Abstract
Simple cladogenetic theory suggests that gene genealogies can be used to detect mixis in a population and delineate reproductively isolated groups within sexual taxa. We have taken this approach in a study of Coccidioides immitis, an ascomycete fungus responsible for a recent epidemic of coccidioidomycosis (Valley fever) in California. To test whether this fungus represents a single sexual species throughout its entire geographic range, we have compared genealogies from fragments of five nuclear genes. The five genealogies show multiple incompatibilities indicative of sex, but also share a branch that partitions the isolates into two reproductively isolated taxa, one centered in California and the other outside California. We conclude that coccidioidomycosis can be caused by two distinct noninterbreeding taxa. This result should aid the future study of the disease and illustrates the utility of the genealogical approach in population genetics.
Keywords: population genetics, microbial evolution, speciation
While the genomes of asexual individuals are inherited intact from one generation to the next, sexual genomes are reshuffled, fragmented, and reconstituted in every generation, and consequently they are mosaics of segments, each with its own individual history (1–4). It follows that genealogies constructed from different parts of asexual genomes will be identical, while those from parts of sexual genomes will all be different, and there will be as many different genealogies as there are nonrecombining parts in the genome. This simple principle suggests that the compatibility of different gene genealogies can be used to infer the presence or absence of mixis in a population, full compatibility among genealogies indicating complete asexuality and incompatibility indicating mixis (5–8). The same argument can be applied in identifying reproductively isolated groups within sexual taxa. Consider the genealogy of a single nonrecombining gene in a sexual population that has very recently split into two reproductively isolated groups. Initially, the genealogy will show individuals from the two groups intermingled. With time, selection and drift within each group will lead to the sorting of preexisting variation, so that first one and then both groups will become monophyletic. The sequence of events during speciation is thus (i) the two groups share polymorphisms, (ii) they do not share polymorphisms, and (iii) they show fixed differences (9, 10). Eventually, such sorting will have occurred at all loci, and the two groups will be identifiable by lack of shared polymorphisms at all loci and branches that are compatible with all gene genealogies. Here, we have taken this approach to study the population structure of the pathogenic fungus Coccidioides immitis.
C. immitis is known from the southwestern United States, Mexico, and Central and South America (11), where it grows in the desert soil as a haploid hypha that segments to produce asexual spores. Inhalation of spores by humans and other mammals may result in infection of the lungs, and the fungus can proliferate to other tissues by endosporulation and further release of spores (12–14). Recently, there has been an epidemic of coccidioidomycosis (Valley fever) in California, with numbers of case reports about 10 times higher than usual (13, 14). As with many other fungi, no sexual state has yet been identified in C. immitis, and very little is known about its genetic structure and geographic differentiation. Yet such knowledge is essential for locating the sources of new outbreaks, defining the relatedness of isolates recovered from different patients, and developing effective vaccines against the fungus. Recently, a molecular genetic analysis of nucleotide polymorphisms among isolates from a single hospital in Tucson, Arizona, provided strong evidence for sexual reproduction (15). To test whether the species is panmictic in its entire range or whether there is geographic differentiation among different populations, we have used a sample of 17 isolates, from California, Texas, Arizona, Mexico, and Argentina, covering the entire known species range (see Table 1).
Table 1.
Distribution of nucleotide polymorphisms in five nuclear gene loci of C. immitis
| Locus | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chitin synthase (369 bp) | Dioxygenase (529 bp) | Orotidine decarboxylase (396 bp) | Serine proteinase (647 bp) | Chitinase (443 bp) | |||||||||||||||||||||||||||||||||
| Site | 1 | 2 | 2 | 3 | 8 | 10 | 10 | 11 | 12 | 2 | 2 | 4 | 4 | 5 | 6 | 6 | 6 | 4 | 5 | 6 | 7 | 8 | 8 | 8 | 8 | 7 | 8 | 9 | 10 | 10 | 10 | 10 | 11 | ||||
| 9 | 1 | 8 | 3 | 7 | 0 | 2 | 7 | 7 | 9 | 9 | 7 | 7 | 0 | 0 | 2 | 4 | 7 | 1 | 3 | 4 | 1 | 8 | 8 | 9 | 3 | 1 | 1 | 2 | 4 | 6 | 7 | 1 | |||||
| 2 | 9 | 8 | 9 | 2 | 5 | 0 | 9 | 2 | 2 | 4 | 3 | 6 | 6 | 6 | 3 | 7 | 7 | 7 | 2 | 4 | 2 | 5 | 7 | 1 | 0 | 1 | 0 | 4 | 1 | 9 | 2 | 1 | |||||
| Nucleic acids | |||||||||||||||||||||||||||||||||||||
| Con | A | G | T | G | C | C | G | C | T | C | A | A | G | C | C | A | A | G | C | C | G | G | A | C | A | C | T | T | C | T | C | C | G | ||||
| Strain | |||||||||||||||||||||||||||||||||||||
| S | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | A | . | . | . | . | . | . | . | ||||
| AZ1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | C | ||||
| AZ2 | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | ||||
| TX1 | . | . | . | . | . | . | . | . | . | . | . | G | . | T | T | . | G | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | ||||
| TX2 | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | G | . | . | . | C | . | A | C | . | . | C | ||||
| MX1 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | ||||
| MX2 | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | ||||
| AG1-5 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | ||||
| CA1 | G | A | C | A | T | T | A | T | C | . | G | G | . | T | T | . | G | C | G | T | T | . | . | T | . | . | . | C | . | . | . | . | C | ||||
| CA2 | G | . | C | A | T | T | A | T | C | . | G | G | . | T | T | . | G | C | G | T | T | . | . | T | . | . | . | C | . | . | T | . | C | ||||
| CA3 | G | A | C | A | . | . | . | . | . | G | G | G | . | T | T | . | G | C | G | T | T | . | . | T | . | . | . | C | . | . | . | . | C | ||||
| CA4 | G | . | C | A | T | T | A | T | C | . | . | G | . | T | T | . | G | C | G | T | T | . | . | T | . | . | . | C | . | . | . | T | C | ||||
| CA5 | G | . | C | A | . | . | . | . | . | . | . | G | . | T | T | . | G | C | G | T | T | . | . | T | . | . | . | C | . | . | T | . | C | ||||
| Amino acids | |||||||||||||||||||||||||||||||||||||
| Con | G | K | L | E | D | I | K | D | N | T | I | L | A | H | Y | L | L | A | T | R | A | R | N | N | I | S | * | T | N | I | N | N | T | ||||
| Sub | . | . | . | . | . | . | . | . | . | S | V | . | . | . | . | . | . | P | S | . | S | S | D | . | V | Y | * | . | K | T | . | . | . | ||||
Numbers indicate positions in the reference sequences; dots indicate nucleotides or amino acids identical to the consensus sequence. Con, consensus; Sub, substitution; S, Silveira, old laboratory strain (California, 1951); AZ1,2, Tucson, AZ; CA1, Kern County, CA; CA2,3, Santa Barbara, CA; CA4,5, San Diego; TX1,2 San Antonio, TX; MX1,2, Monterrey, Mexico; AG1-5, Argentina. All isolates are clinical, except CA4,5 collected from soil, and AG1-5 from a variety of sources; isolates were obtained between 1969 and 1994. ∗, intron.
MATERIALS AND METHODS
All isolates were obtained from the Roche Molecular Systems Culture Collection (Alameda, CA). We have sequenced 350–650 bp fragments from five nuclear genes, three of which have been shown to have antigenic properties in infected patients: (i) CHS1, coding for chitin synthase, an enzyme responsible for the synthesis of chitin, a major component of the fungal cell wall (16); (ii) pyrG, coding for orotidine 5′-monophosphate decarboxylase (OMPD), an enzyme catalyzing a step in pyrimidine biosynthesis (17); (iii) tcrP, coding for a T cell reactive protein with homology to 4-hydroxy-phenyl-pyruvate dioxygenase (4-HPPD), a liver enzyme reported from mammals (18); (iv) a gene for another antigen, part of a serine proteinase enzyme with homology to human chymotrypsin (19); and (v) CTS2, a gene for a complement fixation antigen with homology to bacterial chitinases (20, 21). For chitin synthase, serine proteinase, and dioxygenase, primers were designed based on sequences of C. immitis available in GenBank, accession nos. L28067L28067, M81863M81863, and L38493L38493 (16, 18, 19), respectively; for chitinase, they were designed based on an unpublished sequence of C. immitis kindly supplied by G. Cole (Medical College of Ohio, Toledo) and T. Kirkland (University of California School of Medicine, San Diego); and for orotidine decarboxylase, they were designed based on conserved regions of sequences of Aspergillus niger, A. nidulans, and Penicillium chrysogenum (ref. 17; PCR primer sequences can be obtained from the authors).
The fungus was cultivated under BL3 containment, and DNA was extracted following a heat treatment to kill the pathogen (22). PCR amplifications followed the protocol in ref. 23, but reactions were subjected to 40 cycles of 94/52/72° for 1/1/1 min, and an additional 7 min at 72°. For each gene, a selected fragment was amplified by PCR and then sequenced directly using the Applied Biosystems model 373 automated DNA sequencer, following the manufacturer’s protocol.
RESULTS AND DISCUSSION
Of 2,384 nucleotide sites sequenced, 33 were polymorphic (1.4%), and of these, 8 sites had unique, noninformative alleles, and 11 had nonsynonymous substitutions (Table 1). Levels of polymorphism were similar across loci, but introns (317 bp in three genes) were less polymorphic than coding regions (0.3% vs. 1.5%, P < 0.05). No site had more than two alleles.
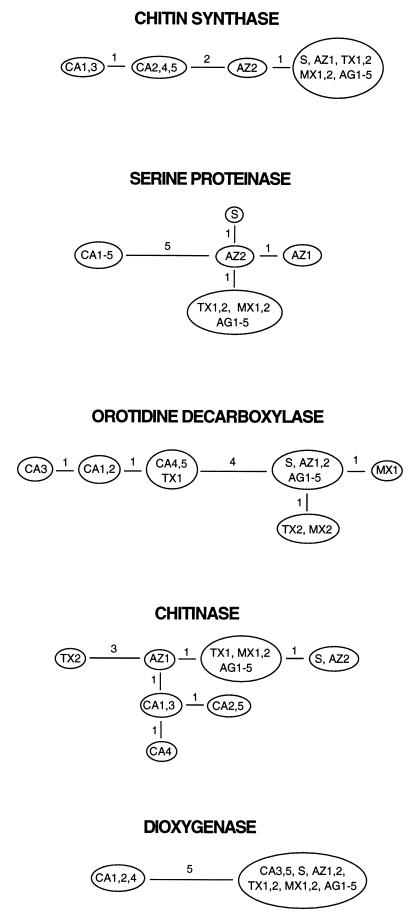
The genealogy of each of the five loci is best described by a single parsimonious tree (Fig. 1). Each tree has minimal length, equal to the number of polymorphic sites in that locus, showing no evidence of parallelisms, reversals, convergences, or recombination within loci (i.e., no homoplasy; homoplasy is present if the number of steps in the most parsimonious tree is larger than the number of alleles at each site minus 1, summed across sites). To test the compatibility of genealogies across loci, we pooled the data from the five loci and looked for homoplasy again. Contrary to the analysis within loci, the most parsimonious tree based on data from all five loci (a total of 25 informative sites) was seven steps longer than the minimum possible (32 vs. 25 steps), indicating extensive incompatibility among genealogies from different loci. Pairwise comparisons of trees from different loci yielded only two fully compatible pairs (i.e., with zero deviation from the minimum length): serine proteinase with chitin synthase, and serine proteinase with dioxygenase. One-step deviations were found in seven pairs, and one pair had a three-step deviation. To further test the compatibility of different trees, we compared the sum of lengths of the five most parsimonious trees constructed from the observed data to the sum of lengths of trees from data sets in which sites were randomly shuffled among the five loci. This test compares the compatibility of sites in different loci with the compatibility of sites in the same locus (26, 27). In sexual species we expect greater incompatibility among distant sites than among neighboring ones, whereas in asexual populations there should be no such difference. Only 6 out of 10,000 such randomizations yielded the observed length of 25 (P = 0.0006; informative sites only; range of sums of tree lengths from randomized data is 25–32; test performed using paup∗4.0d52), showing significant incompatibility among the five loci. This incompatibility indicates sexual reproduction in C. immitis, in agreement with data from a single locale (15). Note that the very low divergence among isolates and complete lack of multiple alleles in any of the sites suggest that homoplasy due to multiple hits is highly unlikely in our data set.
Figure 1.
Genealogies from fragments of five nuclear genes for 17 isolates of C. immitis (data and abbreviations are as in Table 1). Parsimony trees were constructed using hennig86 (24), and numbers of nucleotide changes are shown on each branch. All trees have the minimum possible length (i.e., no homoplasy), indicating no intralocus recombination [if there were free recombination between sites, the probability of observing genealogies of minimal length would be P < 0.01 for each locus (25)].
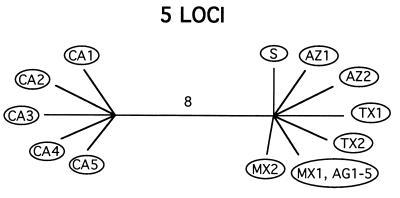
Having demonstrated mixis, we then tested for deviations from panmixia, and in particular whether there might nonetheless be groups of isolates that do not interbreed. For this, we searched for branches in the genealogies that are shared by all five trees. This is equivalent to searching for partitions that divide the isolates into groups that do not share polymorphisms. (The program used to search for partitions can be obtained from the authors.) Note that simple visual inspection of individual gene genealogies may not reveal such partitions, because individual gene genealogies may be missing the branch separating the two groups due to lack of mutations along it. An exhaustive search in our data set uncovered only one such partition, dividing the isolates into two groups, one with 5 and the other with 12 isolates (Fig. 2; Table 1). In addition to not sharing any polymorphisms, these two groups are separated by eight fixed differences distributed among three loci, three of which are replacement substitutions. Furthermore, the partition correlates with the geographic origin of isolates, since all isolates from California group together, except for one old laboratory strain, S, isolated in the San Joaquin Valley (California) in 1951 (Fig. 2). This isolate has previously been shown to differ from other Californian isolates by restriction fragment length polymorphism analysis of genomic DNA (28).
Figure 2.
When the five loci are considered together, the genealogy of the 17 isolates collapses into a single branch, subdividing the population into two reproductively isolated groups. The branch is eight steps long and has a probability of less than 0.001 of occurring by chance. Note the correlation with geography (abbreviations are as in Table 1).
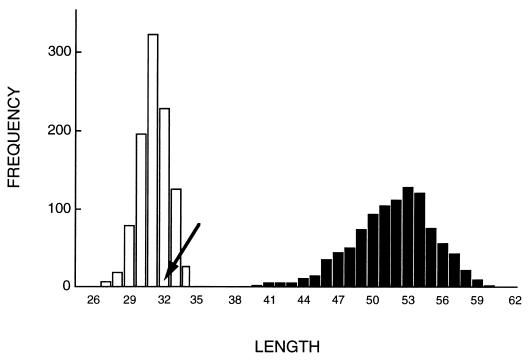
The probability of there being two or more groups of isolates that, by chance, do not share polymorphisms was calculated by randomly shuffling the gene sequences among isolates, leaving the linkage of alleles within loci intact (i.e., the loci were randomized as blocks). In 1,000 such randomizations, no partitions were found that would create groups without shared polymorphisms (P < 0.001). Note that this test is conservative in that it considers only the absence of shared polymorphisms among groups—the fact that there are eight fixed differences between the two groups and the correlation with geographical locale does not enter into the test. Shortest trees fitted to these randomized datasets were longer than the tree fitted to the observed dataset (range = 40–60, n = 1,000, vs. 32, P < 0.001; Fig. 3), also indicating deviation from panmixia. To test whether there is any structure within either the Californian or the non-Californian groups, we repeated the randomizations, this time shuffling sequences among isolates within each of the two groups (i.e., leaving the partition intact). Shortest trees fitted to these randomized sets were no longer than the observed tree, indicating free recombination within each of these two groups (range 27–34, n = 1,000, vs. 32, P = 0.85; Fig. 3).
Figure 3.
Frequency distributions of tree lengths of maximum parsimony trees fitted to randomized data sets (25 informative sites in five loci; 1,000 randomizations in each case). Solid bars, shuffling genotypes among isolates in the entire population. Open bars, shuffling among isolates within each of the two groups. Arrow shows the tree length from the observed data set.
Differentiation between the two taxa of C. immitis can be quantified by comparing the divergence of sequences between taxa with the divergence of sequences within (29). The average pairwise divergence of isolates within the Californian and non-Californian groups is dC = 2.7 × 10−3 (6.7 × 10−3) and dNC = 2.1 × 10−3 (4.2 × 10−3), respectively (coding regions only; values in parentheses based on third-base positions); whereas the average pairwise divergence between groups is dC-NC = 9.5 × 10−3 (21.4 × 10−3), 4-fold larger than the within-group values. These estimates are similar to the divergence of the Adh locus within a global sample of Drosophila melanogaster, and between D. melanogaster and D. simulans or D. mauritiana (29, 30), respectively. Based on these values, the time since the two taxa have been reproductively isolated can be estimated to be 8 Myr (ref. 29; assuming a substitution rate at third-base positions of u = 10−9/bp/yr).
The presence of fixed differences between the two taxa allows us to make a simple test for their identification by combining PCR with restriction fragment length polymorphism analysis, and using enzymes that distinguish between polymorphisms at those sites. We have used two such enzymes (MnlI and ApoI, distinguishing polymorphisms in positions 477 and 744 of serine proteinase, respectively; see Table 1) to rescore the 17 isolates, plus an additional 77 isolates, and thus to delineate in more detail the distribution of the two taxa. In agreement with the previous results, we have found that one taxon is centered in California, being found in Kern (12 isolates), Santa Barbara (3), San Diego (14, including 4 isolates from soil), and Ventura (4) Counties, plus 1 isolate from Washington State (Fig. 4). The other taxon occurs mainly outside California, with isolates from Tucson, Arizona (27), San Antonio, Texas (17), Monterrey, Mexico (6), plus 1 isolate from a rodent in Arizona. Both species were represented in isolates from Argentina (five with the non-Californian genotype, all identical, see Table 1; one with the Californian genotype). The Californian genotype was also found in one patient from San Antonio, but it was later determined that the infection had been acquired in California (31). The non-Californian genotype was also found in three Californian patients, but it is not clear whether these exceptions are due to patients moving between localities or to a real overlap in the range of the two taxa.
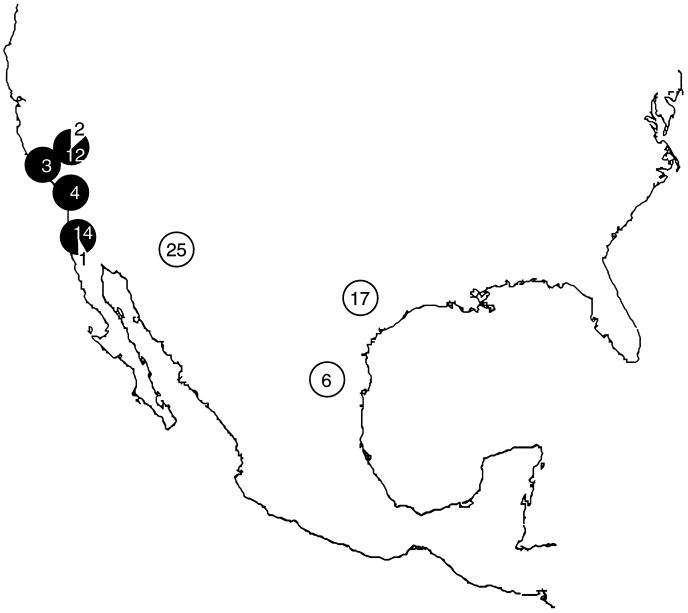
Figure 4.
Geographic distribution of isolates of the two taxa of C. immitis, identified using restriction enzyme analysis of two polymorphisms in serine proteinase distinguishing the two taxa: Kern (14 isolates; 1993–94), Santa Barbara (3), San Diego (15, including 4 isolates from soil; 1975–94 and 1969, respectively), and Ventura (4) Counties, CA; Tucson, Arizona (25; 1979–90); San Antonio, TX (17; 1992–93); and Monterrey, Mexico (6). Solid circles, Californian genotype; open circles: non-Californian genotype. Six isolates from Argentina and one isolate from Washington State are not shown.
CONCLUSIONS
Our analysis has revealed an unexpected subdivision of C. immitis into two sexual but reproductively isolated taxa that are as divergent as two species of Drosophila. This subdivision is correlated with the geographical origin of isolates. Little is known about the life history of this fungus, and these results could help interpret the epidemiology of coccidioidomycosis. For example, there is substantial variation in the symptoms of the disease among infected patients (12), and it would be interesting to know how much of this variation can be explained by differences in the pathogenicity of the two taxa. Our restriction analysis suggests that the recent epidemic in California (13, 14) was mainly caused by one of the two species. Could these two groups also have different host species? Kangaroo rats (Dipodomys spp.), which are believed to be one of the primary hosts for C. immitis (12), also show reproductive isolation between populations that are separated by the Sierra Nevada and the Tehachapi Mountains of California (32). In addition, speciation in many lineages of aridlands mice in western North America (Perognathus, Chaetodipus, and Onychomys spp.) coincides with the development of these mountains, over the last 12 Myr (33). These same mountains may also prevent dispersal of C. immitis by wind. Finally, knowledge that there are two pathogenic taxa rather than one may be important in the development of a vaccine against this fungus (34). Three of the eight fixed differences among the two groups are nonsynonymous substitutions, all in the 650-bp fragment of serine proteinase, an antigenic enzyme that is concentrated in the walls of the parasitic cells during active growth in the host (19).
More generally, the comparison of genealogies of single-copy nuclear genes provides an attractive means of delineating mixis and reproductive isolation, where incompatibility at terminal branches indicates mixis and compatibility at deeper, internal branches indicates reproductive isolation. A nice feature of this approach is that it does not require a predetermined set of populations, but it can help identify such populations, and hence detect barriers to gene flow among populations even in sympatry. Such analyses are particularly useful when, as in C. immitis, mating tests are impossible to perform. When such tests do become possible, we predict that isolates belonging to the reproductively isolated taxa identified above will not cross. Our analysis was simplified by the fact that C. immitis is a haploid, but similar analyses can be done on diploids, in which every individual carries two copies of each gene, and the genealogy of a gene will include every individual twice. Reproductively isolated taxa could be identified as groups sharing no polymorphisms, as in haploids, but with the additional constraint that the two alleles belonging to the same individual must be included in the same group.
Acknowledgments
We thank A. Catanzaro, R. A. Cox, J. Galgiani, T. N. Kirkland, R. Negroni, A. Padhye, M. Rinaldi, M. A. Rodriguez, R. Talbot, and C. R. Zimmerman for providing the isolates of C. immitis; G. L. Koenig for supplying the cultures; G. T. Cole and T. N. Kirkland for allowing us to design primers from unpublished sequences; T. L. Lang for technical assistance; D. L. Swofford for permission to use paup∗; A. Sasaki for discussing the theory of coalescence; and T. J. White for reading the manuscript. This study was supported by the National Institutes of Health.
References
- 1.Hein J. Math Biosci. 1990;98:185–200. doi: 10.1016/0025-5564(90)90123-g. [DOI] [PubMed] [Google Scholar]
- 2.Maynard Smith J, Dowson C G, Spratt B G. Nature (London) 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 3.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddison W. In: Experimental and Molecular Approaches to Plant Biosystematics. Hoch P C, Stephenson A G, editors. St. Louis: Missouri Botanical Gardens; 1995. pp. 273–287. [Google Scholar]
- 5.Woese C R, Gibson J, Fox G E. Nature (London) 1980;283:212–214. doi: 10.1038/283212a0. [DOI] [PubMed] [Google Scholar]
- 6.Dykhuizen D E, Green L. J Bact. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykhuizen D E, Polin D S, Dunn J J, Wilske B, Preac-Mursic V, Dattwyler R J, Luft B J. Proc Natl Acad Sci. 1993;90:10163–10167. doi: 10.1073/pnas.90.21.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hey J, Kliman R M. In: Non-Neutral Evolution: Theories and Molecular Data. Golding B, editor. New York: Chapman & Hall; 1994. pp. 208–216. [Google Scholar]
- 9.Neigel J E, Avise J C. In: Evolutionary Processes and Theory. Nevo E, Karlin S, editors. New York: Academic; 1986. pp. 515–534. [Google Scholar]
- 10.Avise J C, Ball R M J. Oxf Surv Evol Biol. 1990;7:45–67. [Google Scholar]
- 11.Rippon J W. Medical Mycology. Philadelphia: W. B. Saunders; 1988. [Google Scholar]
- 12.Pappagianis D. Curr Top Med Mycol. 1988;2:199–238. doi: 10.1007/978-1-4612-3730-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Pappagianis D, Sun R K, Werner S B, Rutherford G W I, Elsea R W, Miller G B J, Bootwala N, Hopkins R S. Morbid Mortal Wkly Rep. 1993;42:21–24. [Google Scholar]
- 14.Pappagianis D. Clin Infect Dis. 1994;19(S1):S14–S18. doi: 10.1093/clinids/19.supplement_1.14. [DOI] [PubMed] [Google Scholar]
- 15.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan S, Sigler L, Cole G T. Microbiol. 1994;140:1481–1494. doi: 10.1099/00221287-140-6-1481. [DOI] [PubMed] [Google Scholar]
- 17.Radford A. J Mol Evol. 1993;36:389–395. doi: 10.1007/BF00182186. [DOI] [PubMed] [Google Scholar]
- 18.Wyckoff E E, Pishko E J, Kirkland T N, Cole G. Gene. 1995;161:107–111. doi: 10.1016/0378-1119(95)00250-a. [DOI] [PubMed] [Google Scholar]
- 19.Cole G T, Zhu S, Hsu L, Kruse D, Seshan K R, Wang F. Infect Immun. 1992;60:416–427. doi: 10.1128/iai.60.2.416-427.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson S M, Zimmermann C R, Pappagianis D. Infect Immun. 1993;61:3090–3092. doi: 10.1128/iai.61.7.3090-3092.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pishko E J, Kirkland T N, Cole G T. Gene. 1995;167:173–177. doi: 10.1016/0378-1119(95)00654-0. [DOI] [PubMed] [Google Scholar]
- 22.Burt A, Carter D A, Carter G L, White T J, Taylor J W. Fungal Genetics Newsletter. 1995;42:23. [Google Scholar]
- 23.Burt A, Carter D A, White T J, Taylor J W. Mol Ecol. 1994;3:523–525. doi: 10.1111/j.1365-294x.1994.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 24.Farris J S. hennig 86. Stockholm: Nat. Hist. Riksmuseet; 1988. , Version 1.5. [Google Scholar]
- 25.Archie J W. Syst Zool. 1989;38:239–252. [Google Scholar]
- 26.Farris J S, Kallersjo M, Kluge A G, Bult C. Cladistics. 1995;10:315–319. [Google Scholar]
- 27.Huelsenbeck J P, Bull J J, Cunningham C W. Trends Ecol Evol. 1996;11:152–158. doi: 10.1016/0169-5347(96)10006-9. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann C R, Snedker C J, Pappagianis D. J Clin Microbiol. 1994;32:3040–3042. doi: 10.1128/jcm.32.12.3040-3042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 30.Stephens J C, Nei M. J Mol Evol. 1985;22:289–300. doi: 10.1007/BF02115684. [DOI] [PubMed] [Google Scholar]
- 31.Burt, A., Dechairo, B., Koenig, G. L., Carter, D., White, T. & Taylor, J. W. (1997) Mol. Ecol., in press. [DOI] [PubMed]
- 32.Schmidly D J, Wilkins K T, Derr J N. In: Biology of the Heteromyidae. Genoways H H, Brown J H, editors. Vol. 10. Shippensburg, PA: American Society of of Mammalogists; 1993. pp. 319–356. [Google Scholar]
- 33.Riddle B. J Mamm. 1995;76:283–301. [Google Scholar]
- 34.Polonelli L, Poulain D, Cole G T, Conti S, Elad D, Gerloni M, Kirkland T, Matthews R, Segal E, Wyckoff E. J Med Vet Mycol. 1994;32:105–112. doi: 10.1080/02681219480000761. (suppl. 1). [DOI] [PubMed] [Google Scholar]