Abstract
Structural models of inward rectifier K+ channels incorporate four identical or homologous subunits, each of which has two hydrophobic segments (M1 and M2) which are predicted to span the membrane as α helices. Since hydrophobic interactions between proteins and membrane lipids are thought to be generally of a nonspecific nature, we attempted to identify lipid-contacting residues in Kir2.1 as those which tolerate mutation to tryptophan, which has a large hydrophobic side chain. Tolerated mutations were defined as those which produced measurable inwardly rectifying currents in Xenopus oocytes. To distinguish between water-accessible positions and positions adjacent to membrane lipids or within the protein interior we also mutated residues in M1 and M2 individually to aspartate, since an amino acid with a charged side chain should not be tolerated at lipid-facing or interior positions, due to the energy cost of burying a charge in a hydrophobic environment. Surprisingly, 17 out of 20 and 17 out of 22 non-tryptophan residues in M1 and M2, respectively, tolerated being mutated to tryptophan. Moreover, aspartate was tolerated at 15 out of 22 and 15 out of 21 non-aspartate M1 and M2 positions respectively. Periodicity in the pattern of tolerated vs. nontolerated mutations consistent with α helices or β strands did not emerge convincingly from these data. We consider the possibility that parts of M1 and M2 may be in contact with water.
Ion channels perform important physiological functions by forming pores in biological membranes which are otherwise impermeable to electrolytes (1). By necessity these proteins must be integral membrane proteins, with a functional moiety involved in ion permeation residing within the bounds of the membrane itself. This functional necessity presumably requires the ion channel to maintain a specific intramembrane structure, with specific interactions between its individual transmembrane segments. Mutation of residues which are involved in these specific interactions would be expected to disrupt the structure and thereby the function of the protein. Indeed, mutations that prevent the formation of salt bridges in an inward rectifier K+ channel severely altered ion permeation (2). Residues located on the outside of the protein and in contact with the membrane lipids are expected to be hydrophobic (3), but not subjected to interactions that impose constraints over the size or shape of these residues, as in the case of photosynthetic reaction centers (4). Mutation of lipid-exposed residues to other hydrophobic amino acids would therefore be expected to be tolerated, and the tertiary structure of the protein within the membrane should be related to the pattern of tolerated and nontolerated mutations along the primary sequence (5–8). Introduction of a charged residue into a position within the interior of the protein or on the exterior (facing lipids) would not be expected to be tolerated in either case because of the unfavorable hydrophobic environment (9).
Integral membrane proteins whose structures are known to high resolution have membrane-spanning segments which form either α helices (10, 11) or β barrels (12, 13). A simple bundle of α helices arranged perpendicular to the plane of the membrane would be expected to give rise to a pattern whereby tolerant and intolerant residues are segregated to different sides of the helix, if one assumes that residues within a transmembrane segment contact either other protein residues or lipids, unless they are pore-lining and in contact with water. With this rationale in mind, we tested an inward rectifier K+ channel for evidence of a “helical bundle” type of structure.
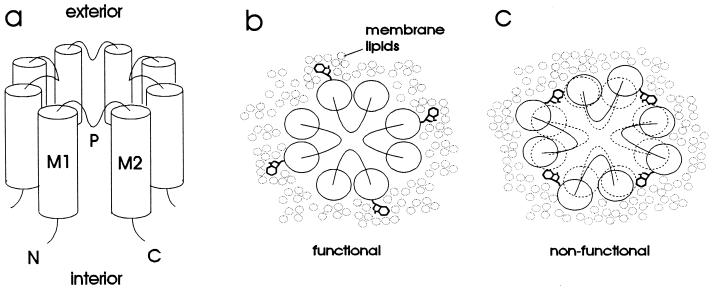
The inward rectifier K+ channels are tetramers (14), with each subunit predicted to have intracellular amino and carboxyl termini, two transmembrane domains designated M1 and M2 (15, 16), and a domain which is thought to line the extracellular part of the pore (H5 or P domain, refs. 17–20) (Fig. 1a). We carried out a scanning mutagenesis study of M1 and M2 in Kir2.1 (IRK1) (16) by testing the effect of altering the size of the side chain of each residue in these two hydrophobic segments (Fig. 1 b and c). Having found that an unexpectedly large number of these mutations yield functional channels, we replaced these residues one at a time with aspartate, and again found a surprisingly large number of such mutations compatible with channel function. These findings are presented here, along with discussions of their possible implications.
Figure 1.
(a) Schematic diagram of proposed inward rectifier structure. The channel is comprised of four subunits, each of which has two transmembrane α helices (M1 and M2) and a pore region (P) that loops part of the way into the transmembrane portion of the protein. Both the amino (N) and carboxyl (C) termini are inside the cell. (b and c) Rationale for tryptophan-scanning mutagenesis, illustrated on a schematic view of the channel from the extracellular side. (b) Substitution of tryptophan into a lipid-exposed position is accommodated by the membrane lipids and therefore does not disrupt the structure of the channel. (c) Substitution of tryptophan into an internal position disrupts the protein structure and therefore renders the channel nonfunctional.
MATERIALS AND METHODS
Mutagenesis and in Vitro Transcription.
Silent point mutations that provided convenient restriction sites for cassette mutagenesis were introduced into Kir2.1 (16) by single strand site-directed mutagenesis (Amersham). This modified Kir2.1 was subcloned into a version of pgemhe (21) in which the BpmI and SapI restriction sites had been removed. Mutants were made by restriction digest of this construct at two unique restriction sites to remove a small segment, treating the digested DNA with alkaline phosphatase and ligating mutagenic cassettes into the gap to reform a circular plasmid. Cassettes were made by annealing complimentary oligonucleotides that had 5′ phosphates and incorporated the desired mutation. Ligation products were transformed into Escherichia coli and isolated by standard methods (22). All mutations were confirmed by sequencing through the cassette insert. Plasmids were then linearized with NheI and capped cRNA was transcribed in vitro with T7 RNA polymerase (mMessage mMachine; Ambion, Austin, TX). The yield and quality of transcripts was assessed by agarose gel electrophoresis and ethidium bromide staining. The concentration of each sample was adjusted by trial and error until the signal intensity on an agarose gel was equal to that of a known amount of an RNA standard.
Oocyte Preparation.
Stages V–VI Xenopus oocytes were prepared by treatment with 2 mg/ml collagenase (Worthington, type CLS3) for 2 hr at room temperature with agitation in 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 5 mM Hepes (pH 7.4). Separated oocytes were then rinsed and stored in 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5 mM Hepes (pH 7.4) (ND-96) at 18°C with 100 μg/ml streptomycin and 60 μg/ml ampicillin. Up to 24 hr after collagenase treatment oocytes were injected (Nanoject; Drummond, Broomall, PA) with 1 ng cRNA (unless otherwise indicated) in 46 nl of clinical injection water. In some cases the oocytes were stored for up to 1 week at 4°C before injection. Two electrode voltage clamp recording was carried out at 21–23°C, 16–48 hr after injection.
Electrophysiology.
Two electrode voltage clamp recordings (CA-1; Dagan Instruments, Minneapolis) were low-pass filtered at 2 kHz. Electrodes were filled with 3 M KCl and had resistances of ≈1 MΩ. During recording the bath solution contained 3 mM MgCl2, 5 mM Hepes (pH 7.4), and 90 mM KCl. Patch clamp recordings (Axopatch 200A; Axon Instruments, Foster City, CA) were low-pass filtered at 1 kHz. The solution in the patch pipettes and bath contained 1 mM CaCl2, 1 mM MgCl2, 10 mM Hepes (pH 7.4), and 150 mM KCl. Data were digitized and stored on disk using a TL-1 DMA interface and pclamp software (Axon Instruments).
RESULTS AND DISCUSSION
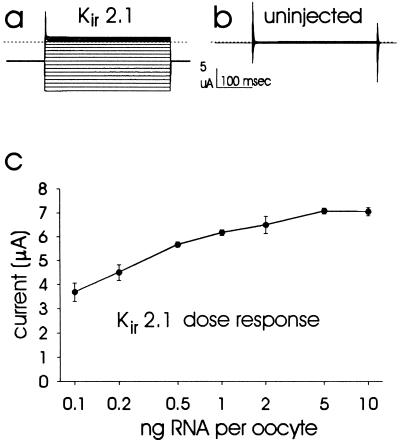
The effect of tryptophan or aspartate mutations of Kir2.1 was examined by two electrode voltage clamp recording from oocytes, each injected with 1 ng of cRNA for the mutant or wild-type Kir2.1 channel. Fig. 2a shows inwardly rectifying current recorded from an oocyte injected with Kir2.1 cRNA. For comparison, Fig. 2b shows a recording under identical conditions from an uninjected oocyte. Mean ± SD currents at −80 mV were −6.64 ± 0.99 μA for Kir2.1-injected and −0.15 ± 0.12 μA for uninjected oocytes. For comparison between wild-type and mutant channels it is important to ensure that the amount of cRNA injected in each case is comparable and that the measured differences in current amplitude could not be accounted for by variations in the cRNA levels. Quantitation of these cRNAs was achieved and verified by loading the same amount of different mutant and wild-type cRNA along with RNA standard on the same agarose/ethidium bromide gel (see Materials and Methods). The amount of cRNA used for each oocyte injection, 1 ng, is near saturation so that a two-fold error in the amount of cRNA injected would account for a <10% change in expressed current (Fig. 2c). We are therefore confident that differences in expression level between mutants and Kir2.1 >10% are not due to differences in the concentrations of cRNA injected, since a two-fold difference was clearly discernible on agarose/ethidium bromide gels.
Figure 2.
(a and b) Typical two-electrode voltage clamp records (a) from an oocyte injected with Kir2.1 and (b) from an uninjected oocyte. The holding potential was −60 mV with 350 msec voltage steps to between 100 mV and −140 mV in −10 mV increments. Currents recorded at different step potentials are superimposed. The dotted lines indicate zero current level. (c) Inward current magnitudes recorded at −80 mV from oocytes injected with a range of concentrations of Kir2.1 cRNA. Data points represent mean ± SEM from five oocytes each, after subtraction of the mean current (inward 0.40 μA) recorded from five uninjected oocytes (SEM = 0.05 μA). Inward currents are presented as positive values for ease of presentation of the dose response.
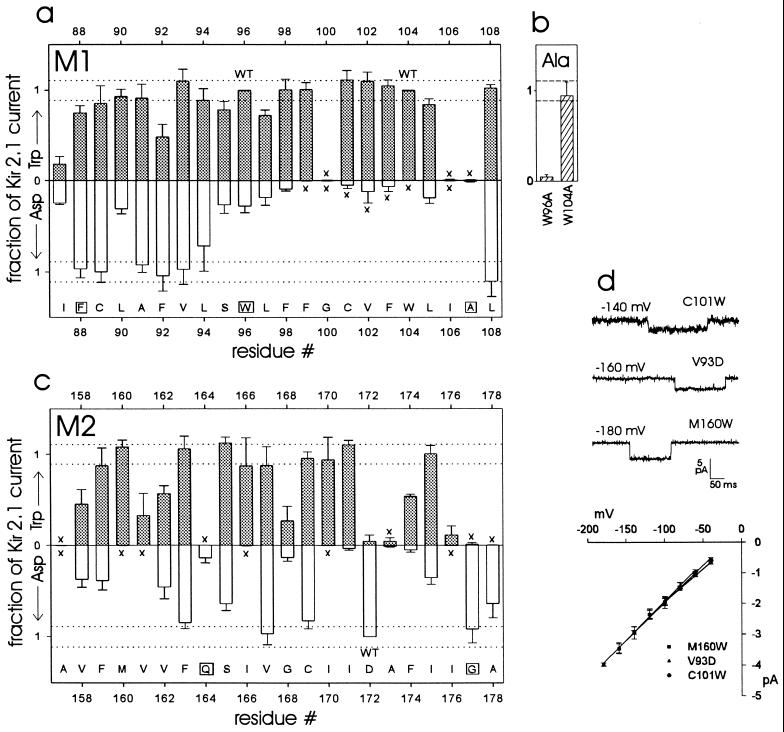
The normalized expressed currents of mutant channels is presented in Fig. 3 a–c (see figure legend for details of the normalization procedure). Altogether, 34 of the 42 nontryptophan residues tolerated tryptophan substitution to some degree, with 21 producing as much current as Kir2.1. Of the two wild-type tryptophan positions in M1, position 104 was more tolerant of alanine than was position 96 (Fig. 3b). Aspartate substitution was tolerated to varying degrees at 30 of the 41 non-aspartate positions, with 11 of these mutants expressing currents equivalent to Kir2.1. Ten of the aspartate-intolerant positions tolerated tryptophan to a greater or lesser extent. Aspartate was tolerated at five of the tryptophan-intolerant positions (although two of these aspartate mutants expressed very small currents). The amino terminal half of M1 was tolerant of aspartate and tryptophan (Fig. 3a), while the rest of M1 did not tolerate aspartate, with the exception of position 108, and to a lesser degree positions 105 and 107. In M2 the aspartate-tolerant positions were more scattered (Fig. 3c).
Figure 3.
(a–c) Expression of mutants by two electrode voltage clamp, compared with Kir2.1. (a) M1. (b) M1 tryptophan to alanine. (c) M2. (a, c) ░⃞, Tryptophan mutants (except for residues 96 and 104, which are tryptophan in Kir2.1); □, Aspartate mutants (except residue 172, which is aspartate in Kir2.1). “WT” indicates wild-type residues. Bars = SD. The aspartate data are inverted for ease of presentation. The abscissa indicates the position of each residue in the amino acid sequence of Kir2.1. The letters along the abscissa are the single-letter code for the wild-type Kir2.1 residue at each position; boxes indicate residues that are invariant in inward rectifiers. The dotted lines are ± SD for all of the normalized Kir2.1 data (total of 77 oocytes from nine frogs); these data fell within a Gaussian distribution by the Kolmogorov–Smirnov normality test (P = 0.20). For each batch of oocytes, the mean current in uninjected oocytes was subtracted from the mean Kir2.1 current and from the mutant currents (all recorded at −80 mV; typically five oocytes each). The leak-subtracted mutant currents were then divided by the leak-subtracted mean Kir2.1 current. In cases of ambiguity we performed a one-way ANOVA, comparing mutant currents with Kir2.1 currents in the same batch of oocytes, after “leak subtraction” as above. C89W, L94W, L94D, F159W, I166W, V167W, and C169D were not significantly different from Kir2.1. S95W, L105W, and F163D were significantly different from Kir2.1 (P < 0.05). In cases of very small currents we compared mutant-injected (unsubtracted) with uninjected oocytes from the same batch; “x” indicates observations where there was no significant difference between mutant-injected and uninjected oocytes (P > 0.05). (d) Patch clamp recording of three mutants in the cell-attached configuration. Channel openings are in the downward direction. Each oocyte was injected with 0.1 ng cRNA. (Lower) Single channel current-voltage relationships, with regression lines fitted to the data points (mean ± SEM) for each mutant. The slope conductances are: V93D 25.2 pS; C101W 21.7 pS; and M160W 24.3 pS.
All of the mutant channels retained their selectivity for K+ over Na+, as determined by recording from oocytes bathed in external solutions containing 90 mM Na+ (data not shown). In addition, all mutant channels exhibited inward rectification similar to wild-type channels, and the three mutants on which we carried out patch clamp recording had single channel properties similar to wild-type Kir2.1 (Fig. 3d) (16). Thus it appears that mutations that did not affect the level of expressed current did not significantly disrupt channel structure. For those mutations that reduced current amplitude, it is possible either that the mutation reduced the function of individual channels or that the mutation compromised the level of channel protein expression, or both. In any case, some of the mutant channels are still expressed and at least partially functional.
The folded conformation of a soluble protein is stabilized in large part by the exclusion of buried hydrophobic residues from the aqueous phase. This effect is not operative for proteins within the hydrophobic interior of the membrane bilayer, so any contacts between different transmembrane segments of the protein or between subunits would likely be very specific to be conserved (23). In the case of the photosynthetic reaction center, the interior of the membrane spanning domain is indeed packed, as in the case of soluble proteins (4). It thus seemed reasonable to assume that mutation of any residue to tryptophan (or mutation of tryptophan to alanine) would severely compromise the structure of the channel if that residue interacts intimately with another part of the protein, because of the large change in side-chain volume (6–8). Mutation to tryptophan of a residue that lines a relatively constricted part of the pore may also disrupt channel function.
It also seemed reasonable to assume that tolerance of a nonconservative mutation to aspartate at any position indicates that it is accessible to the aqueous phase, because of the unfavorable energy cost of burying a charge within the hydrophobic interior of the membrane or within the protein itself. In principle it is possible that the mutant aspartate residue could be protonated due to an increase in its pKa resulting from unusual interactions with surrounding residues (24–26). It seems unlikely, however, that such a special set of circumstances exists for each of the positions that tolerated aspartate. It is also possible in theory that a charged residue could be buried within the membrane due to the overall hydrophobicity of the stretch of primary sequence in which it finds itself (27, 28). If this nonspecific mechanism were operative, then we would expect all of the tryptophan-tolerant positions to also tolerate aspartate, which is not the case.
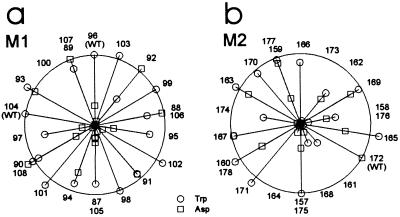
In both M1 and M2 the pattern of tryptophan tolerance does not follow an α helical periodicity (Figs. 4 and 5), and is therefore inconsistent with a simple model of an inward rectifier in which transmembrane α helices are arranged parallel to each other, with membrane lipids surrounding the helical bundle (Fig. 1 a and b). The dearth of α helical periodicity in tryptophan tolerance in M2 of Kir2.1 is in notable contrast to that in M2 of Kir1.1 (ROMK1). Kir2.1’s Met-160, Ser-165, Cys-169, and Ile-171 all tolerate tryptophan, while the equivalent residues in Kir1.1 are all intolerant to tryptophan (8). At two of these positions the amino acid residue is the same in the two clones (Ser-165 in Kir2.1 = Ser-164 in Kir1.1; Ile-171 in Kir2.1 = Ile-170 in Kir1.1). Thus, based on tryptophan tolerance, while it is possible to make a case for M2 being an α helix in Kir1.1, it is not possible to do so for M2 of Kir2.1. This difference is unexpected, since different inward rectifiers within the same gene family are assumed to have similar architecture. One possible explanation may be that Kir1.1 channel subunits are in contact with proteins endogenous to the Xenopus oocyte, whereas Kir2.1 residues corresponding to those Kir1.1 residues involved in such protein–protein interactions are in contact with lipids or water.
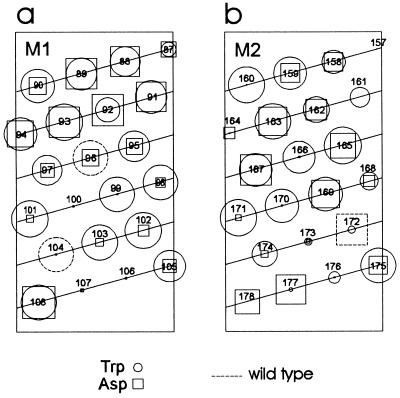
Figure 4.
Data from Fig. 3 presented on a helical wheel plot. (a) M1 and (b) M2. The length of each spoke represents the mutant current magnitude relative to Kir2.1 (represented by the circumference). ○, Tryptophan mutants; □, aspartate mutants. The numbers represent residue positions. “WT” indicates wild-type residues.
Figure 5.
Data from Fig. 3 presented on a helical net plot. (a) M1. (b) M2. ○, Tryptophan mutants; □, aspartate mutants. Dotted outlines represent wild-type residues. The area of each square or circle is proportional to the amount of current expressed. The numbers represent residue positions.
The tolerance of aspartate residues in positions predicted to be within the membrane is a rather surprising result, and raises further questions concerning the conventional model of an ion channel. Given that a position tolerant of both tryptophan and aspartate is in contact with water but not in a narrow part of the pore, these results suggest that most, if not all, of the residues from about position 88 to about position 94 are not in contact with the hydrophobic part of the protein or the membrane. From position 95–107 the aspartate tolerance was much reduced, although the tryptophan tolerance generally remained high. Position 108 tolerated aspartate and tryptophan, indicating that it is also in the aqueous phase. This overall pattern suggests that unless there are “crevices” harboring water (see below), the membrane is spanned by a part of M1 starting at about residue 95 and ending at residue 107, with residues 99, 101–103, and possibly 104 interacting with membrane lipid acyl chains. This stretch is not long enough to be able to span the membrane as an α helix, but would have sufficient length as a β strand (29).
Positions 160, 166, and 170 in M2 tolerate tryptophan but not aspartate and are therefore predicted to interact with membrane lipids. Certain positions in M2 tolerate both tryptophan and aspartate (positions 163, 165, 167, and 169, and to a lesser extent 158, 159, and 162). In contrast to M1 these positions are not consecutive in the sequence, and therefore cannot be thought of as being wholly outside of the membrane. Nevertheless, their tolerance indicates that these residues are accessible to the aqueous phase, but do not line a narrow part of the pore. We speculate that these residues are in contact with water that might be present within the membrane but not in the channel pore. Consistent with this possibility, the S4 segment of the Shaker voltage-gated K+ channel, a transmembrane segment believed to function as a voltage sensor but not as a pore-lining structure, also is exposed to water for more than half of its residues, perhaps via a “crevice” in the protein (30, 31). A similar situation could be imagined in Kir2.1 in between M2 and other intramembrane protein elements.
Asp-172 in Kir2.1 is thought to line the pore (32–34), and tryptophan is tolerated poorly at this position (Fig. 3c). Position 177 and to lesser extents positions 178, 164, 173, and 107 tolerate aspartate but not tryptophan, analogous to position 172, and thus may occupy an environment similar to that of 172. The smaller currents expressed by A107D, Q164D, A173D, and A178D may reflect interactions between the extra negative charge and the permeating ion.
A priori one would expect that intolerant residues are those that are highly conserved among related genes, while variable residues would be expected to be tolerant to mutation. This is true for some of our data, but this expectation is not consistently borne out. A notable exception is Phe-88, which is invariant but could be changed to aspartate with no effect on expression level and to tryptophan with limited effect (Fig. 3a). Alanine and methionine were also tolerated at this position (data not shown). It is possible that this residue is important for a function unrelated to the structure of the pore-forming unit, such as interaction with an auxiliary protein that is not required for current expression in Xenopus oocytes. The tolerance of aspartate at the invariant position Gly-177 (Fig. 3c) is also surprising.
In conclusion, the pattern of tolerated and nontolerated mutations that emerged from this scanning mutagenesis study is surprising in view of proposed models of inward rectifier channel structure and in view of the assumptions upon which we have based our interpretations of the data. Specifically, most of the amino terminal half (amino acids 88–95) of M1 appears to be totally exposed to an aqueous environment whereas most of the carboxyl terminal half (amino acids 98–105) appears to be in contact with lipids. Moreover, half of all M2 positions tolerate aspartate residues, even though only five of these positions (164, 172, 173, 177, and 178) tolerate tryptophan poorly or not at all and therefore could line a narrow part of the pore. Our observations that half of all the residues in M1 and M2, the only presumed membrane-spanning segments in Kir2.1 channel subunits, tolerate aspartate and often tryptophan as well raises the question as to whether some of these residues are in contact with water that might be present within the membrane but not in the channel pore.
Acknowledgments
We thank Dr. E. Liman for pgemhe; Bruce Cohen, Ed Cooper, Virginia Cornish, Chou-Long Huang, Joyce Liao, Zack Ma, Dan Minor, Brigitte Rauman, Eitan Reuveny, Paul Slesinger, Andrew Tinker, Jian Yang, and Biao Zhao for helpful discussions; Sharon Fried, Erica Wolff and Mei Yu for excellent technical assistance. A.C. thanks Eiten Reuveny for a modified version of Kir2.1. This work was supported by National Institute of Mental Health Grant MH48200 and by the Howard Hughes Medical Institute, and was done partly during the tenure of an American Heart Association Research Fellowship Award to A.C.; Y.N.J. and L.Y.J. are Howard Hughes Medical Institute investigators.
References
- 1.Hille B. Ionic Channels of Excitable Membranes. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 2.Yang J, Yu M, Jan Y N, Jan L Y. Proc Natl Acad Sci USA. 1997;94:1568–1572. doi: 10.1073/pnas.94.4.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rees D C, DeAntonio L, Eisenberg D. Science. 1989;245:510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 4.Yeates T O, Komiya H, Rees D C, Allen J P, Feher G. Proc Natl Acad Sci USA. 1987;84:6438–6442. doi: 10.1073/pnas.84.18.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinkle P C, Hinkle P V, Kaback H R. Biochemistry. 1990;29:10989–10994. doi: 10.1021/bi00501a017. [DOI] [PubMed] [Google Scholar]
- 6.Sharp L L, Zhou J, Blair D F. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharp L L, Zhou J, Blair D F. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 8.Choe S, Stevens C F, Sullivan J M. Proc Natl Acad Sci USA. 1995;92:12046–12049. doi: 10.1073/pnas.92.26.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fauchere J-L, Pliska V. Eur J Med Chem. 1983;18:369–375. [Google Scholar]
- 10.Henderson R, Unwin P N. Nature (London) 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 11.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Nature (London) 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 12.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 13.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Nature (London) 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Jan Y N, Jan L Y. Neuron. 1995;15:1441–1447. doi: 10.1016/0896-6273(95)90021-7. [DOI] [PubMed] [Google Scholar]
- 15.Ho K, Nichols C G, Lederer W J, Lytton J, Vassilev P M, Kanazirska M V, Hebert S C. Nature (London) 1993;362:31–38. doi: 10.1038/362031a0. [DOI] [PubMed] [Google Scholar]
- 16.Kubo Y, Baldwin T J, Jan Y N, Jan L Y. Nature (London) 1993;362:127–133. doi: 10.1038/362127a0. [DOI] [PubMed] [Google Scholar]
- 17.MacKinnon R, Yellen G. Science. 1990;250:276–279. doi: 10.1126/science.2218530. [DOI] [PubMed] [Google Scholar]
- 18.Yellen G, Jurman M E, Abramson T, MacKinnon R. Science. 1991;251:939–942. doi: 10.1126/science.2000494. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q, Miller C. Science. 1995;268:304–307. doi: 10.1126/science.7716526. [DOI] [PubMed] [Google Scholar]
- 20.Pascual J M, Shieh C C, Kirsch G E, Brown A M. Neuron. 1995;14:1055–1063. doi: 10.1016/0896-6273(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 21.Liman E R, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Lemmon M A, Flanagan J M, Treutlein H R, Zhang J, Engelman D M. Biochemistry. 1992;31:12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- 24.Fersht A R. J Mol Biol. 1972;64:497–509. doi: 10.1016/0022-2836(72)90513-x. [DOI] [PubMed] [Google Scholar]
- 25.Urry D W, Gowda D C, Peng S, Parker T M, Jing N, Harris R D. Biopolymers. 1994;34:889–896. doi: 10.1002/bip.360340708. [DOI] [PubMed] [Google Scholar]
- 26.Smith S O, Smith C S, Bormann B J. Nat Struct Biol. 1996;3:252–258. doi: 10.1038/nsb0396-252. [DOI] [PubMed] [Google Scholar]
- 27.Wen J, Chen X, Bowie J U. Nat Struct Biol. 1996;3:141–148. doi: 10.1038/nsb0296-141. [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 29.Creighton T E. Proteins: Structures and Molecular Principles. New York: Freeman; 1993. [Google Scholar]
- 30.Larsson H P, Baker O S, Dhillon S, Isacoff E Y. Neuron. 1996;16:387–397. doi: 10.1016/s0896-6273(00)80056-2. [DOI] [PubMed] [Google Scholar]
- 31.Mannuzzu L M, Moronne M M, Isacoff E Y. Science. 1996;271:213–216. doi: 10.1126/science.271.5246.213. [DOI] [PubMed] [Google Scholar]
- 32.Stanfield P R, Davies N W, Shelton P A, Sutcliffe M J, Khan I A, Brammar W J, Conley E C. J Physiol (London) 1994;478:1–6. doi: 10.1113/jphysiol.1994.sp020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wible B A, Taglialatela M, Ficker E, Brown A M. Nature (London) 1994;371:246–249. doi: 10.1038/371246a0. [DOI] [PubMed] [Google Scholar]
- 34.Reuveny E, Jan Y N, Jan L Y. Biophys J. 1996;70:754–761. doi: 10.1016/S0006-3495(96)79615-7. [DOI] [PMC free article] [PubMed] [Google Scholar]