that lacks the oriN sequence. In contrast to the stability of the BEST4175 subgenome, the cell carrying only the oriN plasmid, pBET131, produced plasmid-free cells at a rate of 10% after 10 generations (T.T. & M. Ogura, unpublished data). From these observations we conclude that the 310-kb DNA segment is indispensable for strain BEST4175. This is the first example that the bacterial genome has been physically separated into two replicons and both are stably maintained as the main genome and the subgenome.
We propose to define subgenome as independent replicons that derive from the host genome having essential gene(s). It is discriminated from large-sized plasmids or second chromosomes often found in many bacteria (5–11), based on their a priori presence. The subgenome described above probably lacks the ter function. The main genome retains the terC sequence (28), and probably it functions despite of the shortened right half of the BEST4175 genome by 300 kb. Although it is not clear whether the subgenome has a fortuitous functional terminator, the successful isolation of a B. subtilis terC deletion mutant suggests that the ter sequence may not be required for B. subtilis genome (29).
DISCUSSION
It is possible in principle to dissect long genomic DNA segment by homologous recombination between two DNA repeats located at both ends. To our surprise the establishment of as long as 300 kb of the genomic DNA as an independent replicon requires only an origin of DNA replication and two direct homologous repeats as short as 590 bp. Homologous stretches are ubiquitous in bacterial genomes such as rrn operons (3, 17, 30), transposable elements (31), and short repeated sequences (32). Origin (-like) sequences could be generated by duplication of oriC itself, by internal cryptic origins (33) or lysogenic prophages (our unpublished observations), and they may be provided by infections, horizontal transfer mechanisms, or fusions from natural environment (34). These situations imply that bacterial genomes have a potential to divide if both ori and repeated sequences are present in the same genome. If so, subgenomes (even plasmids or second chromosomes) could likely be formed by natural genome surgery as an entity adapting to a physiological niche of the host bacteria.
Our present result raised a number of other interesting questions that remained to be demonstrated, for example (i) how (large or small) and which region of the genome can be separated from the main genome? (ii) how many subgenomes can the bacterial genome provide? (iii) how does regulation of gene expression change in different genomes? (iv) do subgenomes reintegrate in different loci of the main genome? (v) can the subgenome become a unit to transfer between species? In addition our method allows the isolation of specific regions of the genome as a supercoiled form in CsCl-ethidium bromide centrifugation, thus providing a large DNA of interest in a pure form. This method may make it possible to transfer large DNA segments to various other bacterial species.
The B. subtilis genome seems so plastic as to tolerate alteration of large inversions (4, 23, 27) and excision (this study). The genome surgery to cause excision of specific genomic DNA segment can be in principle applicable to other bacteria and may provide an experimental approach leading to a hypothetical bacterium with a minimal genome (35).
Acknowledgments
We thank Drs. T. Toda, S. Hiraga, H. Yanagawa, and M. Ogura for helpful discussions.
ABBREVIATION
- cccDNA
covalently closed circular DNA
References
- 1.Itaya M. J Bacteriol. 1993;175:741–749. doi: 10.1128/jb.175.3.741-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segall A, Mahan M J, Roth J R. Science. 1988;241:1314–1318. doi: 10.1126/science.3045970. [DOI] [PubMed] [Google Scholar]
- 3.Liu S-L, Sanderson K E. Proc Natl Acad Sci USA. 1995;92:1018–1022. doi: 10.1073/pnas.92.4.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itaya M. Biosci Biotech Biochem. 1994;58:1836–1841. doi: 10.1271/bbb.58.1836. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary M, Mackenzie C, Nereng K S, Sodergren E, Weinstock G M, Kaplan S. J Bacteriol. 1994;176:7694–7702. doi: 10.1128/jb.176.24.7694-7702.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honeycutt R J, McClelland M, Sobral B W S. J Bacteriol. 1993;175:6945–6952. doi: 10.1128/jb.175.21.6945-6952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allardet-Servent A, Michaux-Charachon S, Jumus-Bilak E, Karayan L, Ramuz M. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carles TC, Finan T M. Genetics. 1991;127:5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong K K, McClelland M. J Bacteriol. 1992;174:1656–1661. doi: 10.1128/jb.174.5.1656-1661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabata K, Kosuge T, Nakahara T, Hoshino T. FEBS Lett. 1993;331:81–85. doi: 10.1016/0014-5793(93)80301-a. [DOI] [PubMed] [Google Scholar]
- 11.Zuerner R L, Herrmann J L, Girons I S. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arber W. In: Lambda II. Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. pp. 381–394. [Google Scholar]
- 13.Brooks L K. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Niedhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1987. pp. 1134–1137. [Google Scholar]
- 14.Itaya M, Tanaka T. J Mol Biol. 1991;220:631–648. doi: 10.1016/0022-2836(91)90106-g. [DOI] [PubMed] [Google Scholar]
- 15.Bron S. In: Molecular Biological Methods for Bacillus. Harwood C R, Cutting S, editors. London: Wiley; 1990. pp. 75–138. [Google Scholar]
- 16.Errington J. In: Molecular Biological Methods for Bacillus. Harwood C R, Cutting S, editors. London: Wiley; 1990. pp. 175–220. [Google Scholar]
- 17.Toda T, Itaya M. Microbiology. 1995;141:1937–1945. doi: 10.1099/13500872-141-8-1937. [DOI] [PubMed] [Google Scholar]
- 18.Itaya M, Laffan J J, Sueoka N. J Bacteriol. 1992;174:5466–5470. doi: 10.1128/jb.174.16.5466-5470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandel M, Higa A. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in Molecular Genetics. Plainview, New York: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 21.Spizizen J. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itaya M, Tanaka T. Mol Gen Genet. 1990;223:268–272. doi: 10.1007/BF00265063. [DOI] [PubMed] [Google Scholar]
- 23.Toda T, Tanaka T, Itaya M. Biosci Biotech Biochem. 1996;60:773–778. doi: 10.1271/bbb.60.773. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka T, Koshikawa T. J Bacteriol. 1977;132:699–701. doi: 10.1128/jb.131.2.699-701.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itaya M. Mol Gen Genet. 1993;241:287–297. doi: 10.1007/BF00284680. [DOI] [PubMed] [Google Scholar]
- 26.Saito H, Miura K. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 27.Itaya M, Toda T, Ohshiro Y, Ogura M, Tanaka T. Nucleic Acids Symp Ser. 1995;34:243–244. [PubMed] [Google Scholar]
- 28.Yoshikawa H, Wake R G. In: Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 507–528. [Google Scholar]
- 29.Iismaa T P, Wake R G. J Mol Biol. 1987;195:299–310. doi: 10.1016/0022-2836(87)90651-6. [DOI] [PubMed] [Google Scholar]
- 30.Hill C W, Harvey S, Gray J A. In: The Bacterial Chromosome. Drlica K, Riley M, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. 335–340. [Google Scholar]
- 31.Umeda M, Ohtsubo E. J Mol Biol. 1989;208:601–614. doi: 10.1016/0022-2836(89)90151-4. [DOI] [PubMed] [Google Scholar]
- 32.Lupski J R, Weinstock G M. J Bacteriol. 1992;174:4525–4529. doi: 10.1128/jb.174.14.4525-4529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grandjean V, Nguyen J, Hauck Y, Hirschbein L. Plasmid. 1993;30:1–13. doi: 10.1006/plas.1993.1029. [DOI] [PubMed] [Google Scholar]
- 34.Lorenz M G, Wackernagel W. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itaya M. FEBS Lett. 1995;362:257–260. doi: 10.1016/0014-5793(95)00233-y. [DOI] [PubMed] [Google Scholar]