Abstract
Fragile X syndrome arises from blocked expression of the fragile X mental retardation protein (FMRP). Golgi-impregnated mature cerebral cortex from fragile X patients exhibits long, thin, tortuous postsynaptic spines resembling spines observed during normal early neocortical development. Here we describe dendritic spines in Golgi-impregnated cerebral cortex of transgenic fragile X gene (Fmr1) knockout mice that lack expression of the protein. Dendritic spines on apical dendrites of layer V pyramidal cells in occipital cortex of fragile X knockout mice were longer than those in wild-type mice and were often thin and tortuous, paralleling the human syndrome and suggesting that FMRP expression is required for normal spine morphological development. Moreover, spine density along the apical dendrite was greater in the knockout mice, which may reflect impaired developmental organizational processes of synapse stabilization and elimination or pruning.
Fragile X syndrome is the most common inherited form of human mental retardation after Down Syndrome. It is an X-linked genetic trait with an incidence of 1/2,000 in males (1–3). Phenotypic abnormalities include moderate to severe mental retardation, autistic behavior, macroorchidism, and facial abnormalities (1). The FMR1 gene contains a trinucleotide repeat [(CGG)n] in the 5′ untranslated region that is expanded (≥200 repeats) in fragile X patients (2, 4, 5), resulting in hypermethylation of the FMR1 promoter region and absence of the encoded fragile X mental retardation protein (FMRP). Studies prior to the characterization of fragile X syndrome associated immature dendritic spine morphology (long and thin) with some forms of mental retardation (6–8). Similarly, fragile X cerebral cortical autopsy material exhibits thin, elongated spines and small synaptic contacts (9, 10).
Recently, transgenic Fmr1 mice were produced in which the Fmr1 gene was disrupted in embryonic stem cells using a targeting vector to exon 5 and homologous recombination (11). The resultant homozygous knockout mice express no FMRP. Phenotypically, these mice exhibit increased testicular size and mildly impaired performance on the Morris water maze (11), paralleling human symptoms. To explore further parallels to the human syndrome, we examined the effect of the Fmr1 knockout on dendritic spines in the visual cortex using the Golgi-Cox impregnation technique.
MATERIALS AND METHODS
Male mutant (n = 4) and wild-type FVB strain (n = 4) adult (16-week-old) mice were prepared for Golgi-Cox impregnation. Mice were anesthetized with sodium pentobarbital (85 mg/kg) and perfused transcardially with 60 ml of 0.9% NaCl (pH 7.3). Brains were removed and were cut sagittally along the midline. Tissue blocks were then immersed in 100 ml of Golgi-Cox solution (1% potassium dichromate/1% mercuric chloride/0.8% potassium chromate in distilled water) for 18 days. Tissue blocks were then immersed in 30% sucrose in 0.9% saline for 2 days and subsequently sectioned (200 μm) using a vibratome. Sections were mounted on gelatinized slides, developed, fixed, dehydrated, coverslipped, and assigned codes that did not reveal group identity.
Spine Length Measurements.
Layer V pyramidal neurons from visual cortex exhibiting complete Golgi impregnation and having apical dendrites with a length greater than 100 μm were selected for quantification. The length of each of 30 spines was measured beginning 50 μm from the soma using an eyepiece reticule. Spine lengths were collected from five neurons from five different tissue sections from each of the animals. Overall means for each animal and frequency of each of the five reticule spine length “bins” (30 spines per neuron, 5 neurons per animal) were calculated and used for statistical analysis.
Spine Density Measurements.
Fully impregnated layer V pyramidal neurons from visual cortex were selected for density measurements. Spine density on apical dendrites was obtained in 25 μm segments beginning at the point of contact between the apical dendrite and the soma from five neurons from five sections for each of the animals. The number of spines visible in profile along the dendrite was counted for every 25-μm segment along the entire visible length of apical dendrite (in all cases, at least 100 μm). Spines less than about 0.4 μm in profile length (½ reticule scale division) were not counted; these spines were usually those projecting at oblique angles and partially obscured by overlap with the dendritic shaft. Dendrite diameter was measured at the beginning of each counted segment, and an additional sample of three knockout and two wild-type dendritic diameters was also measured to ensure an adequate sample size for the detection of any difference, as this can affect spine density measures. Overall means for each segment for each animal were analyzed using a two-factor (genotype by dendritic segment) ANOVA. All spine measurements were made by an experimenter blind to the experimental condition of the animals.
RESULTS
Knockout mice displayed significantly longer dendritic spines than wild-type control animals (one-tailed t test; t = 2.25, df = 6, P = 0.033). Fig. 1 shows representative apical dendrites from knockout and wild-type mouse visual cortex, and Fig. 2 presents group means.
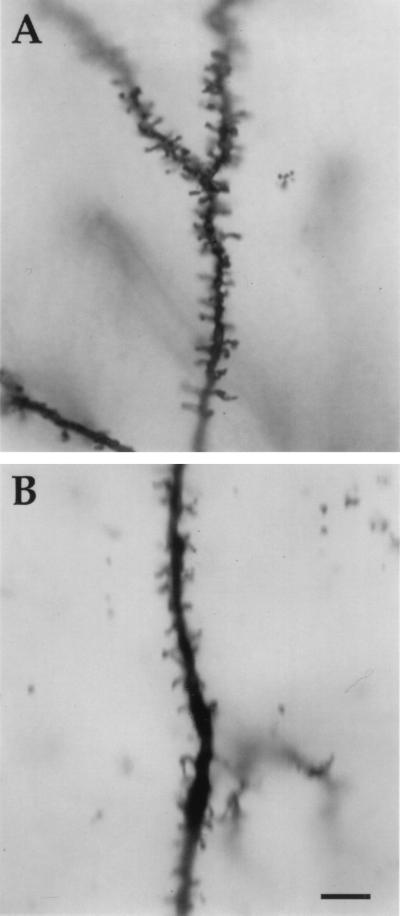
Figure 1.
Golgi-Cox impregnated apical dendrites in transgenic and wild-type mice. (A) Segment of apical dendrite from layer V pyramidal neuron in Fmr1 knockout mouse demonstrating both the increased incidence of long, thin dendritic spines and the increased spine density. (B) Apical dendrite of layer V pyramidal neuron from wild-type mouse. (Bar = 10 μm.)
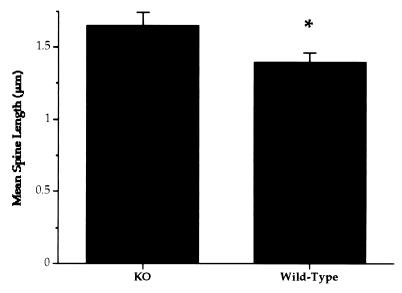
Figure 2.
Mean dendritic length (and SEM) of spines on apical dendrites of layer V pyramidal neurons in visual cortex from knockout (KO) and wild-type mice. Dendritic spines in Fmr1 transgenic mice were significantly longer than those in the wild-type animals (one-tailed t test; t = 2.25, df = 6, P = 0.033).
The Fmr1 knockout mice had both short “mushroom”-shaped dendritic spines and thin, elongated spines; elongated spines were more prevalent in knockout than in wild-type mice, and knockout mice had fewer short spines, as Fig. 3 shows (χ2 = 46.29, 3 df, P < .0005). As Fig. 4 indicates, the density of dendritic spines along the apical dendrites of layer V pyramidal cells was substantially greater in the knockouts than in wild-type mice (F1,33 = 63.3; P < 0.0001). As neither mean dendrite diameter (knockout = 1.55 ± 0.05 μm; wild-type = 1.59 ± 0.05 μm; F1,10 = 0.1048, P > .5) nor the frequency of the longest category of spine length (3.33 μm in Fig. 3) differed between knockout and wild-type mice, correction of the density values for such differences (e.g., ref. 12) was unnecessary (see ref. 13).
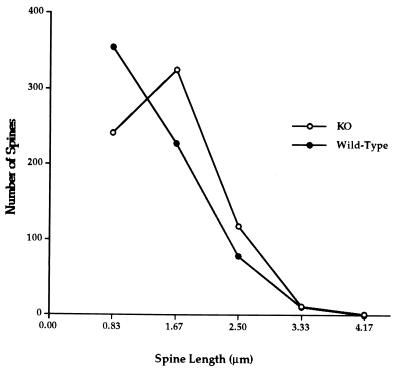
Figure 3.
Number of spines in each reticule-based length category on apical dendrites of layer V pyramidal neurons in visual cortex from knockout (KO) and wild-type mice. Knockout mice have fewer short spines and more long spines (χ2 = 46.29, 3 df, P < .0005).
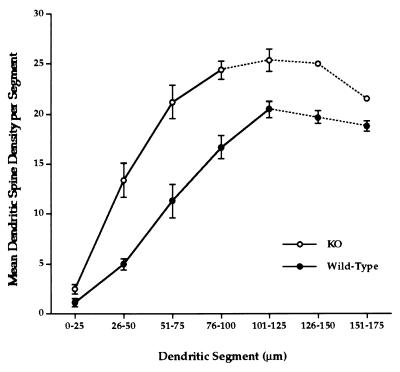
Figure 4.
Mean spine density (and SEM) distributions in fragile X knockout (KO) and wild-type mice. Overall spine density along apical dendrites of layer V pyramidal cells is significantly greater in knockout mice than in wild-type controls (F1,33 = 63.3, P < 0.0001). Dotted lines in graphs indicate reduced numbers of dendrites in the analysis due to some apical dendrites having been truncated by the section plane.
DISCUSSION
The prevalence of thin, elongated dendritic spine morphology in Fmr1 knockout mice is reminiscent of that observed during early synaptogenesis in the developing brain (14) as well as of the morphology seen following sensory deprivation (15, 16). The failure to match the adult wild-type spine length distribution suggests a reduced tendency for spines to mature to an adult morphology. In addition, the increased number of long, thin spines and the increased spine density observed in the knockout mice point to a deficit in the normal selection or “pruning” of synaptic contacts that occurs in development (17). These two deficits could be related or independent effects of the absence of the Fmr1 gene. These results implicate the Fmr1 gene and its associated protein, FMRP, in the developmental processes leading to normal adult dendritic spine morphology and number. Moreover, they suggest that FMRP is required for the synapse stabilization–elimination process whereby sensory systems become organized in development (18).
It is of interest in this regard that we have recently shown, using a synaptoneurosome preparation, that FMRP is synthesized locally at the synapse in response to synaptic activity. FMR1 mRNA is taken up into postsynaptic polyribosomes in response to group 1 metabotropic glutamate receptor activation (19–21), and FMRP levels increase in minutes, as demonstrated using Western blot analysis (21). The present findings suggest that synthesis of FMRP at synapses in response to synaptic activation may play a role in synapse maturation and in the selection–elimination process. Both FMR1 mRNA and FMRP are enriched in nervous tissue (22–25). Although the function of FMRP is unknown, it has been shown to contain three RNA binding regions (two KH domains and one RGG box) (26, 27). FMRP binds approximately 4% of fetal human brain mRNAs (27). It is clear that RNA binding is functionally important because mutations in the RNA-binding region are associated with severe mental retardation (28).
Understanding the role that FMRP and its synthesis at the synapse may play in synapse maturation, selection, and stabilization awaits a clearer understanding of the protein’s cellular functions. Given its RNA binding properties, however, a number of possibilities exist. Among them are: (i) FMRP may facilitate or inhibit protein synthesis by regulating access or targeting of mRNA to polyribosomes, or (ii) FMRP may localize mRNA to specific sites, thus regulating the spatial pattern of protein expression. Thus, FMRP could act as a spatial and/or temporal regulator of activity-dependent protein synthesis at the synapse. We have demonstrated the presence of FMRP in spines using immunohistochemistry (21).
The use of knockout mice to study the normal function of a protein is often criticized (e.g., ref. 29), because expression or functions of other proteins may be altered in response to the loss of the absent protein. Similarly, it is not clear to what extent the observed results of a transgenic knockout are due to the absence of the protein at the time of testing or a result of altered development due to the absence, and resulting compensatory mechanisms, of the protein throughout the life of the organism. These concerns are less relevant in the case of the Fmr1 knockout. In these animals the lack of FMRP mimics the mechanism underlying the human syndrome. Thus, although compensatory mechanisms may occur as a result of an absence of FMRP, it is likely that similar compensation may occur both in the mouse model and the human syndrome.
The altered dendritic spine morphology and density, as well as the observed behavioral deficits and macroorchidism associated with the Fmr1 knockout, suggest that this mouse may be an excellent model for human fragile X syndrome, as well as for providing a new system for studying synaptic maturation and stabilization. In addition, this work, when considered with work demonstrating synaptic control of FMRP synthesis (21), supports the possibility that FMRP may play an essential role during the development of an adult synaptic architecture.
Acknowledgments
We thank Brett Atwater for histological assistance. This paper was supported by the FRAXA Foundation, MH35321, the National Association for Research on Schizophrenia and Affective Disorders, the MacArthur Foundation Psychopathology Development Network, and the Kiwanis Foundation.
ABBREVIATIONS
- FMRP
fragile X mental retardation protein
- FMR1
fragile X mental retardation gene or mRNA
- Fmr1
mouse FMR1 gene
References
- 1.Hagerman R J, Cronister A, editors. Fragile X Syndrome: Diagnosis, Treatment, and Research. 2nd Ed. Baltimore: Johns Hopkins Univ. Press; 1996. [Google Scholar]
- 2.Warren S T, Nelson D L. J Am Med Assoc. 1994;271:536–542. [PubMed] [Google Scholar]
- 3.Brown W T. Am J Hum Genet. 1996;58:903–905. [PMC free article] [PubMed] [Google Scholar]
- 4.Verheij C, Bakker C E, de Graaff E, Keulemans J, Willemsen R, Verkerk A J, Galjaard H, Reuser A J, Hoogeveen A T, Oostra B A. Nature (London) 1993;363:722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- 5.Verkerk A J M H, Pieretti M, Sutcliffe J S, Fu Y H, Kuhl D P A, et al. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 6.Marin-Padilla M. Brain Res. 1972;44:625–629. doi: 10.1016/0006-8993(72)90324-1. [DOI] [PubMed] [Google Scholar]
- 7.Marin-Padilla M. Brain Res. 1974;66:375–391. [Google Scholar]
- 8.Purpura D P. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- 9.Rudelli R D. Acta Neuropathol. 1985;67:289–295. doi: 10.1007/BF00687814. [DOI] [PubMed] [Google Scholar]
- 10.Hinton V J, Brown W T, Wisniewski K, Rudelli R D. Am J Med Genet. 1994;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- 11.Dutch-Belgium Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 12.Feldman M L, Peters A. J Comp Neurol. 1979;188:527–542. doi: 10.1002/cne.901880403. [DOI] [PubMed] [Google Scholar]
- 13.Horner C H, Arbuthnott E. J Anat. 1991;177:179–184. [PMC free article] [PubMed] [Google Scholar]
- 14.Purpura D P. Adv Neurol. 1975;12:91–134. [PubMed] [Google Scholar]
- 15.Tieman S B. J Comp Neurol. 1984;222:166–176. doi: 10.1002/cne.902220203. [DOI] [PubMed] [Google Scholar]
- 16.Friedlander M J, Martin K A, Wassenhove-McCarthy D. J Neurosci. 1991;11:3268–3288. doi: 10.1523/JNEUROSCI.11-10-03268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boothe R G, Greenough W T, Lund J S, Wrege K. J Comp Neurol. 1979;186:473–490. doi: 10.1002/cne.901860310. [DOI] [PubMed] [Google Scholar]
- 18.LeVay S, Wiesel T N, Hubel D H. J Comp Neurol. 1980;191:1–51. doi: 10.1002/cne.901910102. [DOI] [PubMed] [Google Scholar]
- 19.Weiler I J, Greenough W T. Mol Cell Neurosci. 1991;2:305–314. doi: 10.1016/1044-7431(91)90060-2. [DOI] [PubMed] [Google Scholar]
- 20.Weiler I J, Greenough W T. Proc Natl Acad Sci USA. 1993;90:7168–7171. doi: 10.1073/pnas.90.15.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiler I J, Irwin S A, Klintsova A Y, Spencer C M, Brazelton A D, Miyashiro K, Comery T A, Patel B, Eberwine J, Greenough W T. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abitbol M, Menini C, Delezoide A-L, Rhyner T, Vekemans M, Mallet J. Nat Genet. 1993;4:147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- 23.Devys D, Lutz T, Rouyer N, Bellocq J P, Mandel J L. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 24.Hinds H L, Ashley C T, Sutcliffe J S, Nelson D L, Warren S T, Housman D E, Schalling M. Nat Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- 25.Khandjian E W, Fortin A, Thibodeau A, Tremblay S, Cote F, Devys D, Mandel J L, Rousseau F. Hum Mol Genet. 1995;4:783–789. doi: 10.1093/hmg/4.5.783. [DOI] [PubMed] [Google Scholar]
- 26.Ashley C T, Wilkinson K D, Reines D, Warren S T. Science. 1993;262:563–565. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 27.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 28.De Boulle K, Verkerk A J M H, Reyniers E, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van Den Bos F, De Graaff E, Oostra B A, Willems P J. Nat Genet. 1993;3:1–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 29.Routtenberg A. Nature (London) 1995;374:314–315. doi: 10.1038/374314b0. [DOI] [PubMed] [Google Scholar]