Abstract
Local translation of proteins in distal dendrites is thought to support synaptic structural plasticity. We have previously shown that metabotropic glutamate receptor (mGluR1) stimulation initiates a phosphorylation cascade, triggering rapid association of some mRNAs with translation machinery near synapses, and leading to protein synthesis. To determine the identity of these mRNAs, a cDNA library produced from distal nerve processes was used to screen synaptic polyribosome-associated mRNA. We identified mRNA for the fragile X mental retardation protein (FMRP) in these processes by use of synaptic subcellular fractions, termed synaptoneurosomes. We found that this mRNA associates with translational complexes in synaptoneurosomes within 1–2 min after mGluR1 stimulation of this preparation, and we observed increased expression of FMRP after mGluR1 stimulation. In addition, we found that FMRP is associated with polyribosomal complexes in these fractions. In vivo, we observed FMRP immunoreactivity in spines, dendrites, and somata of the developing rat brain, but not in nuclei or axons. We suggest that rapid production of FMRP near synapses in response to activation may be important for normal maturation of synaptic connections.
Changes in synaptic connectivity are likely to be a key mechanism by which nervous system organization is permanently changed by experience. Local translation of some proteins in dendrites is increasingly considered to be important for changes in synaptic structure and receptor composition. Certain mRNAs are known to be targeted to dendrites (1, 2), and polyribosomal aggregates are observed in or near dendritic spines, more frequently at newly forming synapses (3–5). Dendrites have been shown to be equipped with components necessary for protein synthesis (6), and synthesis of proteins has been demonstrated directly in synaptoneurosomes (7, 8) and in preparations of dendrites isolated from hippocampal neurons in culture (9). Local translation of transfected reporter-tagged mRNA has been demonstrated in transected dendrites (10).
Protein translation induced by metabotropic receptor stimulation has previously been proposed to play a role in long-term potentiation (LTP), a model for synaptic plasticity (1, 11, 12). LTP induction also alters levels of specific mRNAs in tissue slices (13), and isotope-tagged leucine is taken up in dendritic regions of hippocampal slices in response to stimulation (14). We have demonstrated that phosphoinositide-linked metabotropic glutamate receptors (mGluR1), known to trigger a phosphorylation cascade, cause certain mRNAs to associate rapidly with protein translation complexes in synaptoneurosomes (15); this process is modulated by ionotropic receptors (16). Furthermore, we have shown that depolarization by 40 mM K+ or stimulation by phosphoinositide receptor-specific mGluR agonists increases [35S]methionine incorporation into trichloroacetic acid-precipitable polypeptides (15, 17), indicating that de novo protein synthesis at the synapse occurs as a result of these activities.
Protein translation near synapses may play a role in activity-dependent modification of synapses; for example, synaptic maturation and stabilization during development appear to result, in part, from patterns of presynaptic activation (18). Similar mechanisms have been postulated to affect activity-specific modification of adult synapses (19).
We report here that the mRNA for fragile X mental retardation protein (FMRP) rapidly associates with synaptic polyribosomal complexes in synaptoneurosomes after stimulation by a specific mGluR agonist. Moreover, immunostaining of the synaptosomal proteins at short intervals after stimulation shows increased FMRP expression relative to unstimulated samples, indicating rapid synthesis of FMRP in response to synaptic activation.
MATERIALS AND METHODS
Materials.
mGluR specific agonists 1S,3R-trans-ACPD [(1S,3R)-1-aminocyclopentane-1,3-dicarboxylic acid] and DHPG (3,5-dihydroxyphenylglycine) were obtained from Tocris Neuramin (Bristol, U.K.). Isotopically labeled nucleotides were obtained from Amersham. Biotinylated secondary antibodies were supplied by Kirkegaard & Perry Laboratories; Vector Laboratories supplied avidin–biotin complex kits. Pierce supplied SuperSignal CL-HRP kits. Antibody to glial fibrillary acid protein (GFAP) was from ICN; horseradish-conjugated goat anti-mouse antibody was from Sigma. FMRP specific antibody 1C3–1a (20) was purchased from Chemicon. All other reagents were from Sigma.
Sense and antisense oligonucleotide probes were made to a portion of the human FMR1 sequence (nucleotides 118–162) that has 100% homology to the published sequence (21) of FMR1 and mouse Fmr1 (GenBank accession no. L23971L23971) and 67% homology with FXR1, an autosomally encoded protein with substantial homology to FMRP (22). Another 48 mer synthetic oligonucleotide (sense and antisense) was made to the 3′ coding region of the human FMR1 sequence (GenBank accession number X69962X69962, nucleotides 2023–2070), a stretch with 92% homology to mouse Fmr1, and no homology to any known fragile X-related gene family members, including FXR1 and FXR2.
Synaptoneurosomes.
Synaptoneurosomes, a preparation enriched in resealed presynaptic terminals still attached to resealed postsynaptic processes, were prepared as described (17, 23); briefly, pooled occipital–parietal cortices of 4–8 decapitated juvenile (12–15 d) Long-Evans rats were homogenized in chilled homogenizing buffer (125 mM NaCl/2 mM potassium acetate/100 mM sucrose/50 mM Hepes, pH 7.5/2 mM dithiothreitol/0.2 mg/ml heparin). The suspension was filtered through nylon filters of decreasing pore size (final filter, 10 μm pore), and divided into aliquots for stimulation. Each preparation was standardized by reference to an unstimulated control aliquot. The experiments were completed within 40 min of animal sacrifice.
Polysome Gradients.
Synaptoneurosomes were lysed by addition of Triton X-100 (final concentration, 1.2%), deoxycholate to 0.6%, cycloheximide to 100 μg/ml, in 100 mM Tris (pH 7.6) in the presence of RNase inhibitors. The lysates were layered over 1 M sucrose in a polysome buffer (RKB; 0.01 M Tris, pH 7.6/1.5 mM MgCl2/1 mM potassium acetate/2 mM 2-mercaptoethanol), and centrifuged at 400,000 × g for 11 min in a Beckman TL-100 ultracentrifuge. The resultant polysomal pellets were resuspended in 80 mM Tris (pH 8), 80 mM NaCl, 3 mM MgCl2, 1.2% Triton N-101 (RP; 24, 25). Equal amounts of polysomal pellet RNA were layered on a 12-ml, 15–45% continuous sucrose gradient in 20 mM Tris (pH 9), 80 mM NaCl, 3 mM MgCl2, 0.05% 2-mercaptoethanol with 7–10 units RNasin and 1 mg/ml heparin, and centrifuged for 90 min at 41,000 rpm in a SW41 rotor (most non-polyribosome-associated RNAs are thus not included in the gradient). Samples were collected with an ISCO spectrophotometer-coupled gradient fraction collector, diluted with an equal volume of 12× SSC/14.8% formaldehyde, heated 15 min at 60°C, and frozen in dry ice (26, 27). Samples were dotted on nylon membrane with a Schleicher & Schuell dot blot apparatus, UV crosslinked, and hybridized to oligonucleotides labeled with [32P]dCTP using terminal deoxynucleotide transferase; cDNA inserts were labeled by random hexamer priming with Klenow enzyme.
For probing with oligonucleotides, blots were hybridized in a solution containing 10% dextran sulfate, 1 × SSPE, 2 × Denhardt’s solution, 2% SDS, 200 μg/ml salmon sperm DNA, 200 μg/ml yeast tRNA, and 400 μg/ml poly(A); hybridization occurred overnight at 56°. Blots were washed 2 × 5 min and 2 × 30 min at room temperature (RT) in 1× SSPE, 0.5% SDS, 0.1% skim milk; 2 × 30 min at RT in 1% SDS, 0.2× SSPE; 30 min at 40° in 0.5% SDS, 0.1× SSPE.
For fractionation of ribosome-associated proteins, lysates were centrifuged through 1 M sucrose (as above) and polysomes were washed in buffer containing 50 mM Tris (pH 7.5), 1 mg/ml heparin, 20 mM EDTA, 2 mM EGTA, with 0.1 mg/ml PMSF, 10 μg/ml leupeptin, 20 μg/ml aprotinin, and 100 μM sodium orthovanadate. This was followed with sequential washes in buffers containing 0.5, 1, or 2 M K+ in 50 mM Tris (pH 7.5), with the same protease inhibitors. The eluates were concentrated with Centricon-30 microconcentrators, separated on 8% SDS polyacrylamide gels, blotted to nitrocellulose, and stained with antibody to FMRP.
To measure FMRP expression in synaptoneurosome preparations, a t = 0 sample was removed from a homogeneous suspension, which was then split into two samples. Aliquots were removed from the untreated sample at t = 2 min and t = 5 min. For the treated sample, 10−4 M DHPG was added at t = 0 and aliquots were taken at t = 2 min and t = 5 min. Samples were lysed by the addition of Triton X-100 (1% final concentration) in 50 mM Tris (pH 8), with 50 mM NaCl, 100 μg/ml PMSF, 10 μg/ml leupeptin, and 20 μg/ml aprotinin.
Protein samples were separated on an 8% SDS polyacrylamide gel and blotted to nitrocellulose. The membrane was blocked overnight with 4% nonfat dry milk in Tris-buffered saline (0.1% Tween). The blot was incubated for 1 hr with FMRP-specific antibody (1C3, dilution 1:600), then reacted with horseradish-peroxidase-conjugated goat anti-mouse γ-chain antibody (1:4,500). The immunocomplexes were revealed with chemiluminescence. The blots were then reincubated with specific antibody against GFAP and developed in the same manner.
For quantitative analysis of immunoblots, films were scanned using a flatbed scanner. FMRP and GFAP signals were quantified using nih image 6.0. Three circles with a radius of less than the width of the FMRP band to be measured were placed on each band, and the “mean gray” value for the pixels within each circle was obtained. These measurements were averaged for each band, and the “mean gray” background for an area spanning all of the lanes was subtracted from each. The final value for each band was then divided by the average “mean gray” value (obtained in the same fashion) of the GFAP band from the same lane.
Electron Microscopy.
Rats (P12–P15) were anesthetized (sodium pentobarbital) and intracardially perfused with 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M sodium phosphate buffer (PB; pH 7.2). Brains were fixed 1 hr to overnight at 4°C. Methods used for preembedding immunocytochemistry were described previously (28, 29). Briefly, coronal 40–50 μm sections of dorsal hippocampus, cerebral cortex, and cerebellum were cut on the vibratome into cold PB. Nonspecific binding sites were blocked with 3% normal horse serum (NHS), with 1% BSA and 0.1% Triton X-100 in PB. Sections were incubated in primary antibody [1C3, 1:500 in PB with 1% NHS and 1% BSA (HBPB)] for 24 hr, followed by 1 hr of incubation in biotinylated anti-mouse secondary antibody (1:100) diluted in HBPB. The tissue was washed in HBPB, incubated in avidin-biotin-peroxidase complex for 1 hr, then in 0.05% DAB and 0.01% hydrogen peroxide in PB. After terminating the reaction the tissue was fixed for 10 min in 2% glutaraldehyde in PB, postfixed in 1% osmium tetroxide, dehydrated, and flat-embedded in resin. Ultrathin sections (60 nm) were stained with 4% uranyl acetate in methanol and 0.25% lead citrate and visualized in a CM200 Philips electron microscope (Philips Electronic Instruments, The Netherlands).
RESULTS
We screened 24 clones from a cDNA library made from distal processes of cultured hippocampal neurons (30) against dot blots of total brain RNA and synaptoneurosomal RNA, and determined that 10 of them were enriched in synaptoneurosomal preparations. Previously, by fractionating synaptoneurosomal lysates on sucrose density gradients (17), we had observed that a subset of mRNAs shows a changed distribution along the polyribosome profile, from a broad distribution in an unstimulated preparation to concentration in the small polyribosome fractions immediately after stimulation. We rescreened these 10 synapse-associated cDNAs against dot blots of synaptoneurosomal polyribosomal gradients to test for participation in neurotransmitter-triggered association with translation complexes. Of these, eight mRNAs (as well as the mRNA for a constitutively expressed gene, CoA reductase) were not affected by neurotransmitter stimulation; however, an mRNA with homology to the fragile X mental retardation gene family was one of two associating with ribosomes at the synapse in response to stimulation (the other mRNA has not yet been characterized). Using this cDNA, we isolated a 1.8-kb clone (termed 8–10b) from a rat hippocampal cDNA library (2 × 106 independent clones; average insert size, 1.5 kb); this longer clone likewise was shown to redistribute into small polysomes immediately after neurotransmitter stimulation. Clone 8–10b has extensive amino acid homology (Fig. 1) with both FMRP and FXR1 [a fragile X-related protein that also contains the RNA-binding KH domains (22)]. The 8–10b clone contains a 43-aa insert between two KH domain elements that is not present in FMRP or FXR1.
Figure 1.
Amino acid homologies of FMR1, FXR1 and 2, and the as yet incomplete sequence of clone 8–10b. The consensus sequence for human and mouse FMR1 is taken from Ashley et al. (21). The sequences for FXR1 and FXR2 are from GenBank accession numbers HSU25165 and HSU31501, respectively. Boxes indicate KH and RGG domains of interest; a dotted line indicates either gaps in matching sequences or portions of the 8–10b sequence not yet available.
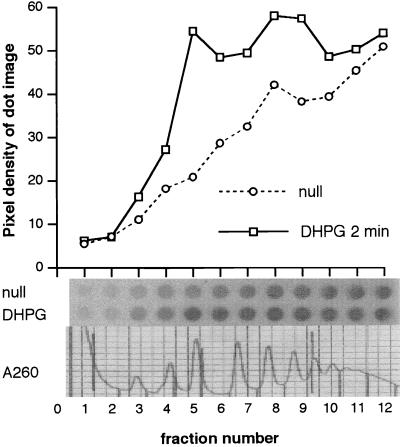
Consequently, sense and antisense oligonucleotides designed from the FMR1 sequence (nucleotides 118–162) were used to probe polysome gradient-derived dot blots of stimulated and unstimulated synaptoneurosomal lysates. Hybridization with the antisense probe shows that rat mRNA homologous to FMR1 was concentrated in polyribosomes 1 min after stimulation by DHPG (31), a specific agonist for phosphoinositide-coupled (group 1) metabotropic glutamate receptors (Fig. 2; data from one of eight replications). Although the presence of FMR1 mRNA is increased in polysomes of all sizes, the increase is most striking in the 80S and small polyribosome region of the profile. Hybridization of a parallel set of dot blots from the same experiment with a control sense probe showed no difference between stimulated and unstimulated lysates. The process was abolished in the presence of the protein synthesis inhibitor puromycin (0.2 mM; data not shown). The 3′ FMR1 oligonucleotide probe (nucleotides 2023–2070), which has no homology to other fragile X-related family members, also revealed a shift of mRNA into polyribosomes following mGluR1 stimulation (five replications; data not shown).
Figure 2.
A representative sucrose gradient-derived dot blot probed with oligonucleotides designed from human FMR1 (nucleotides 118–162) shows that FMR1 mRNA moves into small polyribosomes shortly after mGluR stimulation by 10−2 M DHPG; unstimulated and stimulated aliquots were processed in parallel. Abscissa: dots from serial samples, starting at the top of a 15–45% continuous sucrose gradient. Ordinate: pixel density of dot image, measured on a Fujix BAS 1000 phosphorimager. The original dot images are shown at the base of the figure, along with a tracing of the OD260 polysomal profile.
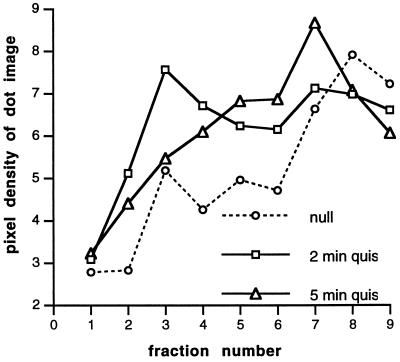
The initial association of FMR1 mRNA with small polyribosomes is succeeded by association with larger polyribosomes, as shown by a comparison of polysome profiles (Fig. 3) of samples taken after 2 min and 5 min of exposure to another metabotropic glutamate agonist treatment [10−5 M quisqualate in the presence of 4 × 10−3 M 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX)]. This suggests that a larger proportion of longer polypeptide chains is present after 5 min of stimulation.
Figure 3.
A polysome gradient dot blot from samples stimulated for 2 or 5 min with 10−5 M quisqualate (in the presence of 4 × 10−3 M CNQX to eliminate the ionotropic glutamate receptor response) was probed with the same oligonucleotide (nucleotides 118–162). Abscissa and ordinate are as in Fig. 2.
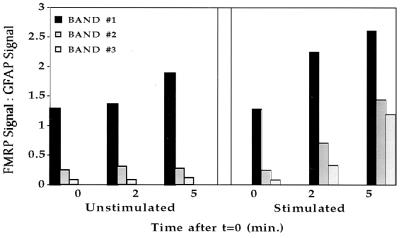
By using trichloroacetic acid precipitation of [35S]methionine-labeled samples taken at short intervals from stimulated and nonstimulated synaptoneurosomal suspensions, we repeatedly observed a temporary acceleration of ongoing protein translation as a result of stimulation (17). Western blot analysis was now used to demonstrate specifically that the rat homologue of FMRP is present in protein preparations from synaptoneurosomes. Fig. 4 shows that there was a steady increase of FMRP relative to GFAP (a glial protein that is not expected to be translated in this preparation) in both stimulated and unstimulated synaptoneurosomes in the course of 5 min of observation, suggesting that FMRP is being synthesized continuously in these preparations. Moreover, after stimulation with the mGluR1 agonist DHPG, FMRP translation was substantially accelerated (Fig. 5).
Figure 4.
Western blot of protein from synaptoneurosomes lysed at indicated intervals (min) after t = 0, either untreated (control) or stimulated by 10−4 M DHPG. (Upper) Staining by anti-FMRP. (Lower) Same blot restained with anti-GFAP.
Figure 5.
Quantitative analysis of the densitometric data obtained from Fig. 4, as described. The three bands stained by anti-FMRP were evaluated separately and standardized to the GFAP protein band.
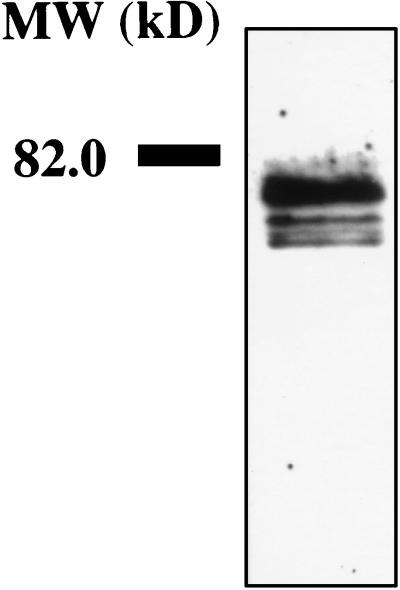
FMRP has been shown to be associated with ribosomes and to be part of the fraction eluted at 0.5 M K+ (32). To test for polysome-associated FMRP, polyribosomes were concentrated from a stimulated synaptoneurosome suspension; following a wash with buffer containing heparin, polyribosome-associated proteins were eluted with 0.5 M K+, separated on SDS/PAGE, blotted, and stained with monoclonal antibody. Fig. 6 shows the presence of FMRP in the polyribosome-associated proteins near synapses.
Figure 6.
Polyribosome-associated proteins eluted with 0.5 M K+ buffer were separated on an 8% SDS polyacrylamide gel, blotted to nitrocellulose, stained with anti-FMRP, and visualized by enhanced chemiluminescence.
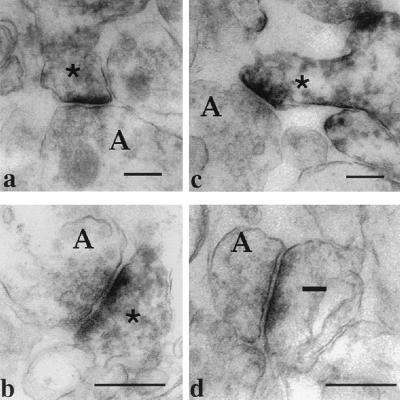
Preembedding immunolabeling for electron microscopy revealed FMRP immunoreactivity within spines, in dendrites and in somata, but not in axons, neuronal nuclei, or glia of developing rat brain. Immunoreactivity is located in spines near postsynaptic densities (Fig. 7).
Figure 7.
Electron micrographs demonstrating FMRP localization in dendritic spines in cerebral cortex and hippocampus. (a and b) Dendritic spines in s. radiatum of CA1 area of hippocampus. (c) Dendritic spine in the visual cortex containing dark immunoperoxidase precipitate (∗). Presynaptic axonal terminals (A) are not labeled. (d) Unlabeled (—) spine in the visual cortex. Bars = 500 nm.
DISCUSSION
We have shown that mRNA complementary to human FMR1 and murine Fmr1 moves into close association with polyribosomes isolated from rat cortex synaptoneurosomes within 1 min after mGluR1 stimulation. This suggested that translation of this protein could occur near synapses; in accordance with this we observed a slight increase in FMRP expression over time in synaptoneurosomal preparations, as demonstrated by staining with FMRP-specific monoclonal antibody. This indicates that there is a low constitutive level of translation in synapses. This translation is markedly accelerated during the first 5 min after stimulation by the mGluR1 agonist DHPG, which suggests that in vivo synthesis of FMRP might be under the control of afferent activation.
Fragile X syndrome is one of the most common inherited forms of mental retardation in humans, with an estimated population incidence of 1/2,000 in males (33). It is inherited as an X-linked genetic trait with reduced penetrance (34); phenotypic traits for the syndrome include moderate to severe mental retardation, autistic behavior, macroorchidism, and facial abnormalities (35, 36). The FMR1 gene contains a trinucleotide repeat [(CGG)n] in the 5′ untranslated region; expansion of the repeat length beyond 200 usually leads to a hypermethylation of the promotor region and transcriptional suppression of the FMR1 gene (37–41). The protein product, FMRP, is enriched in nervous tissue (20, 42–44). Although its function is still unknown, it has been demonstrated that it contains three regions (two KH domains and one RGG box) that are known to bind RNA (32, 37, 45) and ribosomes (46). A patient with a point mutation in one of the KH domains exhibited severe mental retardation symptoms (47), strongly suggesting that RNA binding is vital to the function of the protein.
It is, by now, widely accepted that specific mRNAs are transported to dendrites and translated locally (1, 9). Both subcellular localization of translation (10) and localized translational control (48) have been shown for a subset of mRNAs in neurons. FMRP contains both nuclear localization signals (NLS) and nuclear export signals (49–51), and it has recently been shown that FMRP can be found in ribonucleoprotein particles (52), suggesting that nascent FMRP could enter the nucleus to assemble into mRNP particles prior to export back into the cytoplasm. In the case of CaMKIIα, transfection of constructs containing NLS has shown that protein is found both near the soma and in distal (but not proximal) dendrites, implying that nuclear localization machinery is overruled in the more distal regions of the dendrites (53). Thus, the presence of a nuclear localization signal does not mean that FMRP is restricted to the region of the soma; indeed, FMRP is predominantly localized in cytoplasm (20, 42).
FMRP may be involved in dendritic spine maturation, as both patients and Fmr1 knockout mice exhibit an immature spine morphology. Autopsy results indicate that forebrain synapses in fragile X patients exhibit a thin, elongated morphology in Golgi preparations and a reduced synaptic contact size in electron microscopy, both of which are characteristic of immature or experience-deprived synapses in the cerebral cortex (54, 55). Studies of visual cortical synapses in monocularly deprived brains indicate a similar failure of morphological synaptic maturation, suggesting the importance of experience-driven neuronal/synaptic activity to the synaptic stabilization process (56, 57). It has recently been shown that fragile X knockout mice share this immature synapse morphology (58). This suggests that synaptically regulated synthesis of FMRP may be involved in dendritic spine maturation, and it is not unreasonable to postulate that this process may be impaired in cases of fragile X syndrome.
Although the specific function of FMRP that leads to spine abnormalities has not been determined, FMRP has been shown to bind to ribosomes (46, 52). Polyribosomal aggregates are seen in dendritic spines, particularly during development and synaptogenesis, and we have previously shown that protein translation occurs in developing synapses in response to neurotransmitter stimulation (17). We have now shown that a rat homologue of FMRP is produced at synapses and its translation is accelerated upon mGluR1 stimulation. This protein is observed in dendrites and dendritic spines in normal P12–P14 rats in the hippocampus, cerebral cortex, and cerebellum. We propose that FMRP may be involved in regulating translation of protein at the synapse, and that its absence might impede this synthesis and consequently impair synaptic maturation.
Acknowledgments
A.K. is grateful to Drs. Maryann Martone and Mark Ellisman for training at the National Center for Microscopy and Imaging Research in San Diego (National Institutes of Health Grant RR04050 to Mark Ellisman). We thank Dr. Edouard Khandjian for valuable advice. This work was supported by grants from the MacArthur Foundation, the Kiwanis International Spastic Paralysis Research Foundation, and the Illinois Chapter of the American Association on Mental Retardation; a grant from the FRAXA Foundation; National Institutes of Health Grants MH35321 and AG10154; and U.S. Public Health Service Training Grant HD0777.
ABBREVIATIONS
- FMRP
fragile X mental retardation protein
- GFAP
glial fibrillary acid protein
- DHPG
3,5-dihydroxyphenylglycine
References
- 1.Steward O, Banker G. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 2.St. Johnston D. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 3.Steward O, Levy W. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steward O, Falk P. J Neurosci. 1986;6:412–423. doi: 10.1523/JNEUROSCI.06-02-00412.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martone M E, Pollock J A, Jones Y Z, Ellisman M H. J Neurosci. 1996;16:7437–7446. doi: 10.1523/JNEUROSCI.16-23-07437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiedge H, Brosius J. J Neurosci. 1996;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wedege E, Luqmani Y, Bradford H. J Neurochem. 1977;29:527–537. doi: 10.1111/j.1471-4159.1977.tb10702.x. [DOI] [PubMed] [Google Scholar]
- 8.Rao A, Steward O. J Neurosci. 1991;11:2881–2895. doi: 10.1523/JNEUROSCI.11-09-02881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torre E R, Steward O. J Neurosci. 1992;12:762–772. doi: 10.1523/JNEUROSCI.12-03-00762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crino P B, Eberwine J. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 11.Fazeli M S, Corbet J, Dunn M J, Bliss T V P. J Neurosci. 1993;13:1346–1353. doi: 10.1523/JNEUROSCI.13-04-01346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiler I J, Hawrylak N, Greenough W T. Behav Brain Res. 1994;66:1–6. doi: 10.1016/0166-4328(94)00116-w. [DOI] [PubMed] [Google Scholar]
- 13.Mackler S A, Brooks B P, Eberwine J H. Neuron. 1992;9:539–548. doi: 10.1016/0896-6273(92)90191-f. [DOI] [PubMed] [Google Scholar]
- 14.Feig S, Lipton P. J Neurosci. 1993;13:1010–1021. doi: 10.1523/JNEUROSCI.13-03-01010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiler I J, Greenough W T. Proc Natl Acad Sci USA. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiler I J, Childers W S, Greenough W T. J Neurochem. 1996;66:197–210. doi: 10.1046/j.1471-4159.1996.66010197.x. [DOI] [PubMed] [Google Scholar]
- 17.Weiler I J, Greenough W T. Mol Cell Neurosci. 1991;2:305–314. doi: 10.1016/1044-7431(91)90060-2. [DOI] [PubMed] [Google Scholar]
- 18.LeVay S, Wiesel T N, Hubel D H. J Comp Neurol. 1980;191:1–51. doi: 10.1002/cne.901910102. [DOI] [PubMed] [Google Scholar]
- 19.Abraham W C, Bear M F. Trends Neurosci. 1996;19:126–130. doi: 10.1016/s0166-2236(96)80018-x. [DOI] [PubMed] [Google Scholar]
- 20.Devys D, Lutz Y, Rouyer N, Bellocq J-P, Mandel J-L. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 21.Ashley C T, Sutcliffe J S, Kunst C B, Leiner H A, Eichler E E, Nelson D L, Warren S T. Nat Genet. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 22.Siomi M C, Siomi H, Sauer W H, Srinivasan S, Nussbaum R L, Dreyfuss G. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollingsworth E B, McNeal E T, Burton J L, Williams R, Daly J, Creveling C. J Neurosci. 1985;5:2240–2253. doi: 10.1523/JNEUROSCI.05-08-02240.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry R P, LaTorre J, Kelley D E, Greenberg J R. Biochim Biophys Acta. 1972;262:220–226. doi: 10.1016/0005-2787(72)90236-5. [DOI] [PubMed] [Google Scholar]
- 25.Kaspar R L, Gehrke L. J Immunol. 1994;153:277–286. [PubMed] [Google Scholar]
- 26.White B A, Bancroft F C. J Biol Chem. 1982;257:8569–8572. [PubMed] [Google Scholar]
- 27.Singleton C K, Delude R L, McPherson C E. Dev Biol. 1987;119:433–441. doi: 10.1016/0012-1606(87)90047-9. [DOI] [PubMed] [Google Scholar]
- 28.Somogyi P, Halasy K, Somogyi P, Storm-Mathisen J, Ottersen O P. Neurosci. 1986;19:1045–1050. doi: 10.1016/0306-4522(86)90121-1. [DOI] [PubMed] [Google Scholar]
- 29.Sesack S R, Aoki C, Pickel V M. J Neurosci. 1994;14:88–106. doi: 10.1523/JNEUROSCI.14-01-00088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyashiro K, Dichter M, Eberwine J H. Proc Natl Acad Sci USA. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoepp D D, Johnson B G, Monn J A. J Neurochem. 1996;66:1981–1985. doi: 10.1046/j.1471-4159.1996.66051981.x. [DOI] [PubMed] [Google Scholar]
- 32.Siomi H, Choi M, Siomi M C, Nussbaum R, Dreyfuss G. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 33.Brown W T. Am J Hum Genet. 1996;58:903–905. [PMC free article] [PubMed] [Google Scholar]
- 34.Heitz D, Devys D, Imbert G, Kretz C, Mandel J L. J Med Genet. 1992;29:794–801. doi: 10.1136/jmg.29.11.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren S T, Nelson D L. J Am Med Assoc. 1994;271:536–542. [PubMed] [Google Scholar]
- 36.Hagerman R J, Cronister A, editors. Fragile X Syndrome: Diagnosis, Treatment, and Research. 2nd Ed. Baltimore: Johns Hopkins Univ. Press; 1996. [Google Scholar]
- 37.Verkerk A J M H, Pieretti M, Sutcliffe J S, Fu Y H, Kuhl D P A, et al. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 38.Fu Y H, Kuhl J P A, Pizzuti A, Pieretti M, Sutcliffe J S, Richards S, Verkerk A J M H, Holden J J A, Fenwick R G, Warren S T, Oostra B A, Nelson D L, Caskey C T. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 39.Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley J C, Warren W T, Schlessinger D, Sutherland G R, Richards R I. Science. 1991;252:1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- 40.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M F, Mandel J L. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 41.Smeets H J M, Smits A P T, Verheij C E, Theelen J P G, Willemsen R, van de Burgt I, Hoogeveen A T, Oosterwijk J C, Oostra B A. Hum Mol Genet. 1995;4:2103–2108. doi: 10.1093/hmg/4.11.2103. [DOI] [PubMed] [Google Scholar]
- 42.Verheij C, Bakker C E, deGraaff E, Keulemans J, Willemsen R, Verkerk A J M H, Galjaard H, Reuser A J J, Hoogeveen A T, Oostra B A. Nature (London) 1993;363:722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- 43.Abitbol M, Menini C, Delezoide A L, Rhyner T, Vekemans M, Mallet J. Nat Genet. 1993;4:147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- 44.Hinds H L, Ashley C T, Sutcliffe J S, Nelson D L, Warren S T, Housman D E, Schalling M. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- 45.Musco G, Stier G, Joseph C, Castiglione Morelli M A, Nilges M, Gibson T J, Pastore A. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 46.Khandjian E W, Corbin F, Woerly S, Rousseau F. Nat Genet. 1996;12:1–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- 47.De Boulle K, Verkerk A J M H, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van Den Bos F, De Graaff E, Oostra B A, Willems P J. Nat Genet. 1993;3:1–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 48.Bian F, Chu T, Schilling K, Oberdick J. Mol Cell Neurosci. 1996;7:116–133. doi: 10.1006/mcne.1996.0009. [DOI] [PubMed] [Google Scholar]
- 49.Sittler A, Devys D, Weber C, Mandel J-L. Hum Mol Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 50.Fridell R A, Benson R E, Hua J, Bogerd H P, Cullen B R. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 51.Eberhart D E, Malter H E, Feng Y, Warren S T. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 52.Tamanini F, Meijer N, Verheij C, Willems P J, Galjaard H, Oostra B A, Hoogeveen A T. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 53.Mayford M, Baranes D, Podsypanina K, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rudelli R D, Brown W T, Wisniewski K, Jenkins E C, Laure-Kamionowska M, Connell F, Wisniewski H M. Acta Neuropathol. 1985;67:89–295. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- 55.Hinton V J, Brown W T, Wisniewski K, Rudelli R D. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- 56.Tieman S B. J Comp Neurol. 1984;222:166–176. doi: 10.1002/cne.902220203. [DOI] [PubMed] [Google Scholar]
- 57.Friedlander M J, Martin K A C, Wassenhove-McCarthy D. J Neurosci. 1991;11:3268–3288. doi: 10.1523/JNEUROSCI.11-10-03268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Comery T A, Harris J B, Willems P J, Oostra B A, Irwin S A, Weiler I J, Greenough W T. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]