Abstract
Rotavirus contains two outer capsid viral proteins, the spike protein VP4 and major capsid component VP7, both of which are implicated in cell entry. We show that VP4 and VP7 contain tripeptide sequences previously shown to act as recognition sites for integrins in extracellular matrix proteins. VP4 contains the α2β1 integrin ligand site DGE. In VP7, the αxβ2 integrin ligand site GPR and the α4β1 integrin ligand site LDV are embedded in a novel disintegrin-like domain that also shows sequence similarity to fibronectin and the tie receptor tyrosine kinase. Microorganism sequence homology to these ligand motifs and to disintegrins has not been reported previously. In our experiments, peptides including these rotaviral tripeptides and mAbs directed to these integrins specifically blocked rotavirus infection of cells shown to express α2β1 and β2 integrins. Rotavirus VP4-mediated cell entry may involve the α2β1 integrin, whereas VP7 appears to interact with αxβ2 and α4β1 integrins.
Rotaviruses are the major cause of severe acute diarrheal illness of infants and young children worldwide, and are important pathogens in most avian and mammalian species. The spike viral protein VP4 and major outer capsid glycoprotein VP7 independently elicit neutralizing, protective antibodies and are determinants of virulence. VP4 is an 88-kDa protein that determines host cell tropism and carries P (protease-sensitive) serotype specificity. VP4 is proteolytically cleaved for an increase in virus infectivity and for rapid virus internalization, into two subunits, VP8* (28 kDa) and VP5* (60 kDa) (1, 2). VP8* contains the hemagglutinin domain (amino acids 93–208) (3), overlapping an antigenic region at amino acids 79–192 (4). VP5* contains a putative cell fusion region at amino acids 384–401 related by sequence to the potential fusion region of Sindbis virus (5), and antigenic regions encompassing amino acid 305, the fusion region, and amino acids 428–458 (4). VP7 carries G (glycoprotein) serotype specificity and contains six antigenic regions, A–F (6, 7). Immunodominant regions A and C form a single, conformation-dependent site.
Most evidence suggests that VP4 is the major rotavirus cell attachment protein. Many animal, but not human, rotaviruses require cellular sialic acid for hemagglutination, cell binding, and infectivity. Rhesus rotavirus mutants that retained infectivity but no longer required sialic acid showed a mutation in gene 4, encoding VP4 (8). Baculovirus-expressed rhesus rotavirus VP4 was shown to compete with infectious virus for binding to murine enterocyte lysate (9). In addition, virus-like particles containing both rhesus rotavirus VP4 and VP7, but not VP7 alone, bound to MA104 cells (10). Finally, the cell binding and infectivity of rhesus/murine and simian/human virus reassortants differing in their requirements for cellular sialic acid correlated with the parental origin of gene 4 (11).
Earlier studies implicated VP7 as a rotavirus cell attachment protein. Two laboratories reported that a protein in infected cell lysates identified as VP7 by antiserum blocking and adsorption with mAbs bound to MA104 cell monolayers (2, 12). It was later suggested that this protein was the nonstructural protein NSP2, rather than VP7 (13). It remains possible that VP7 has a minor role in cell attachment (11).
Rotavirus infection of cells appears to be a multi-step process, involving binding, protease cleavage, and pH-independent entry, in which only a proportion of bound virions productively infect the cells. On the available evidence, it is likely that rotavirus enters cells by direct membrane penetration rather than endocytosis, possibly involving VP4 and VP7. Apart from glycoconjugates, particular cellular components involved in rotavirus binding and entry into cells have not been identified.
One commonly used family of viral receptors and essential entry molecules on cells is the integrin family. These αβ heterodimeric, transmembrane glycoproteins are the major receptors by which cells attach to extracellular matrix components such as fibrinogen, fibronectin, and collagen. A number of integrin recognition sites in these ligands have been defined. The first of these, the RGD sequence present in fibronectin, is also found in VP1 of foot-and-mouth disease virus and is used for attachment to the integrin αvβ3 (14). Additionally, the RGD sequence in the adenovirus type 2 penton base interacts with αvβ3 and αvβ5 to facilitate virus entry (15).
We examined rotavirus sequences for the presence of RGD and other known integrin ligand binding sites. Here we report that both outer capsid proteins contain at least one potential site, and that two of the integrins to which these ligands bind are expressed on cell lines permissive to rotavirus infection. In our experiments, peptides including these sites and mAbs directed to the implicated integrins specifically blocked simian rotavirus SA11 and human rotavirus RV-5 infection of these cell lines.
MATERIALS AND METHODS
Cells and Viruses.
RV-5 rotavirus, serotype G2P1B, and SA11 virus, serotype G3P[2], were grown in MA104 monkey kidney cells (16). Caco-2 human colonic adenocarcinoma cells were obtained from R. Whitehead (Ludwig Institute for Cancer Research, Parkville, Victoria, Australia) at passage 30 and grown as described (17).
Sequence Analysis.
The similarities of SA11 VP4 and VP7 amino acid sequences to the integrin recognition sites DGE(A), GPR, and LDV; to the tie receptor tyrosine kinase (TK); and to disintegrins were found manually. The sequences were aligned, and pairwise percentage similarities were calculated using the clustal w multiple sequence alignment program (18). The integrin recognition sites were also aligned with this program to the rotavirus gene sequences deposited in the GenBank database (July 31, 1996). The rotavirus VP4 and VP7 sequences were also examined without success for the following integrin ligand motifs: RGD, NGR, RRETAWA, REDV, SDGR, YIGSR, RGES, RSGIY, RSGD, YIGSE, DRDES, and SRYD (19, 20).
Peptides.
Peptides were at least 95% pure by HPLC unless indicated otherwise and were custom-made by Chiron [RDGEE (>60% pure), RAGEE, LDVT (>70% pure), and LRVT], or purchased from Auspep (GPRP and RGES) or Sigma [GPRP, GPGG, GHRP, and RGD]. For assay, the peptides were dissolved in distilled water, brought to pH 7.5 with 7% NaHCO3, then diluted in DMEM, under sterile conditions at all times.
mAbs.
Several mouse mAbs to human integrin subunits were generously provided by colleagues. These were as follows: AK7 and RMAC11 directed against the α chain (CD49b) of α2β1 (CD49b/CD29; VLA-2) (21, 22) from M. Berndt (Baker Medical Research Institute, Prahran, Victoria, Australia) and T. D’Apice (St. Vincent’s Hospital, Melbourne), respectively; TS2/7 directed against the α chain of α1β1 (CD49a/CD29; VLA-1), B-5G10 directed against the α chain of α4β1 (CD49d/CD29; VLA-4) (23), and A-1A5 directed against the common β chain of the VLA family of integrins (CD29) (24) from M. Hemler (Dana–Farber Cancer Institute, Boston); MHM24 directed to the α chain of αLβ2 (CD11a/CD18; LFA-1), OKM1 and 2LPM19c directed to the α chain of αMβ2 (CD11b/CD18; Mac-1) (25) and 99.1.1.1.1 directed to the α chain of αXβ2 (CD11c/CD18; p150, 95) (26) from P. Cameron, G. Stent, and S. Sonza (Macfarlane Burnet Centre for Medical Research, Fairfield, Victoria, Australia). P4G9 and P4C2 directed against α4, and P4C10 and 8A2 directed against β1, were obtained from Life Technologies. MHM23 directed against β2 was purchased from Dako, and control mAb MOPC21 was from Organon Teknika–Cappel. mAbs AK7 and RV-4:1 were purified from mouse ascites fluids by protein G affinity chromatography. The remaining antibodies were tested as ascites fluids and/or hybridoma cell supernatant fluids. Control and test mAbs were matched for isotype and protein concentration. Control mAbs were directed to irrelevant antigens, or the rotavirus antigens VP4 and VP7, and did not neutralize the test rotavirus by fluorescent focus reduction assay or react with it by enzyme immunoassay (16).
Fluorescent Focus Reduction Assay.
The fluorescent focus reduction assay used was adapted from the neutralization assay described previously (16).
Peptide dilutions in DMEM were added in triplicate to washed, confluent MA104 or Caco-2 cell monolayers in microtiter trays for 1 h before viral adsorption, removed, then replaced with fresh peptide solution of the same dilution, to which trypsin-activated, diluted SA11 (purified triple-layered) or RV-5 (culture supernatant) (16) was added. Virus was adsorbed for 1 h then removed and replaced with DMEM. All incubations were carried out at 37°C in 5% CO2. At 16 h post-infection, virus-infected cells were stained by indirect immunofluorescence, and infected cells were counted as described (16).
mAb dilutions in DMEM were added to cells as above at 37°C for 2 h before virus adsorption and were removed before virus was added. The remainder of the assay is described above.
Flow Cytometry.
Confluent MA104 or Caco-2 cell monolayers (4–5 days old) were treated with trypsin-EDTA then allowed to recover in growth medium for 30 min at 37°C before staining of 5 × 105 cells by indirect immunofluorescence and analysis using a FACSort machine (Becton Dickinson). Cell binding by serial dilutions of anti-α2 mAb AK7 and control mAb RV-4:1 (both at 10 μg/ml) was detected in a two-step stain with fluorescein isothiocyanate-conjugated, affinity-purified sheep anti-mouse F(ab′)2 fragments (Silenus, Melbourne), whereas binding of serial dilutions of anti-β2 mAb MHM23 and control MOPC21 (both at 70 μg/ml) was detected in a three-step stain with biotinylated, affinity-purified sheep anti-mouse immunoglobulins (Silenus) and phycoerythrin-conjugated streptavidin (Becton Dickinson). Each experiment was repeated at least once.
RESULTS
Integrin Ligand Motifs Found in Rotavirus.
The following three integrin recognition sites were found in rotavirus by sequence comparisons. The amino acid sequence DGE(A), corresponding to amino acids 435–438 of the type I collagen sequence, serves as a recognition site in collagen for the α2β1 integrin on colonic adenocarcinoma cells, platelets, and other cells (27). The sequence GPR in the N-terminal domain of the Aα chain of fibrinogen is recognized by the integrin αxβ2 on tumor necrosis factor-stimulated neutrophils (28). The sequence LDV, in the first connecting segment (CS1) of the independently spliced IIICS domain of fibronectin, is the minimal essential sequence for a major site of adhesion of fibronectin to the α4β1 integrin on a range of cell types (29).
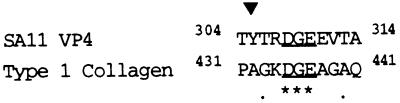
Simian rotavirus strain SA11 contains DGE in the VP5* subunit of VP4 (Fig. 1). The sequence at this position is highly conserved among mammalian rotaviruses. In group A rotaviruses, from sequences deposited in the GenBank database, 87.0% of strains (n = 54) shows the RDGE sequence, including human strain RV-5. The other 13% of strains (n = 6) contains DGI (human virus 116E), DEE (human strain K8), DGV (bovine strains A44, B223, and KK3), and DDE (equine strain L338). Group B and C rotaviruses contain the sequences RDG and RAG, respectively, at this position. RDG is one of only two amino acid triplets in VP4 that are conserved between group A and B rotaviruses.
Figure 1.
Alignment of SA11 VP4 sequence to the α2β1 integrin ligand site (underlined) in type 1 collagen. The sequences show 27% identity. Stars indicate identical amino acids, dots indicate conserved amino acids, and both symbols refer to the relatedness of the named sequence to SA11. ▾, The amino acid positions at which mutations were found in escape mutants selected with neutralizing mAbs (6, 7, 30).
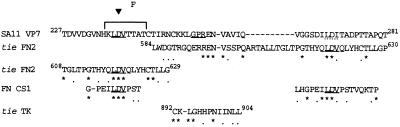
All mammalian rotaviruses contain GPR in VP7 at amino acids 253–255, in a longer stretch of sequence (amino acids 251–259) that is 98.6% conserved among the 71 VP7 sequences in the GenBank database. This region also shows similarity to a portion of the TK domain of the tie receptor TK (Fig. 2) (31). GPR is also present in the rotavirus inner core protein VP3 but is unlikely to be active as VP3 is not available in the infectious virion for interaction with integrins.
Figure 2.
Alignment of SA11 VP7 (amino acids 227–281) with the second fibronectin type III repeat (FN2; amino acids 584–630) and the TK domain (amino acids 892–904) of the tie receptor TK (31), and the fibronectin CS1 region. The stars, dots, and triangle are defined in the Fig. 1 legend. Antigenic regions are shown by a square bracket. SA11 (amino acids 245–281) shows 24% similarity and 19% identity to tie FN2 (amino acids 584–630). An overlapping set of SA11 residues (amino acids 227–248) shows 31% identity to a portion of this tie FN2 region (amino acids 608–629). The tie TK domain (amino acids 892–904) shows 39% identity to SA11 (amino acids 249–261). SA11 (amino acids 263–279) shows 29% identity to fibronectin CS1.
The VP7 of SA11 rotavirus contains LDV in antigenic region F and the related sequence LDI at amino acids 269–271. In SA11, these LDV/LDI motifs are flanked by regions of sequence similar to the flanking sequence in the fibronectin CS1 region (Fig. 2). These regions in SA11 VP7 also show relatedness to longer, continuous stretches of sequence in the second fibronectin type III repeat (FN2) of the tie receptor TK (Fig. 2) (31). From GenBank sequences, all mammalian group A rotaviruses show LDI (n = 65) or IDI (n = 6) at residues 269–271 and contain glycine and proline in positions identical to fibronectin CS1. Only 43.7% of mammalian group A rotaviruses shows LDV (77.4%), LDI (12.9%), or LK/SV in antigenic region F as the remainder of viruses have a potential glycosylation site at amino acids 238–240.
Interestingly, NSP2 of some rotaviruses contains LDV or the related sequence LEI at amino acids 282–284. NSP2 may not be present in the virion for interaction with cell surface integrins, but attachment of integrins in infected cell lysates to the LDV of NSP2 could explain the hitherto puzzling binding of NSP2 from these virus strains (13).
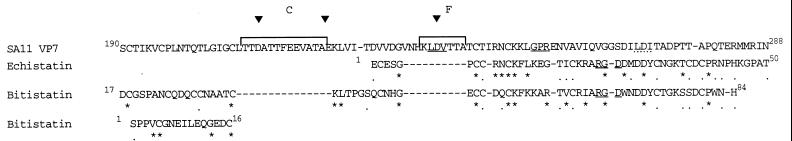
A 99-aa section of SA11 VP7, incorporating GPR, LDV, and LDI, shows sequence relatedness to snake venom disintegrins, particularly echistatin and bitistatin (Fig. 3). Disintegrins bind to platelet integrins, often via RGD (33). Four of nine cysteine residues conserved among disintegrins (33), the disintegrin-like domains of snake venom metalloproteases and members of the ADAM (A Disintegrin And Metalloprotease domain) gene family (34) are also present in SA11 VP7. The substitutions in SA11 for four of the remaining five cysteines are conservative. In this alignment, antigenic regions C and F were excluded as they showed no sequence relatedness to the disintegrins.
Figure 3.
SA11 VP7 (amino acids 190–288) is aligned with the complete sequences of two representative snake venom disintegrins, echistatin and bitistatin (32), which show 48% similarity by use of the clustal w program and 48% identity to each other. The illustrated region of SA11 is 24% similar and 19% identical to echistatin and 20% similar and 14% identical to bitistatin. Two adjacent regions of bitistatin both align to amino acids 190–207 of SA11 VP7. Stars, dots, square brackets, and triangles are defined in the legends of Figs. 1 and 2.
Synthetic Peptides Containing Integrin Ligand Motifs Block Rotavirus Infection of Cells.
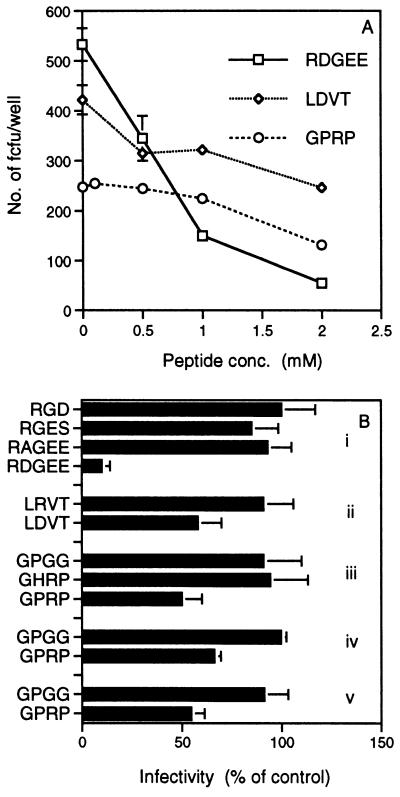
To determine whether the integrin ligand sites present in rotavirus have functional significance, synthetic peptides corresponding to the viral sequence and containing the ligand sites were tested for their ability to block virus infection of cells (Fig. 4). The peptide RDGEE blocked SA11 virus infectivity in a dose-dependent fashion, to 90% of the negative control (no peptide) at a 2 mM concentration (Fig. 4 A and Bi). Control peptides RAGEE, RGES, and RGD did not significantly affect virus infectivity (Fig. 4Bi). Peptides GPRP and LDVT also specifically blocked SA11 infectivity in a dose-dependent manner, but to a lesser degree. At a 2 mM concentration, GPRP and LDVT reduced infectivity by 50% and 42%, respectively (Fig. 4 A, Bii, and Biii). Control peptides GHRP, GPGG, and LRVT did not show significant levels of blocking (Fig. 4 Bii and Biii). GPRP blocked RV-5 virus infection of MA104 and Caco-2 cells to 34% (Fig. 4Biv) and 45% (Fig. 4Bv), respectively.
Figure 4.
Inhibition of SA11 and RV-5 rotavirus infection of cells by synthetic peptides that correspond to VP4 and VP7 sequences and contain integrin recognition sites. Values plotted are the mean of three replicates from a representative experiment. Error bars show the SEM. Many error bars are too small to be visible on the graphs. Each experiment was repeated at least three times and gave similar results. (A) Dose-dependent inhibition of SA11 infectivity by RDGEE, LDVT, and GPRP. Results (y axis) are expressed as the number of fluorescent cell-forming units (fcfu) per well. Each fluorescent cell-forming unit is assumed to result from infection of a cell with a single virus particle. Negative control peptides tested are listed in B. At each peptide concentration, counts of negative control peptide fluorescent cell-forming units per well varied by less than 5% from the zero peptide count. (B) Peptides RDGEE, LDVT, and GPRP, but not sequence-related control peptides, inhibited virus infectivity. Data are shown for peptides at a concentration of 2 mM. The number of fluorescent cell-forming units in virus-infected wells not treated with any peptide represents 100% infectivity. (i–iii) SA11 in MA104 cells. (iv) RV-5 in MA104 cells. (v) RV-5 in Caco-2 cells.
Antiintegrin mAbs Block Rotavirus Infection of Cells.
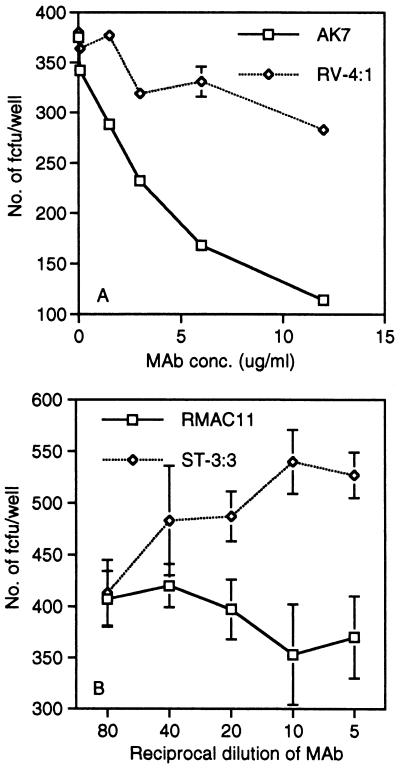
Anti-α2 mAbs that block cell adhesion to collagen (AK7 and RMAC11) (22, 35) also blocked SA11 rotavirus infectivity, to 60% and 30%, respectively, at 12 μg/ml (Fig. 5). RMAC11 maps to the same epitope region of the I domain of α2 as do mAbs that block echovirus type 1 infection of cells (35). The AK7 mAb also blocked human rotavirus RV-5 infection of MA104 and Caco-2 cells in a dose-dependent fashion, to 42 ± 9% and 39 ± 1% (mean ± SD), respectively, at 12 μg/ml (data not shown).
Figure 5.
Treatment of MA104 cells with anti-α2 mAbs AK7 (A) and RMAC11 (B), but not with irrelevant mAbs RV-4:1 (A; to human rotavirus RV-4 VP7 antigenic region A) and ST-3:3 (B; to human rotavirus ST-3 VP4), inhibits SA11 infection.
SA11 infectivity was blocked by the anti-β2 mAb, MHM23, to 46% at 40 μg/ml but not by control mAb MOPC21 (Fig. 6A), or by anti-αL mAb MHM24 (36), anti-αM mAbs 2LPM19c and OKM1 (25), or anti-αX mAb 99.1.1.1.1 (data not shown). MHM23 also blocked RV-5 infectivity in MA104 cells to a similar degree (data not shown), and in Caco-2 cells, to 72% at 38 μg/ml (Fig. 6B). Three of five other β2 mAbs that we have tested also blocked SA11 infectivity, to low levels, but none of 12 to αL or αM showed blocking (37).
Figure 6.
SA11 infectivity in MA104 cells (A) and RV-5 infectivity in Caco-2 cells (B) are blocked by anti-β2 mAb MHM23 but not by control mAb MOPC21.
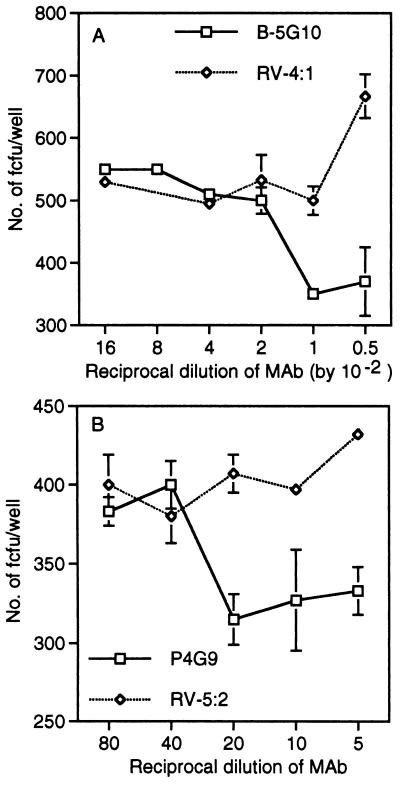
Two anti-α4 mAbs also blocked SA11 virus infection (Fig. 7). mAbs B-5G10 and P4G9, which map to the C and A epitope regions of α4, respectively (23), reduced infectivity by up to 40% (B-5G10) and 23% (P4G9). Mild trypsin treatment of cells affects α4 integrin expression (38). The blocking by P4G9, but not that by B-5G10, was eliminated when 1 mg/ml trypsin was included in the virus diluent (data not shown). P4G9 is capable of blocking integrin ligand binding, whereas B-5G10 does not, although B-5G10 blocks the aggregation of leukocytic cells induced by P4G9 (23). mAb P4C2, which blocks binding of α4β1-bearing cells to fibronectin and maps to the epitope B2 region of α4 (23), did not affect SA11 virus infectivity (data not shown). Thus, in contrast to the α2 mAbs, blocking of rotavirus infectivity by α4 mAbs did not correlate with the ability of the mAbs to block cellular adhesion.
Figure 7.
Treatment of MA104 cells with anti-α4 mAbs B-5G10 (A) and P4G9 (B) but not with irrelevant mAbs RV-4:1 (A), and RV-5:2 (B; to human rotavirus RV-5 VP4) inhibits SA11 infection.
The anti-α1 mAb TS2/7, mAbs to the β1 subunit [whether adhesion-blocking (P4C10) or adhesion-enhancing (8A2, A-IA5)] (24), and their isotype- and concentration-matched irrelevant controls did not affect SA11 virus infectivity (data not shown).
Therefore, whereas the α subunits were implicated in rotavirus interaction with β1 integrins, the β2 subunit was implicated in a rotavirus–β2 integrin interaction.
Peptide and mAb Blocking of SA11 Infectivity Is Additive.
Combinations of two or three of the blocking peptides or mAbs showed increased infectivity blocking (Fig. 8). RDGEE+LDVT showed increased blocking (53%) over RDGEE alone (15%), and RDGEE+LDVT+GPRP showed increased blocking (66%) over LDVT+GPRP (29%), RDGEE+GPRP (30%), GPRP (19%), and RDGEE (Fig. 8A). Higher levels of blocking were shown by mAbs AK7+MHM23 (59%) and AK7+MHM23+B-5G10 (53%) over AK7 (34%) or MHM23 (33%) alone.
Figure 8.
Inhibition of SA11 infectivity by combinations of peptides (A) and mAbs (B). Each test peptide (RDGEE, LDVT, and GPRP) was used at 0.5 mM, with control peptide GHRP at 0.5 mM for two test peptides, and GHRP at 1.0 mM for one test peptide, to give an total peptide concentration of 1.5 mM. Similarly, each test mAb was used at 20 μg/ml, with 20 μg/ml control mAb RV-4:1 for two test mAbs, and 40 μg/ml control mAb RV-4:1 for one test mAb.
MA104 and Caco-2 Cells Express α2β1 and β2 Integrins.
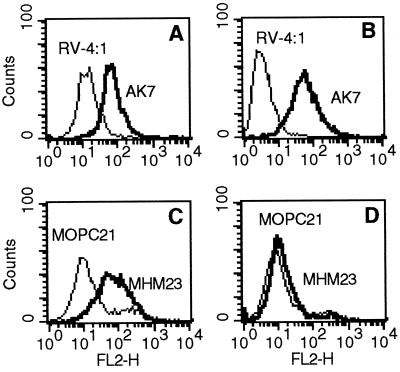
Flow cytometric analysis of trypsinized cells stained with mAbs AK7 and P4C10 showed that both MA104 and Caco-2 cell lines express α2β1 (Fig. 9) (β1 data not shown). Trypsinized MA104 and Caco-2 cells also express β2 at low levels (Fig. 9 C and D). Expression of α4 on either of these cell lines could not be detected by flow cytometry with mAb P4G9 in the three-step stain on trypsinized cells, although P4G9 did stain trypsinized cells of the α4-expressing, rhabdomyosarcoma line RD (data not shown).
Figure 9.
Flow cytometric analysis of α2 and β2 integrin surface expression on MA104 and Caco-2 cells. Results are shown for optimal, saturating concentrations of mAbs AK7 (10 μg/ml) and MHM23 (70 μg/ml). (A and C) MA104 cells express both α2 and β2 integrins. (B and D) Caco-2 cells express α2 integrin and a low level of β2 integrins.
DISCUSSION
The known properties of the rotavirus outer capsid proteins VP4 and VP7 suggest that they both interact with the host cell plasma membrane. Consistent with this, we present evidence that each outer capsid protein interacts with different integrins in the early stages of infection of cells. As the levels of blocking of infectivity were greatest, and the concentrations required were lowest, with the α2β1 ligand peptide and mAbs directed to the α2 subunit, it is likely that the interaction of VP4 with α2β1 is the most significant, at least in the cell lines studied here. This is in agreement with the major role for VP4 in cell attachment. The possible minor role for VP7 in cell attachment is also consistent with its interaction with the β2- and α4-containing integrins. This interaction with integrins may help explain the importance of VP7 as a neutralizing antibody target. It is likely that some antibodies to VP7 and VP4 may neutralize rotavirus by preventing their interaction with integrins.
The concentrations of peptides and mAbs required for blocking of infectivity, and the levels of blocking achieved, were similar to those observed with adenovirus, foot-and-mouth disease virus, and coxsackievirus A9 (14, 15). As adenovirus uses integrins for cellular penetration, rather than for attachment as do the picornaviruses, it is not possible to determine from the degree of blocking whether integrins are involved in rotavirus attachment or in penetration. Direct demonstration of rotavirus binding to integrins has not yet been achieved. Confirmation of the roles of these integrins in rotavirus entry into cells requires analysis of virus replication and yields from cells transfected with each integrin.
The observed sequence relatedness between disintegrins and the C terminus region of VP7 invites functional and structural comparisons. In the three-dimensional structure of echistatin, the protein chain folds to form a rigid core stabilized by four cysteine cross-links, from which protrudes a hairpin loop containing the RGD sequence at its tip (33). In addition to the structural constraints provided by the capsid structure, by analogy with snake venom disintegrins, the disintegrin-like domain of rotavirus VP7 may be stabilized by cysteine links, from which GPR- and LDV-presenting loops extend.
As well as their β subunit disintegrin-like domains, many members of the ADAM (A Disintegrin And Metalloprotease domain) gene family, including fertilin, possess a putative fusion peptide in their α subunit. A peptide analogue of the fertilin α fusion peptide interacts with membranes and induces fusion (39). This fusion peptide is similar in sequence to a potential fusion peptide of the E2 glycoprotein of rubella virus. Thus, the disintegrin-like domain of rotavirus VP7 and the fusion region of VP4 may interact with cells and integrins similarly to the fusion peptide and disintegrin-like domain of the α and β subunits of fertilin. The observed physical interactions between VP4 and VP7 (7, 40) are also consistent with this hypothesis.
A neutralizing mAb that maps to the rotavirus fusion region binds to the distal end of the VP4 spike (40). This fusion region is likely to be in close proximity to the DGE sequence as the cysteine residues at amino acid positions 318 and 379 form a disulfide bond (41). As a virus-neutralizing mAb mapped to amino acid 305 (30) (Fig. 1), it appears that the integrin ligand sequence on the VP4 spike is antigenic and accessible for molecular interactions.
In vivo, rotaviruses generally infect only the mature villus epithelial cells of the small intestine. The α2β1 integrin is expressed on the apical surface of crypt and villus epithelial cells throughout the human intestine (42). In vitro, many adherent cell lines support limited group A rotavirus growth, but fully permissive cell lines are limited to MA104 and colonic adenocarcinoma types (Caco-2 and HT-29) (17). Our flow cytometric analysis showed that both MA104 and Caco-2 cell lines express α2β1. The HepG2 cell line, which is semipermissive for rotaviruses (17), also expresses this integrin, as do most adherent cell lines tested (38). β2 integrins have been detected in 4 of 15 rectal surface epithelial cell biopsies (43). As shown in this study, MA104 and Caco-2 cells also express the β2 integrin subunit at low levels. Thus, cells susceptible to rotavirus infection, which have been tested, express both the α2β1 integrin and the β2 integrin subunit.
Expression of α4 on epithelial cells in the human intestine has not been detected (42). Our studies with a more sensitive three-step stain also failed to show α4 expression in MA104 and Caco-2 cells, although this may have been due to destruction of the mAb epitope by trypsin (38). Further studies, including analysis of α4 mRNA expression, are needed to confirm the absence of α4 on these cell lines and on intestinal cells. The absence of α4 from MA104 cells appears inconsistent with the ability of the peptide LDVT and anti-α4β1 mAbs to block rotavirus infection. However, similar observations with Rous sarcoma virus and its receptor, an avian low density lipoprotein receptor, have been made. Specific antisera to this protein inhibited infection but did not identify it in susceptible avian cells (44). Alternatively, as high levels of anti-α4 mAbs were needed for infectivity blocking, and virus-blocking and integrin function-blocking did not correlate, virus-blocking may have little physiologic relevance.
The widespread expression of α2β1 in the intestine does not appear to explain by itself the trophism of rotavirus for the mature enterocyte. Other receptors, such as glycoconjugates and β2 integrins, may contribute to this specificity. Integrin expression may alter during differentiation (42). Post-entry blocks to infection may also be present in vivo in immature enterocytes.
The adenovirus penton base also contains LDV, 13–50 residues from the RGD sequence. The avian adenovirus type 10 lacks this RGD sequence (45) but contains the α2β1 integrin ligand site DGE two residues from the usual RGD position. Thus, some adenoviruses may use the α2β1 or α4β1 integrins in cellular interactions. It will be of great interest to analyze the role of these viral integrin ligands experimentally in the pathogenesis of these and other viral infections.
Integrin binding and clustering mediated by ligand is critical for the activation of intracellular signals, including elevation of the intracellular calcium concentration (46). The cytopathic effect of rotavirus is due in part to an increase in intracellular calcium levels, which is first detectable at 5 h after infection (47). Although at least some of this effect is due to synthesis of viral proteins (48), it is also possible that integrin signaling contributes to rotavirus-induced cell death.
The α4 and β2 integrins combine to coordinate leukocyte migration into tissues, and α4 integrins are important in lymphocyte-dependent intestinal immunity (49). The rotavirus VP7 integrin ligand sites LDV and GPR also may be involved in modulating anti-viral immune responses.
The efficacy of rotavirus vaccines trialled to date against severe disease has been variable. Identification of integrins as factors in rotavirus cell entry will assist development of new agents of control, which may include disintegrin or integrin ligand analogues and biologically dead vaccines.
Acknowledgments
We thank S. Sonza and M. Hewish for their advice and critical evaluation of the manuscript. B.S.C. is supported by a grant from the National Health and Medical Research Council of Australia.
ABBREVIATION
- TK
tyrosine kinase
References
- 1.Clark S M, Roth J R, Clark M L, Barnett B B, Spendlove R S. J Virol. 1981;39:816–822. doi: 10.1128/jvi.39.3.816-822.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukuhara N, Yoshie O, Kitaoka S, Konno T. J Virol. 1988;62:2209–2218. doi: 10.1128/jvi.62.7.2209-2218.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuentes Panana E M, Lopez S, Gorziglia M, Arias C F. J Virol. 1995;69:2629–2632. doi: 10.1128/jvi.69.4.2629-2632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorziglia M, Larralde G, Ward R L. J Virol. 1990;64:4534–4539. doi: 10.1128/jvi.64.9.4534-4539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackow E R, Shaw R D, Matsui S M, Vo P T, Dang M N, Greenberg H B. Proc Natl Acad Sci USA. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyall-Smith M L, Lazdins I, Tregear G W, Holmes I H. Proc Natl Acad Sci USA. 1986;83:3465–3468. doi: 10.1073/pnas.83.10.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazdins I, Coulson B S, Kirkwood C, Dyall-Smith M, Masendycz P J, Sonza S, Holmes I H. Virology. 1995;209:80–89. doi: 10.1006/viro.1995.1232. [DOI] [PubMed] [Google Scholar]
- 8.Mendez E, Arias C F, Lopez S. J Virol. 1993;67:5253–5259. doi: 10.1128/jvi.67.9.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass D M, Mackow E R, Greenberg H B. Virology. 1991;183:602–610. doi: 10.1016/0042-6822(91)90989-o. [DOI] [PubMed] [Google Scholar]
- 10.Crawford S E, Labbe M, Cohen J, Burroughs M H, Zhou Y J, Estes M K. J Virol. 1994;68:5945–5952. doi: 10.1128/jvi.68.9.5945-5952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludert J E, Feng N, Yu J H, Broome R L, Hoshino Y, Greenberg H B. J Virol. 1996;70:487–493. doi: 10.1128/jvi.70.1.487-493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabara M, Gilchrist J E, Hudson G R, Babiuk L A. J Virol. 1985;53:58–66. doi: 10.1128/jvi.53.1.58-66.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bass D M, Mackow E R, Greenberg H B. J Virol. 1990;64:322–330. doi: 10.1128/jvi.64.1.322-330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berinstein A, Roivainen M, Hovi T, Mason P W, Baxt B. J Virol. 1995;69:2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 16.Coulson B S, Fowler K J, Bishop R F, Cotton R G. J Virol. 1985;54:14–20. doi: 10.1128/jvi.54.1.14-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamoto N, Ramig R F, Matson D O, Estes M K. Virology. 1991;184:729–737. doi: 10.1016/0042-6822(91)90443-f. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwamoto Y, Robey F A, Graf J, Sasaki M, Kleinman H K, Yamada Y, Martin G R. Science. 1987;238:1132–1134. doi: 10.1126/science.2961059. [DOI] [PubMed] [Google Scholar]
- 20.Humphries M J, Akiyama S K, Komoriya K, Yamada K M. J Cell Biol. 1986;103:2637–2647. doi: 10.1083/jcb.103.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell P J, Faull R, Russ G R, D’Apice A J. Immunol Cell Biol. 1991;69:103–110. doi: 10.1038/icb.1991.16. [DOI] [PubMed] [Google Scholar]
- 22.Gamble J R, Matthias L J, Meyer G, Kaur P, Russ G, Faull R, Berndt M C, Vadas M A. J Cell Biol. 1993;121:931–943. doi: 10.1083/jcb.121.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiffer S G, Hemler M E, Lobb R R, Tizard R, Osborn L. J Biol Chem. 1995;270:14270–14273. doi: 10.1074/jbc.270.24.14270. [DOI] [PubMed] [Google Scholar]
- 24.Chan B M, Hemler M E. J Cell Biol. 1993;120:537–543. doi: 10.1083/jcb.120.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diamond M S, Garcia Aguilar J, Bickford J K, Corbi A L, Springer T A. J Cell Biol. 1993;120:1031–1043. doi: 10.1083/jcb.120.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Doherty U, Steinman R M, Peng M, Cameron P U, Gezelter S, Kopeloff I, Swiggard W J, Pope M, Bhardwaj N. J Exp Med. 1993;178:1067–1078. doi: 10.1084/jem.178.3.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staatz W D, Fok K F, Zutter M M, Adams S P, Rodriguez B A, Santoro S A. J Biol Chem. 1991;266:7363–7367. [PubMed] [Google Scholar]
- 28.Loike J D, Sodeik B, Cao L, Leucona S, Weitz J I, Detmers P A, Wright S D, Silverstein S C. Proc Natl Acad Sci USA. 1991;88:1044–1048. doi: 10.1073/pnas.88.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komoriya A, Green L J, Mervic M, Yamada S S, Yamada K M, Humphries M J. J Biol Chem. 1991;266:15075–15079. [PubMed] [Google Scholar]
- 30.Taniguchi K, Maloy W L, Nishikawa K, Green K Y, Hoshino Y, Urasawa S, Kapikian A Z, Chanock R M, Gorziglia M. J Virol. 1988;62:2421–2426. doi: 10.1128/jvi.62.7.2421-2426.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partanen J, Armstrong E, Makela T P, Korhonen J, Sandberg M, Renkonen R, Knuutila S, Huebner K, Alitalo K. Mol Cell Biol. 1992;12:1698–1707. doi: 10.1128/mcb.12.4.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gould R J, Polokoff M A, Friedman P A, Huang T-F, Holt J C, Cook J J, Niewiarowski S. Proc Soc Exp Biol Med. 1990;195:168–171. doi: 10.3181/00379727-195-43129b. [DOI] [PubMed] [Google Scholar]
- 33.Blobel C P, White J M. Curr Opin Cell Biol. 1992;4:760–765. doi: 10.1016/0955-0674(92)90098-w. [DOI] [PubMed] [Google Scholar]
- 34.Wolfsberg T G, Primakoff P, Myles D G, White J M. J Cell Biol. 1995;131:275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamata T, Puzon W, Takada Y. J Biol Chem. 1994;269:9659–9663. [PubMed] [Google Scholar]
- 36.Champe M, McIntyre B W, Berman P W. J Biol Chem. 1995;270:1388–1394. doi: 10.1074/jbc.270.3.1388. [DOI] [PubMed] [Google Scholar]
- 37.Coulson B S. In: Leucocyte Typing VI: White Cell Differentiation Antigens. Kishimoto T, Goyert S, Kikutani H, Masow D, Miyasaka M, Moretta L, Ohno T, Okumura K, Shaw S, Springer T A, Sugamura K, Sugawara H, Kr, von dem Bourne AEG, Zola H, editors. New York: Garland; 1997. , in press. [Google Scholar]
- 38.Hemler M E, Huang C, Takada Y, Schwarx L, Strominger J, Clabby M. J Biol Chem. 1987;262:11478–11485. [PubMed] [Google Scholar]
- 39.Muga A, Neugebauer W, Hirama T, Surewicz W K. Biochemistry. 1994;33:4444–4448. doi: 10.1021/bi00181a002. [DOI] [PubMed] [Google Scholar]
- 40.Prasad B V, Burns J W, Marietta E, Estes M K, Chiu W. Nature (London) 1990;343:476–479. doi: 10.1038/343476a0. [DOI] [PubMed] [Google Scholar]
- 41.Patton J T, Hua J, Mansell E A. J Virol. 1993;67:4848–4855. doi: 10.1128/jvi.67.8.4848-4855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beaulieu J F. J Cell Sci. 1992;102:427–436. doi: 10.1242/jcs.102.3.427. [DOI] [PubMed] [Google Scholar]
- 43.Hussain L A, Kelly C G, Rodin A, Jourdan M, Lehner T. Clin Exp Immunol. 1995;102:384–388. doi: 10.1111/j.1365-2249.1995.tb03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bates P, Young J A, Varmus H E. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 45.Vrati S, Brookes D E, Strike P, Khatri A, Boyle D B, Both G W. Virology. 1996;220:186–199. doi: 10.1006/viro.1996.0299. [DOI] [PubMed] [Google Scholar]
- 46.Clark E A, Brugge J S. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 47.Michelangeli F, Ruiz M C, del Castillo J R, Ludert J E, Liprandi F. Virology. 1991;181:520–527. doi: 10.1016/0042-6822(91)90884-e. [DOI] [PubMed] [Google Scholar]
- 48.Tian P, Estes M K, Hu Y, Ball J M, Zeng C Q, Schilling W P. J Virol. 1995;69:5763–5772. doi: 10.1128/jvi.69.9.5763-5772.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arroyo A G, Yang J T, Rayburn H, Hynes R O. Cell. 1996;85:997–1008. doi: 10.1016/s0092-8674(00)81301-x. [DOI] [PubMed] [Google Scholar]