Abstract
S-adenosyl-l-methionine (SAM)-dependent O-methyltransferases (OMTs) catalyze the methylation of hydroxycinnamic acid derivatives for the synthesis of methylated plant polyphenolics, including lignin. The distinction in the extent of methylation of lignins in angiosperms and gymnosperms, mediated by substrate-specific OMTs, represents one of the fundamental differences in lignin biosynthesis between these two classes of plants. In angiosperms, two types of structurally and functionally distinct lignin pathway OMTs, caffeic acid 3-O-methyltransferases (CAOMTs) and caffeoyl CoA 3-O-methyltransferases (CCoAOMTs), have been reported and extensively studied. However, little is known about lignin pathway OMTs in gymnosperms. We report here the first cloning of a loblolly pine (Pinus taeda) xylem cDNA encoding a multifunctional enzyme, SAM:hydroxycinnamic Acids/hydroxycinnamoyl CoA Esters OMT (AEOMT). The deduced protein sequence of AEOMT is partially similar to, but clearly distinguishable from, that of CAOMTs and does not exhibit any significant similarity with CCoAOMT protein sequences. However, functionally, yeast-expressed AEOMT enzyme catalyzed the methylation of CAOMT substrates, caffeic and 5-hydroxyferulic acids, as well as CCoAOMT substrates, caffeoyl CoA and 5-hydroxyferuloyl CoA esters, with similar specific activities and was completely inactive with substrates associated with flavonoid synthesis. The lignin-related substrates were also efficiently methylated in crude extracts of loblolly pine secondary xylem. Our results support the notion that, in the context of amino acid sequence and biochemical function, AEOMT represents a novel SAM-dependent OMT, with both CAOMT and CCoAOMT activities and thus the potential to mediate a dual methylation pathway in lignin biosynthesis in loblolly pine xylem.
Lignin is a major cell wall polymer of vascular plants that provides mechanical strength to the stem and protects cellulose fibers from chemical and biological degradation. However, during wood pulping, lignin must be degraded to extract cellulose fibers for paper making. Both gymnosperm and angiosperm tree species are used for wood pulp production, with gymnosperms being the preferred species because of their superior fiber properties. However, due to a lesser degree of methylation of the constituent monomers, gymnosperm lignin is more condensed and therefore more difficult to degrade than angiosperm lignin (1–4). Thus, methylation of hydroxylated monomeric lignin precursors represents a critical step in lignin biosynthesis that influences the final lignin structure and profoundly affects the economics of wood pulp production. For this reason, methylating enzymes, O-methyltransferases (OMTs), involved in the lignin biosynthetic pathway, particularly those in angiosperms, have been extensively studied (for reviews, see refs. 5–7) and, recently, have been regarded with considerable interest as targets for biotechnological manipulation of lignin biosynthesis (7–11).
The lignin pathway OMTs in angiosperms are bifunctional, using caffeic acid and 5-hydroxyferulic acid (5), and are generally referred to as CAOMT (caffeic acid 3-O-methyltransferases). However, OMTs in gymnosperms, which have only been studied in seedlings, are believed to be monofunctional, catalyzing caffeic acid but deficient in catalytic activity with 5-hydroxyferulic acid (5). Although it has not been unambiguously proven at the molecular level because no gymnosperm OMT has been cloned, at the enzyme level, the concept of bifunctional CAOMT in angiosperms versus monofunctional OMT in gymnosperms has been widely accepted for the past 20 years (5). Overall, these OMTs use free hydroxycinnamic acids (5–7, 12, 13) as opposed to another class of angiosperm OMT, caffeoyl CoA 3-O-methyltransferase (CCoAOMT) (14–16) involved in lignin biosynthesis in Zinnia (17, 18), which uses caffeoyl CoA and 5-hydroxyferuloyl CoA esters (17). At both nucleotide and predicted amino acid sequence levels, Zinnia CCoAOMT cDNA does not share any significant similarity with the cDNA encoding a Zinnia bifunctional CAOMT (18). In fact, all known plant CCoAOMTs (19–21) show significant sequence dissimilarity with CAOMTs. Furthermore, in vitro expression of cloned cDNA unequivocally demonstrated that, while Zinnia CAOMT mediates the methylation of caffeic acid and 5-hydroxyferulic acid, it exhibits no activity with caffeoyl CoA (18). Consequently, it was proposed that two classes of structurally and functionally distinct OMTs are involved in the methylation of monomeric lignin precursors at hydroxycinnamic acid and hydroxycinnamoyl CoA levels, respectively (17, 18). Because both CAOMT and CCoAOMT have already been identified for various other angiosperm dicots, the existence of two independent methylation pathways in Zinnia further raises the question of whether these enzymes are also involved in gymnosperms. Thus, the potential variation in the CCoAOMT-mediated methylation pathway, along with the ambiguous distinctness in CAOMT-based methylation flow for lignin precursor synthesis between gymnosperms and angiosperms, needs to be further clarified for a better understanding of the fundamental difference in lignin biosynthesis between these two classes of plants. Moreover, the verification of either or both methylation pathways for lignin precursor synthesis in gymnosperms is particularly important to the establishment of effective approaches to future genetic engineering of methoxyl-enriched lignin in gymnosperms.
We report here the isolation of a loblolly pine (Pinus taeda) differentiating xylem cDNA encoding a multifunctional OMT, hydroxycinnamic acids/hydroxycinnamoyl CoA esters OMT (AEOMT), and characterization of its biochemical function in yeast. In contrast to the monofunctional OMT in loblolly pine seedling and to the two methylation systems mediated independently by CAOMT and CCoAOMT in Zinnia, loblolly pine xylem AEOMT exhibits both CAOMT and CCoAOMT activities, catalyzing the methylation of caffeic acid, 5-hydroxyferulic acid, caffeoyl CoA, and 5-hydroxyferuloyl CoA with similar specific activities. A dual methylation pathway is therefore proposed for lignin biosynthesis in loblolly pine xylem, and its implication in biotechnological manipulation of lignin biosynthesis in gymnosperm tree species is discussed.
MATERIALS AND METHODS
Plant Material.
Differentiating secondary xylem was collected from vegetatively propagated 3-year-old loblolly pine (genotype no. 1932 grown in the Forest Experimental Station of International Paper Co., Bainbridge, GA) and stored immediately in liquid nitrogen until use for isolating protein, RNA, and DNA.
cDNA Cloning of AEOMT.
Poly(A)+ RNA isolated from xylem of loblolly pine was used to construct a cDNA library in λZAP II as described (22). A pool of λZAP II phage cDNAs was isolated from the amplified cDNA library (23) and used as the template cDNA for PCR. Two degenerate primers (5′-GGMATYCCATTYAACAAGGC-3′, 8-fold degeneracy, and 5′-TCYTKYTSDGTYCTCTCTTT-3′, 96-fold degeneracy; see the sequences with arrows below them in Fig. 1) based on a highly conserved C-terminal domain in plant CAOMT cDNAs were synthesized and used for PCR cloning of target DNA fragments according to the conditions described previously (22). A PCR product of about 500 bp was cloned into pCRII (Invitrogen) and sequenced. Initial screening of 20,000 plaque-forming units from the cDNA library using this 500-bp PCR product as a probe resulted in the identification of 20 positive clones, four of which were purified and sequenced in both directions. Two of these four cDNAs were found to be identical full-length cDNAs and were designated as AEOMT. The sequence analysis was performed using the gcg software package.
Figure 1.
The nucleotide and the deduced amino acid sequence of loblolly pine AEOMT cDNA. The translational initiation codon (with a bold line above it) indicates the plant-specific favorable context (27, 28). The putative polyadenylylation signals in the 3′ noncoding region (29) are double underlined. The N-terminal sequence confirmed by amino acid sequencing is italicized and underlined, and the degenerate PCR primers are indicated by arrows. The numbers indicate the number of nucleotides and amino acids (boldface tape).
Expression of Recombinant AEOMT in Escherichia coli.
PCR was used to introduce a NdeI site at the 5′ end and a HindIII site at the 3′ end of the AEOMT clone in pBluescript, and the PCR product was cloned into pET-23b+ vector containing the histidine tag (His⋅Tag, Novagen). The accuracy of the clone at the 5′ and 3′ ends was confirmed by DNA sequencing. The central core of recombinant cDNA in pET vector was replaced by the corresponding portion of the authentic AEOMT cDNA cloned from the cDNA library. The resulting plasmid was transferred into E. coli BL21DE3. The growth and induction of bacterial cells with isopropyl β-d-thiogalactoside were performed according to the manufacturer’s protocols (Novagen). After harvesting by centrifugation, the cell pellet containing AEOMT inclusion bodies was processed to affinity-purify the AEOMT protein using His⋅Bind Resin (Novagen). N-terminal sequencing of this purified recombinant AEOMT protein was carried out by Edman degradation using an automated protein sequencer (Beckman; model 2020). Polyclonal antibodies against this purified recombinant AEOMT protein were raised in rabbits by Alpha Diagnostic International (San Antonio, TX). Protein concentration was determined by the Bradford assay (24) using BSA as a standard. SDS/PAGE and Western blot analysis were conducted as described (13).
Expression of Recombinant AEOMT in Pichia pastoris.
AEOMT cDNA in pBluescript was digested with SmaI and ApaI and ligated into PmlI and ApaI sites of pPICZ A (Invitrogen) to ensure the expression of AEOMT using AEOMT-specific start and stop codons. pPICZ A vector containing AEOMT cDNA was transferred into E. coli JM109, and transformed cells were selected on 25 μg/ml Zeocin as an antibiotic selection marker (Invitrogen). Forty micrograms of plasmid DNA was linearized by digestion with SacI, and 25 μg of purified plasmid DNA was transferred into P. pastoris GS115 by homologous recombination using LiCl transformation protocol (Invitrogen). One hundred transformants were selected to screen for Mut+ phenotype. The growth and induction of the positively transformed yeast cells were carried out according to the manufacturer’s protocols (Invitrogen). After harvesting by centrifugation, the cells were resuspended in buffer containing 0.2 M Tris⋅HCl (pH 7.5), 0.5 μg/ml leupeptin, 5 mM MgCl2, 5 mM mercaptoethanol, and 5% glycerol, and were disintegrated with glass beads by vortexing for 10 times (30 sec on/30 sec off). Cell debris was pelleted by centrifugation, and the supernatant was used directly for protein concentration determination, SDS/PAGE, Western blotting, and enzyme activity. pPICZ A vector without AEOMT cDNA insert was used as a negative control.
Chemicals.
S-Adenosyl-l-methionine (SAM)-14Me, (0.56 mCi/mmol; 1 Ci = 37 GBq) was obtained from Amersham. Apigenin, chlorogenic acid, naringenin, and quercetin were obtained from Aldrich. Taxifolin was obtained from Sigma. 5-Hydroxyferulic acid (mp 169°C) was chemically prepared according to Neish (25). Caffeoyl CoA, feruloyl CoA, and 5-hydroxyferuloyl CoA esters were synthesized via acyl N-hydroxysuccinimide esters as described (26). The authenticity of the intermediate esters was confirmed by 1H NMR and MS, and that of the final CoA esters was confirmed by UV and TLC [on cellulose, Rf values were as follows: caffeoyl CoA, 0.39; feruloyl CoA, 0.53; 5-hydroxyferuloyl CoA, 0.28, in n-butanol/glacial acetic acid/water (5:2:3)].
Biochemical and Molecular Analysis.
The crude protein extracts from secondary developing xylem of loblolly pine (22) and yeast cells described above were used for enzyme assays with caffeic acid, 5-hydroxyferulic acid, or flavonoids as substrates as described (13), with the following modifications. Briefly, 50 μl of reaction mixture contained 50 mM Tris⋅HCl (pH 7.5), 1 mM MgCl2, 4 nmol of phenolic substrate, 2 nmol of SAM-14Me, and 7–15 μg of protein. Enzyme assays using caffeoyl CoA or 5-hydroxyferuloyl CoA were performed as described (16) with some modifications. Briefly, 50 μl of reaction mixture contained 50 mM Tris⋅HCl (pH 7.5), 200 μM MgCl2, 2 mM DTT, 10% glycerol, 2 nmol of hydroxycinnamoyl CoA, 2 nmol of SAM-14Me, and 7–15 μg of protein. Control assays used boiled crude protein extracts. Genomic DNA and total RNA isolation from loblolly pine xylem and Southern and Northern blot analyses using AEOMT cDNA as a probe were performed as described (22).
Identification of Methylated Products by TLC.
For TLC analysis of the products from hydroxycinnamic acids, the standard enzyme reaction described above was scaled up 4-fold (with 50–100 μg of total protein) and cold SAM was used. After extraction with ethyl ether, the solvent was evaporated, and the residue was dissolved in 5 μl of ethanol and spotted on a cellulose plate (Whatman) in 1 M boric acid/sodium borate, pH 7, along with the authentic hydroxycinnamic acids. For TLC analysis of the products from hydroxycinnamoyl CoA esters, the same enzyme reaction mixtures described above were used, except that no glycerol was added. After an initial extraction with 500 μl of ethyl acetate, 30 μl of 5 M NaOH was added to the aqueous phase containing the CoA thioesters to hydrolyze the esters at 40°C for 15 min, as described (16). The mixture was acidified with 30 μl of 6 M HCl, and the hydrolysis products, i.e., the hydroxycinnamic acids, were extracted with 500 μl of ethyl acetate, the solvent was evaporated, and the residue was dissolved in 5 μl of ethanol and separated by TLC as describe above. Rf values were as follows: caffeic acid, 0.85; ferulic acid, 0.45; 5-hydroxyferulic acid, 0.68; sinapic acid, 0.31.
RESULTS AND DISCUSSION
cDNA Cloning of AEOMT and Genome Organization of Loblolly Pine AEOMT.
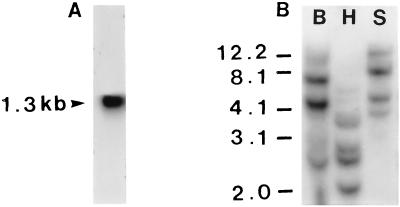
We have repeatedly observed in vitro OMT activities with caffeic acid, 5-hydroxyferulic acid, caffeoyl CoA, and 5-hydroxyferuloyl CoA in crude protein extracts of secondary developing xylem of loblolly pine (Table 1). To our knowledge, this is the first report revealing that a gymnosperm species exhibits in vitro CAOMT and CCoAOMT activities with not only gymnosperm-related monomeric lignin precursors but also angiosperm-associated monomeric lignin precursors. We first speculated that, like in Zinnia, loblolly pine could also contain two classes of lignin pathway OMTs catalyzing independently the methylation of caffeic acid and caffeoyl CoA. We therefore initiated the cloning of loblolly pine OMTs, with the initial focus on PCR cloning of CAOMT. By aligning deduced amino acid sequences of eight different plant CAOMTs, a highly conserved domain of about 170 aa residues near the C-terminal was identified and used to design two degenerate primers for PCR with a pool of loblolly pine xylem cDNAs as the template. An amplified cDNA fragment of about 500 bp was cloned and sequenced, revealing ≈60% similarity with various plant CAOMT cDNA sequences encoding this conserved C-terminal domain. Using this 500-bp cDNA fragment as a probe, a full-length cDNA (AEOMT, Fig. 1) was isolated from loblolly pine xylem cDNA library; this cDNA was 1279-bp long with a 1143-bp coding region, predicting a 381-aa protein with a calculated Mr of 42,012 (Fig. 1). To produce recombinant AEOMT that can be readily purified for protein sequencing, AEOMT cDNA was expressed in E. coli to produce AEOMT–His⋅Tag fusion protein in the form of inclusion bodies. The AEOMT fusion protein was purified to apparent homogeneity with His⋅Tag sequence-specific affinity column, and its purity was demonstrated by a single protein band of about 43 kDa, as estimated by SDS/PAGE. The molecular mass of the fusion protein was expected to be slightly higher than the calculated Mr of AEOMT due to the histidine-tagged amino acid residues. The N-terminal peptide of the purified AEOMT was sequenced and matched to the deduced amino acid sequence (italicized and underlined sequence in Fig. 1), confirming the authenticity of the cloned AEOMT. A Northern blot of loblolly pine xylem RNA was probed with AEOMT cDNA that hybridized to a message of about 1.3 kb, corresponding well with the length of the AEOMT clone (Fig. 2A). Southern blot analysis of loblolly pine genomic DNA at high stringency suggested that an AEOMT gene family may be present in the loblolly pine nuclear genome (Fig. 2B).
Table 1.
Substrate specificity of AEOMT in crude extracts of loblolly pine xylem and yeast, and antibody inhibition of AEOMT activity
| Substrate | Loblolly pine xylem extracts
|
Yeast recombinant AEOMT
|
Loblolloy pine xylem extracts + antibody
|
Yeast recombinant AEOMT + antibody
|
||||
|---|---|---|---|---|---|---|---|---|
| Spec. act. | Rel. act. | Spec. act. | Rel. act. | Spec. act. | % inhibition | Spec. act. | % inhibition | |
| Caffeic acid | 441 ± 37 | 100 | 310 ± 32 | 100 | 317 ± 40 | 28 | 214 ± 15 | 31 |
| 5-Hydroxyferulic acid | 346 ± 25 | 78 | 235 ± 26 | 76 | 286 ± 16 | 17 | 198 ± 24 | 16 |
| Caffeoyl CoA | 755 ± 33 | 171 | 268 ± 28 | 86 | 446 ± 32 | 41 | 105 ± 3 | 61 |
| 5-Hydroxyferuloyl CoA | 657 ± 31 | 149 | 210 ± 12 | 68 | 331 ± 24 | 50 | 95 ± 13 | 55 |
Spec. act., specific activity (in pmol/min/mg); Rel. Act., relative activity (in %). The specific activities were mean ± SD (n = 6 independent assays). Crude extract protein (7 μg) was incubated with antiserum or preimmune antiserum (14 μg of protein) for 10 min at room temperature before addition of enzyme assay components. Preimmune serum had no effect on AEOMT activity. All controls used boiled protein extracts.
Figure 2.
(A) Northern blot analysis of total RNA (20 μg) from loblolly pine xylem, probed with 32P-labeled AEOMT cDNA. (B) Southern blot analysis of loblolly pine genomic DNA (50 μg per lane) digested with BamHI (lane B), HindIII (lane H), and SacI (lane S), probed with 32P-labeled AEOMT cDNA. One kb DNA ladder (GIBCO) was used as size markers, and values are given in kilobases.
Using a similar strategy, we then explored the possibility of cloning loblolly pine CCoAOMT cDNA. However, attempts to amplify CCoAOMT cDNA fragments from loblolly pine cDNA library and xylem mRNA by PCR and reverse transcriptase–PCR, respectively, using plant CCoAOMT-specific degenerate primers were unsuccessful. Also, no signal could be detected in loblolly pine genomic DNA Southern and xylem RNA Northern blots which were probed with Stellaria CCoAOMT cDNA (20) at various levels of stringency. This is surprising because all known plant CCoAOMTs exhibit over 90% identity at both nucleotide and amino acid sequence levels, and a loblolly pine homologue should have been detected if it existed in loblolly pine xylem. These results imply that, if there is a CCoAOMT in loblolly pine, its cDNA sequence must be highly distinct from other plant CCoAOMT cDNAs. Another possibility is that our original speculation of CAOMT and CCoAOMT being two independent enzymes in loblolly pine xylem was incorrect, and the activities of these enzymes observed in loblolly pine xylem extracts could be originated from a common OMT, such as AEOMT.
Comparative Sequence Analysis Suggests That AEOMT Is a Novel SAM-Dependent OMT.
To test whether AEOMT protein sequence is related to angiosperm dicot CAOMTs and CCoAOMTs, multiple sequence alignment of all these plant proteins (eight CAOMTs and nine CCoAOMTs) available in the GenBank database was conducted. It revealed that the deduced AEOMT protein shows 39–41% overall sequence identity (59–62% similarity) with all CAOMT proteins and exhibits insignificant identity (15–17%) with CCoAOMT proteins. The most unique region in AEOMT protein is a stretch of 40 aa (amino acids 1–40 in Fig. 1) at the N terminus that is not conserved between AEOMT and CAOMTs. The following ≈160 aa (amino acids 44–211 in Fig. 1) show only weak similarity with CAOMTs. It is in a stretch of about 170 aa (amino acids 213–381 in Fig. 1) toward the C-terminal half that a reasonable amino acid conservation (42–50% identity/65–72% similarity) can be found between AEOMT and CAOMTs. These analyses indicated that, although AEOMT is not sequence-related to CCoAOMT, AEOMT is partially similar to but clearly distinguishable from plant CAOMTs. However, AEOMT might not be qualified as a member of CAOMT for the reason that all CAOMTs generally share 70–85% overall sequence identity.
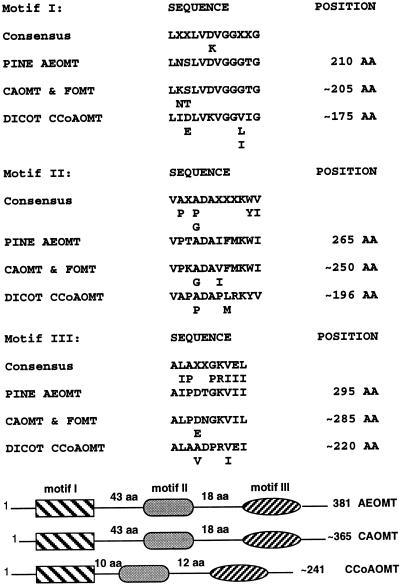
Similar to the case of CAOMTs, all dicot CCoAOMTs show more than 90% overall sequence identity and 95% similarity. In all CCoAOMTs, an amino acid sequence motif GXXXGYS (30) was found to be conserved and was proposed to be a SAM binding domain (17). However, this motif cannot be the only SAM binding domain because it is absent in all SAM-dependent plant CAOMTs. Moreover, two SAM-dependent flavonoid OMTs (FOMTs; GenBank accession nos. L10211L10211 and U16794U16794) also do not have the GXXXGYS motif. Instead, three different motifs were found to be conserved in all known plant CAOMTs, CCoAOMTs, and FOMTs, and in loblolly pine AEOMT, as shown in Fig. 3. Although motifs I and III have been previously suggested as possible SAM binding domains in plant CAOMTs (12), the presence of motifs I, II, and III in all these plant OMTs was identified for the first time. Since CAOMTs, CCoAOMTs, and FOMTs mediate the methylation of distinct substrates, their common conserved amino acid sequence motifs are likely to be the binding domains for their common substrate, SAM. It is interesting to note that the spatial arrangement of these three motifs in AEOMT is identical to that of the three conserved motifs in CAOMTs, and there seems to be an overall similar arrangement of these three motifs in all OMTs examined (Fig. 3), indicating a spatial relationship that might be important in the native form of these motifs for binding the common substrate SAM. Therefore, in addition to GXXXGYS in plant CCoAOMTs, these three consensus motifs could be significant SAM binding domains in all these known plant OMTs, providing evidence that AEOMT represents an SAM-dependent OMT. Because it is not sequence-related to CCoAOMT and has a sequence distinguishable from CAOMT, AEOMT can be considered to represent a novel class of SAM-dependent OMTs. Subsequent to the deposition of this AEOMT in the GenBank database, an OMT sequence from monterey pine was recently deposited (accession no. U70873U70873); this sequence shows 98% similarity with loblolly pine AEOMT and could thus be regarded as another member of this class.
Figure 3.
The three conserved motifs and their positions in the loblolly pine AEOMT, dicot CAOMT, FOMT, and CCoAOMT. The diagrammatic representation below the description of motifs indicates the spatial arrangement of three motifs in relation to the total number of amino acids present in each type of OMT.
AEOMT Functionally Acts as Both CAOMT and CCoAOMT.
Because AEOMT could be the common enzyme responsible for the observed CAOMT and CCoAOMT activities in extracts of loblolly pine xylem as we speculated, we decided to use a variety of OMT substrates to characterize the biochemical function of AEOMT protein in yeast P. pastoris. These substrates include those related to lignin biosynthesis, such as caffeic acid, caffeoyl CoA, 5-hydroxyferulic acid, and 5-hydroxyferuloyl CoA, and those associated with flavonoid biosynthetic pathways, such as apigenin, naringenin, quercetin, and taxifolin. Six independent assays for each of the lignin pathway-associated substrates were performed, and strong methylation activities of AEOMT with all these substrates were observed in crude extracts of P. pastoris expressing AEOMT cDNA (Table 1), whereas no such activity could be detected in extracts of P. pastoris containing vector only. It appears that both xylem and yeast-expressed AEOMTs have a slight preference for substrates that are related to guaiacyl lignin. The identity of both yeast and plant AEOMT-mediated reaction products from caffeic acid, caffeoyl CoA, 5-hydroxyferulic acid, and 5-hydroxyferuloyl CoA were confirmed to be their methylated counterparts, i.e., ferulic acid, feruloyl CoA, sinapic acid, and sinapoyl CoA, respectively, as analyzed by TLC (Fig. 4). However, the yeast-expressed AEOMT was completely inactive with all FOMT substrates tested, such as apigenin, naringenin, quercetin, and taxifolin, and with chlorogenic acid, a phenolic substrate for other nonlignin products (5). These results provide unequivocal evidence that AEOMT represents a common SAM-dependent OMT with both CAOMT and CCoAOMT activities for mediating the methylation pathways specifically in lignin biosynthesis in loblolly pine xylem. It is surprising that AEOMT does not share any meaningful protein sequence identity with any known plant CCoAOMT protein but uses efficiently the CCoAOMT substrates. On the other hand, despite the existence of some sequence similarity between AEOMT and FOMT proteins, AEOMT does not methylate FOMT substrates. These surprising observations have also been shown recently for some other enzymes. For example, an Arabidopsis thaliana defense-related protein ELI3, originally predicted to be a mannitol dehydrogenase (MAD) (31) based solely on a convincingly high overall amino acid sequence identity of 68% to a known MAD protein from celery (Apium graveolens), was confirmed to be a benzyl alcohol dehydrogenase (BAD) that exhibited completely different substrate specificity from MAD enzyme (32). Furthermore, a lignin pathway cinnamyl alcohol dehydrogenase (CAD) protein from Eucalyptus gunnii (33) shares over 50% overall amino acid sequence identity with this A. thaliana BAD but was found inactive with the BAD substrates (32). Thus, the results of AEOMT functional identification shown above further demonstrate that inference of enzyme function based mainly on the deduced amino acid sequence similarity to known proteins can sometimes be misleading.
Figure 4.
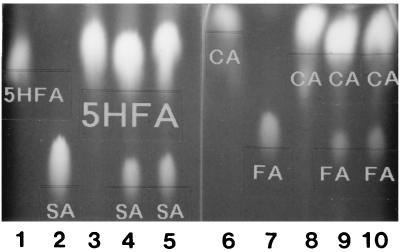
TLC analysis of loblolly pine xylem and yeast-expressed AEOMT enzyme reaction products. Lanes: 1, authentic 5-hydroxyferulic acid (5HFA) only; 2, authentic sinapic acid (SA) only; 3, control (boiled plant extracts + 5HFA); 4, plant crude extracts (50 μg of total protein) + 5-hydroxyferulic acid; 5, yeast crude extracts (100 μg of total protein) + 5-hydroxyferulic acid; 6, authentic caffeic acid (CA) only; 7, authentic ferulic acid (FA) only; 8, control (boiled plant extracts + caffeic acid); 9, plant crude extracts (50 μg of total protein) + caffeic acid; 10, yeast crude extract (100 μg of total protein) + caffeic acid. Data are shown for products sinapic acid (lanes 4 and 5) and ferulic acid (lanes 9 and 10) from 5-hydroxyferulic acid and caffeic acid, respectively. 5-Hydroxyferulic acid shown in lanes 4 and 5 and caffeic acid in lanes 9 and 10 were unreacted substrates. 5-Hydroxyferulic acid (lane 3) and caffeic acid (lane 8) were not converted in control assays. Methylated products from hydroxycinnamoyl CoA esters were hydrolyzed to release the corresponding hydroxycinnamic acids that were separated by TLC and gave results as shown above.
To our knowledge, gymnosperm OMTs have only been previously studied by Higuchi and associates (34, 35) using seedlings as the protein source. They showed that the specific activity of loblolly pine seedling OMT catalyzing the methylation of caffeic acid was 3.3 times that mediating the methylation of 5-hydroxyferulic acid (35). However, our current study demonstrated that the specific activity of AEOMT in crude extracts of secondary developing xylem of loblolly pine was only 1.3 times higher with caffeic acid than with 5-hydroxyferulic acid (Table 1). A 1.3 times higher catalytic efficiency for methylating caffeic acid over 5-hydroxyferulic acid was also confirmed for the yeast-expressed AEOMT (Table 1), revealing that xylem AEOMT is functionally distinct from loblolly pine seedling OMT. These results and genomic Southern analysis (Fig. 2B) further illustrate the polymorphism of OMT in loblolly pine. Consequently, the difference in biochemical function between xylem AEOMT and seedling OMT probably reflects their distinct physiological roles in vivo. In any case, the current results suggest that if 5-hydroxyferulic acid is available in vivo in loblolly pine secondary xylem, it could also be efficiently methylated by AEOMT to synthesize precursors for potential production of angiosperm-like lignin in woody tissue of loblolly pine.
Although having a similar specific activity with these four monomeric lignin precursors, the yeast-expressed AEOMT seemed to exhibit a slightly better catalytic activity with hydroxycinnamic acids than with hydroxycinnamoyl CoA esters (Table 1). In contrast, in extracts of loblolly pine secondary xylem, hydroxycinnamoyl CoA esters were methylated more efficiently than their corresponding hydroxycinnamic acids (Table 1). This difference between yeast-expressed AEOMT and plant enzyme systems in using hydroxycinnamoyl CoA esters, along with the identical efficiency between these two enzyme systems in converting hydroxycinnamic acids, suggests the possible existence of another OMT enzyme in loblolly pine xylem, which also uses these CoA esters. In all events, the results of this study strongly support the notion that AEOMT represents a new class of SAM-dependent OMT that is distinct from CAOMTs and CCoAOMTs with respect to both biochemical function and amino acid sequence. In this regard, it should be stressed that the concept of CAOMT and CCoAOMT mediating two independent methylation pathways in lignin biosynthesis is not a general one; it could be valid for Zinnia (17, 18) but might not be applicable to loblolly pine.
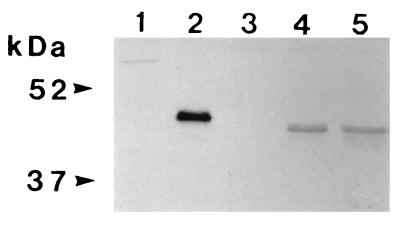
The purified AEOMT–His⋅Tag fusion protein was used as an antigen to prepare anti-AEOMT polyclonal antibodies to further characterize AEOMT proteins. In a Western blot analysis using this antibody as a probe, both yeast-expressed AEOMT and loblolly pine xylem proteins exhibited a single major immunoreactive band with an estimated molecular mass of 42 kDa, which corresponded well with the predicted molecular mass of the AEOMT polypeptide and was as expected, slightly smaller than E. coli-expressed AEOMT–His⋅Tag fusion protein (43 kDa) (Fig. 5). The anti-AEOMT antibody was then used to further characterize the biochemical function of AEOMT. It was found that, while AEOMT activity with different substrates was inhibited to different degrees, both plant and yeast-expressed AEOMT activities with the same substrate were inhibited to an essentially identical level (Table 1). Overall, a lower level of inhibition of AEOMT activity with hydroxycinnamic acids than with hydroxycinnamoyl CoA esters was observed (Table 1). This could suggest that, in the case of hydroxycinnamic acids, the structural conformation of AEOMT in extracts of plant and yeast allows an interaction between protein and substrates but might not be favorable for an efficient protein-antibody binding, resulting in a moderate inhibition of the enzyme activities toward these acids. In contrast, the significant inhibition of the activity toward CoA esters could then imply an effective affinity between antibodies and amino acid domains that are associated with the conversion of CoA esters. Taken together, these results suggest the existence of unique amino acid domains in the AEOMT protein for mediating independently the methylation of hydroxycinnamoyl CoA esters and hydroxycinnamic acids. However, more direct evidence needs to be established to substantiate these points.
Figure 5.
Western blot analysis of AEOMT. Lanes: 1, total protein (15 μg) from E. coli containing vector only (E. coli control); 2, purified insoluble AEOMT fusion protein (0.5 μg) from E. coli expressing AEOMT cDNA, with an approximate size of 43 kDa; 3, total soluble protein (30 μg) from yeast containing vector only (yeast control); 4, AEOMT protein (≈42 kDa) in total soluble protein (30 μg) from yeast expressing AEOMT cDNA; 5, AEOMT protein (≈42 kDa) in total soluble protein (14 μg) from secondary developing xylem of loblolly pine. Molecular mass markers are to the left.
AEOMT Could Be Involved in a Dual Methylation Pathway Associated with Lignin Biosynthesis in Loblolly Pine.
Our results demonstrate that AEOMT cDNA from secondary developing xylem of loblolly pine encodes a multifunctional SAM-dependent OMT with both CAOMT and CCoAOMT activities, catalyzing efficiently the methylation of caffeic acid, caffeoyl CoA, 5-hydroxyferulic acid, and 5-hydroxyferuloyl CoA. Therefore, we propose an AEOMT-mediated dual methylation pathway in lignin biosynthesis in loblolly pine differentiating xylem, with caffeic acid being a divergent point. In one pathway, caffeic acid is methylated through the mediation of AEOMT to form ferulic acid, which is then converted to feruloyl CoA as catalyzed by 4-coumarate:CoA ligase (22). Feruloyl CoA is eventually converted to coniferyl alcohol, as originally proposed by Higuchi (5). In the other pathway, caffeic acid is CoA-ligated by the action of 4-coumarate:CoA ligase (22) to form caffeoyl CoA, which is further methylated by AEOMT to feruloyl CoA which is then converted to coniferyl alcohol as described above. Thus, it is likely that, under specific plant control mechanisms, lignin precursors can be channeled into either or both pathways, providing flexible mechanisms for coordinating the biosynthesis of lignin in loblolly pine xylem.
The cloning of multifunctional AEOMT opens new avenues for investigating the fundamental differences in lignin biosynthesis between gymnosperms and angiosperms. The catalytic activity of AEOMT with both CAOMT and CCoAOMT substrates is especially intriguing, considering the apparent sequence dissimilarity between AEOMT and CAOMT or CCoAOMT. This multiple biochemical function of AEOMT is particularly important in view of its potential effectiveness in facilitating genetic engineering of angiosperm-like lignin in gymnosperms. Because of the previous ambiguity of the role of gymnosperm OMTs, it has always been believed that both angiosperm CAOMT and ferulic acid 5-hydroxylase are needed to be introduced into gymnosperms to engineer angiosperm-like lignin (12, 13). It was further suggested that such genetic engineering would be more efficient with the introduction of angiosperm CCoAOMT (17, 18). Clearly, our current results point out that, at the methylation level, no additional OMT other than the indigenous AEOMT is needed for the potential production of angiosperm-like lignin in woody tissue of loblolly pine or gymnosperms in general.
Acknowledgments
We thank Drs. G. K. Podila and H. Suzuki for helpful discussions, Drs. R. Smeltzer and D. Carraway (both from the International Paper Co.) for helping collect loblolly pine xylem tissue, Drs. S. SanFransisco and D. Knaff (both from Texas Tech University) for protein sequencing, and C. Webb for the artwork. This research was supported by a grant from the National Science Foundation (IBN 9118386).
ABBREVIATIONS
- OMT
O-methyltransferase
- CAOMT
caffeic acid 3-O-methyltransferase
- CCoAOMT
caffeoyl CoA 3-O-methyltransferase
- AEOMT
hydroxycinnamic acids/hydroxycinnamoyl CoA esters OMT
- SAM
S-adenosyl-l-methionine
- FOMT
flavonoid OMT
Footnotes
References
- 1.Sarkanen K V. In: Lignins: Occurrence, Formation, Structure and Reaction. Sarkanen K V, Ludwig C H, editors. New York: Wiley–Interscience; 1971. pp. 95–155. [Google Scholar]
- 2.Grisebach H. In: The Biochemistry of Plants. Conn E E, editor. Vol. 7. New York: Academic; 1981. pp. 457–478. [Google Scholar]
- 3.Chang H M, Sarkanen K V. Tappi. 1973;56:132–136. [Google Scholar]
- 4.Chiang V L, Funaoka M. Holzforschung. 1990;44:309–313. [Google Scholar]
- 5.Higuchi T. In: Biosynthesis and Biodegradation of Wood Components. Higuchi T, editor. New York: Academic; 1985. pp. 141–160. [Google Scholar]
- 6.Whetten R, Sederoff R. Plant Cell. 1995;7:1001–1013. doi: 10.1105/tpc.7.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudet A M, Lapierre C, Grima-Pettenati J. New Phytol. 1995;129:203–226. doi: 10.1111/j.1469-8137.1995.tb04292.x. [DOI] [PubMed] [Google Scholar]
- 8.Dwivedi U N, Campbell W H, Yu J, Datla R S S, Bugos R C, Chiang V L, Podila G K. Plant Mol Biol. 1994;26:61–71. doi: 10.1007/BF00039520. [DOI] [PubMed] [Google Scholar]
- 9.Ni W, Paiva N L, Dixon R A. Transgenic Res. 1994;3:120–126. [Google Scholar]
- 10.Atanassova R, Favet N, Martz F, Chabbert B, Tollier M T, Monties B, Fritig B, Legrand M. Plant J. 1995;8:465–477. [Google Scholar]
- 11.Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier M T, Petit-Conil M, Leple J C, Pilate G, Cornu D, Monties B, Van Montagu M, Inze D, Boerjan W, Jouanin L. Plant J. 1995;8:855–864. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugos R C, Chiang V L, Campbell W H. Plant Mol Biol. 1991;17:1203–1215. doi: 10.1007/BF00028736. [DOI] [PubMed] [Google Scholar]
- 13.Bugos R C, Chiang V L, Campbell W H. Phytochemistry. 1992;31:1495–1498. doi: 10.1016/0031-9422(92)83093-e. [DOI] [PubMed] [Google Scholar]
- 14.Matern U, Wendorff H, Hamerski D, Pakusch A E, Kneusel R E. Bull Liaison Groupe Polyphenols. 1988;14:173–184. [Google Scholar]
- 15.Kühnl T, Koch U, Heller W, Wellmann E. Plant Sci. 1989;60:21–25. [Google Scholar]
- 16.Pakusch A E, Kneusel R E, Matern U. Arch Biochem Biophys. 1989;271:488–494. doi: 10.1016/0003-9861(89)90299-3. [DOI] [PubMed] [Google Scholar]
- 17.Ye Z-H, Kneusel R E, Matern U, Varner J E. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Z-H, Varner J E. Plant Physiol. 1995;108:459–467. doi: 10.1104/pp.108.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt D, Pakusch A E, Matern U. J Biol Chem. 1991;266:17416–17423. [PubMed] [Google Scholar]
- 20.Zhang X-H, Dickson E E, Chinnappa C C. Plant Physiol. 1995;180:429–430. doi: 10.1104/pp.108.1.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng H, Campbell W H. Plant Physiol. 1995;108:1749. [Google Scholar]
- 22.Zhang X-H, Chiang V L. Plant Physiol. 1997;113:65–74. doi: 10.1104/pp.113.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Neish A C. Can J Biochem Physiol. 1959;37:1431–1438. [PubMed] [Google Scholar]
- 26.Stockigt J, Zenk M H. Z Naturforsch. 1975;30:352–358. doi: 10.1515/znc-1975-5-609. [DOI] [PubMed] [Google Scholar]
- 27.Joshi C P. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. J Cell Biol. 1991;115:887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi C P. Nucleic Acids Res. 1987;15:9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidgren J, Svensson L A, Liljas A. Nature (London) 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 31.Williamson J D, Stoop L M H, Massel M O, Conkling M A, Pharr D M. Proc Natl Acad Sci USA. 1995;92:7148–7152. doi: 10.1073/pnas.92.16.7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somssich I E, Wernert P, Kiedrowski S, Hahlbrock K. Proc Natl Acad Sci USA. 1996;93:14199–14203. doi: 10.1073/pnas.93.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grima-Pettenati J, Feuillet C, Goffner D, Bordereis G, Boudet A M. Plant Mol Biol. 1993;21:1085–1095. doi: 10.1007/BF00023605. [DOI] [PubMed] [Google Scholar]
- 34.Shimada M, Fushiki H, Higuchi T. Phytochemistry. 1972;11:2657–2662. [Google Scholar]
- 35.Kuroda H, Shimada M, Higuchi T. Phytochemistry. 1975;14:1759–1763. [Google Scholar]