Abstract
Anionic lipids play a variety of key roles in biomembrane function, including providing the immediate environment for the integral membrane proteins that catalyze photosynthetic and respiratory energy transduction. Little is known about the molecular basis of these lipid–protein interactions. In this study, x-ray crystallography has been used to examine the structural details of an interaction between cardiolipin and the photoreaction center, a key light-driven electron transfer protein complex found in the cytoplasmic membrane of photosynthetic bacteria. X-ray diffraction data collected over the resolution range 30.0–2.1 Å show that binding of the lipid to the protein involves a combination of ionic interactions between the protein and the lipid headgroup and van der Waals interactions between the lipid tails and the electroneutral intramembrane surface of the protein. In the headgroup region, ionic interactions involve polar groups of a number of residues, the protein backbone, and bound water molecules. The lipid tails sit along largely hydrophobic grooves in the irregular surface of the protein. In addition to providing new information on the immediate lipid environment of a key integral membrane protein, this study provides the first, to our knowledge, high-resolution x-ray crystal structure for cardiolipin. The possible significance of this interaction between an integral membrane protein and cardiolipin is considered.
Biological protein/lipid membranes partition cells and organelles and support a wide range of important metabolic processes, including energy transduction, solute transport, protein transport, signal transduction, and motility. Anionic phospholipids such as phosphatidyl glycerol and cardiolipin play an important role in a variety of these processes, through both their contribution to the physical properties of the lipid phase of the membrane and their interactions with proteins (1–3). Anionic phospholipids have a particularly important function in energy-transducing membranes such as the bacterial cytoplasmic membrane and the inner mitochondrial membrane (2), where they exert effects on a variety of cellular processes (see refs. 1–3 for reviews). In particular, cardiolipin (diphosphatidyl glycerol; Fig. 1A) has been shown to be a key factor in the maintenance of optimal activity of a number of major integral membrane proteins. Perhaps the best known of these interactions is the requirement displayed by cytochrome c oxidase for cardiolipin; even after extensive purification, the enzyme contains at least one tightly bound cardiolipin that cannot be removed without destroying enzyme activity (4, 5). However, no cardiolipin was identified in the available atomic structures for cytochrome c oxidase (6–8), although several molecules of phosphatidyl glycerol and phosphatidyl ethanolamine were resolved in a 2.8-Å structure for the bovine heart enzyme (6), and one phosphatidyl choline was resolved in a 2.8-Å structure for the Paracoccus denitrificans enzyme (8).
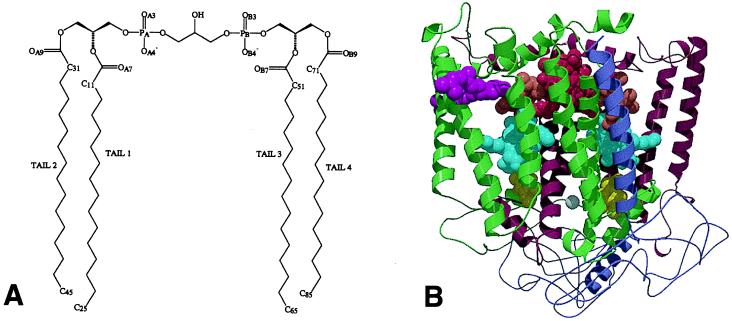
Figure 1.
Molecular structures. (A) Cardiolipin (diphosphatidyl glycerol) with four acyl chains, each with 16 carbons. (B) The Rb. sphaeroides reaction center. The complex consists of an L (maroon), M (green), and H (purple) subunit. These encase a dimer of bacteriochlorophyll (red), two accessory bacteriochlorophylls (sienna), two bacteriopheophytins (cyan), two ubiquinones (yellow), a spheroidenone carotenoid (pink), and a nonheme iron atom (grey). The isoprenoid side chains of the bacteriochlorophyll, bacteriopheophytin, and ubiquinone cofactors have been omitted for clarity.
The photoreaction center is an integral membrane protein complex that uses light energy to pump electrons across the cytoplasmic membrane of photosynthetic bacteria such as Rhodobacter (Rb.) sphaeroides (9, 10). A great deal is known about the structure and function of the bacterial reaction center, including atomic structures for the reaction centers from Rhodopseudomonas (Rps.) viridis (11) and Rb. sphaeroides (12–14) (Fig. 1B). The Rb. sphaeroides reaction center consists of three subunits and ten cofactors (12–14). The L and M subunits both have five transmembrane α helices and are related by an axis of pseudo 2-fold symmetry that runs perpendicular to the plane of the membrane. The H subunit has a single transmembrane helix and a large cytoplasmic domain. The bacteriochlorophyll dimer, monomeric bacteriochlorophyll, bacteriopheophytin, and ubiquinone cofactors consist of a redox active head group and an isoprenoid side chain (omitted for clarity in Fig. 1B). The complex also contains a nonheme iron atom and a single carotenoid molecule that plays a role in photoprotection. The complex catalyzes light-driven transmembrane electron transfer (9, 10), converting light energy into electrochemical potential energy with a high quantum yield. The basis of this highly efficient energy conversion is the separation of electrical charge across the membrane on a very fast time scale, initially between the bacteriochlorophyll dimer (P) located near the periplasmic face of the protein and the bacteriopheophytin most closely associated with the L subunit (HL; Fig. 1B). The P+HL− radical pair is formed in 3–5 ps after the absorption of light energy by P, and the electron is passed on to the QA ubiquinone (Fig. 1B Right) on the cytoplasmic side of the membrane, forming the P+QA− radical pair in 200 ps.
Although the reaction center is structurally well characterized, being one of the most heavily studied integral membrane proteins, almost nothing is known in detailed structural terms about how the protein interacts with its lipid bilayer environment. This is also the case for other integral membrane proteins for which high-resolution structures are now available, including a variety of channels and porins (15–23), transporters (24, 25), toxins (26), electron-transport proteins (27–29), light-harvesting antenna complexes (30–32), and bacteriorhodopsin (33–36). Modeled detergent molecules have been reported in some of these studies (16, 24, 30, 31, 37, 38), but bound lipids have been reported in only a small number of studies (6, 8, 33), including the two structures for cytochrome oxidase detailed above.
In the present study, the x-ray crystal structure of a mutant (AM260W) Rb. sphaeroides reaction center has been determined to a resolution of 2.1 Å. A feature was observed in the electron-density map of the mutant reaction center in a region close to the transmembrane helix of the H subunit. This feature is assigned to a molecule of cardiolipin, and the details of the ionic and hydrophobic interactions between the cardiolipin and the surface of the reaction center are described. The possible role of the bound lipid is discussed.
Materials and Methods
Mutagenesis.
The mutation Ala M260 to Trp (AM260W) was introduced into the pufM gene of the reaction center by using mismatch oligonucleotides. The template for mutagenesis was plasmid pALTCB-1 (39), which consisted of a SacI-BamHI restriction fragment encompassing codons 233–307 of the pufM gene cloned into the plasmid pALTER-1 (Promega). The resulting changes in the sequence of the pufM gene were confined to the target M260 codon (GCC to TGG) and were confirmed by DNA sequencing. The mutated SacI-BamHI restriction fragment was then transferred to plasmid pRKEH10D for expression in the double deletion/insertion mutant strain DD13, as described in detail previously (40–42). The resulting strain, named AM260W, lacked both the LH1 and LH2 antenna complexes and contained the mutant reaction center as the sole pigment protein complex.
Cell Growth and Purification of Reaction Centers.
Rb. sphaeroides strain AM260W was grown under semiaerobic/dark conditions at 34°C in M22+ medium as described previously (42, 43). Intracytoplasmic membranes were isolated by breakage of harvested cells in a French pressure cell, as described in ref. 42, followed by ultracentrifugation at ≈250,000 × g for 1.5 h. Intracytoplasmic membranes were resuspended to a concentration of approximately 10 absorbance units cm−1 at 800 nm in 20 mM Tris⋅HCl (pH 8.0).
Reaction centers were isolated from resuspended intracytoplasmic membranes by using the detergent lauryldimethylamine oxide (LDAO), as described in a recent publication (43). Purification of the reaction centers was achieved by two passes of the solubilized material through a DE52 (Whatman) anion exchange column, followed by further anion exchange separation on a Sepharose Q column (Pharmacia) and gel filtration by using a Superdex 200 preparative grade column (Pharmacia), as described in detail elsewhere (43). Pooled reaction centers from the gel filtration column typically had an absorbance ratio A280/A800 = 1.3 and at this purity were suitable for crystallization.
Purified reaction centers were concentrated by ultrafiltration, first in a stirred cell (Amicon) under nitrogen gas and finally in Centricon concentrators (Amicon). After this initial concentration, the reaction centers were washed with the buffer required for crystallization, which consisted of 10 mM Tris⋅HCl (pH 8.0)/0.1% LDAO and were reconcentrated by using Centricon concentrators. This washing was repeated at least five times to ensure good buffer exchange, and the reaction centers were brought to a final concentration of 60 absorbance units cm−1 at 800 nm, again in 10 mM Tris⋅HCl (pH 8.0)/0.1% LDAO.
Crystallization of Reaction Centers.
Trigonal crystals, space group P3121, were grown by sitting drop vapor diffusion from droplets containing 10 mg/ml reaction center, 0.1% vol/vol LDAO, 3.5% wt/vol 1,2,3-heptanetriol, 0.5 M trisodium citrate, and 10 mM Tris⋅HCl (pH 8.0). The drops were equilibrated against a reservoir of 1.1 M trisodium citrate. Trigonal crystals appeared within 1 to 4 wk and grew as prisms of variable size, ranging from 0.5 mm to 1.5 mm in the longest dimension. The crystals had unit cell dimensions of a = b = 142.0 Å, c = 186.8 Å.
Data Collection and Analysis.
X-ray diffraction data were collected at room temperature at beam line 9.6 of the Daresbury Synchrotron Radiation Source, United Kingdom, by using a Quantum-4 ADSC detector, and were processed by using the denzo and scalepack packages (44). A total of 124,853 unique reflections were recorded by using two crystals, giving data that were 97.6% complete between 30.0 and 2.1 Å and 82.9% complete in the outer shell (2.15 to 2.1 Å), with an overall multiplicity of 4.2 and an overall Rmerge of 5.7%. The Rmerge for the outer shell was 28.1%. Rigid body refinement was performed by using xplor 3.1 (45) by using the coordinates of the wild-type reaction center (43) as a starting model, followed by restrained maximum likelihood refinement in refmac (46), with waters fitted by arpp (47). The R factor was 16.9% with a free R factor of 18.6%. Full refinement details, together with a detailed description of the structure of the AM260W reaction center, will be published elsewhere (K.E.M., P.K.F., J.P.R., N.W.I., R.J.C., and M.R.J., unpublished work). In the figures, structures were illustrated by using the programs molscript (48), raster3d (49), o (50), and xtalview (51).
Results and Discussion
Construction and Properties of the AM260W Reaction Center.
The QA ubiquinone is located at the cytoplasmic end of the branch of reaction center cofactors that is responsible for catalyzing transmembrane electron transfer. One of the two keto oxygens of the head group of the QA ubiquinone forms a hydrogen bond with the backbone amide nitrogen of residue Ala M260. This residue was mutated to tryptophan with a view to causing exclusion of the QA ubiquinone and so preventing light-driven transmembrane electron transfer in the reaction center. Spectroscopic studies, a full account of which has been given recently (52), showed that the AM260W reaction center does not possess QA function, and in particular picosecond time-scale transient absorption spectroscopy showed that light-driven electron transfer is blocked at the state P+HL−.
To characterize further the AM260W reaction center, the complex was purified and crystallized, and x-ray diffraction data were collected according to the procedures described in Methods. The x-ray data obtained confirmed the finding of spectroscopic studies, that the AM260W reaction center lacks the QA ubiquinone, and showed that changes in structure associated with the mutation were confined to the vicinity of the M260 residue and the binding pocket of the QA ubiquinone. A detailed account of these structural changes will be given elsewhere (K.E.M., P.K.F., J.P.R., N.W.I., R.J.C., and M.R.J.; unpublished work).
Electron Density at the Surface of the Reaction Center.
The data set for the AM260W reaction center were of a significantly higher quality than have been obtained by us previously for the wild-type or mutant reaction centers (43, 53, 54). This was because of a combination of factors, including the particularly good quality of the AM260W crystals, an increase in flux of beamline 9.6 at the Daresbury Synchrotron Radiation Source at the time that the data were collected, and the use of a charge-coupled device detector system, which meant that the time taken to collect a full data set was comparatively short (approximately 2 hr), and so gradual loss of high-resolution reflections because of radiation damage was minimized. In addition, low-resolution (10.0–30.0 Å) reflections were collected and used in the refinement.
In the electron density map of the AM260W reaction center, a feature was observed in a region close to the H subunit transmembrane helix that could not be attributed to the protein (Fig. 2A). This feature was attributed to a molecule of cardiolipin, and a model of cardiolipin built into the density is shown (Fig. 2A). This electron density feature was well removed from the regions of the protein involved in crystal contacts, and there were no other electron density features at the surface of the reaction center that could be reliably modeled as either lipid or detergent.
Figure 2.
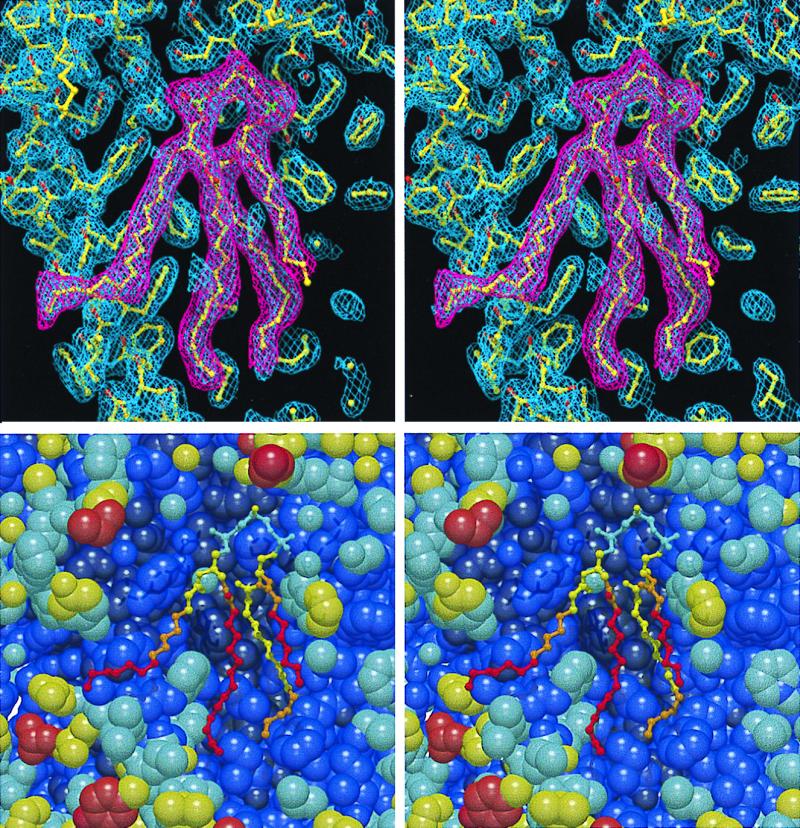
Stereo views of the model of cardiolipin at the surface of the AM260W reaction center. (A) refmac 2 mFo–DFc map (blue) of the electron density attributed to cardiolipin and that of the background protein, with the fitted structure of the protein and the cardiolipin. Overlaid is a mFo–DFc map (magenta) of the density attributed to the cardiolipin. Only those parts of the acyl chains resolved in the electron density were modeled (see text). (B) Stick model of cardiolipin and space-fill model of the surrounding protein, colored according to crystallographic temperature factor. Dark blue (0–30); blue (30–40); sky blue (40–50); yellow (50–60); orange (60–70); red (70–100).
Cardiolipin (diphosphatidyl glycerol) has a polar head group composed of three glycerol molecules connected by two phosphodiester linkages, with four acyl chains connected to the primary and secondary hydroxyl groups of the terminal glycerol moieties of the head group (Fig. 1A). In Rb. sphaeroides, the acyl chains of cardiolipin can vary in length and in the degree of unsaturation (55), and environmental changes can alter the composition (1). In cardiolipins from Rb. sphaeroides, the most common acyl chain has 18 carbon atoms and a single unsaturated bond (i.e., 18:1), although both shorter and longer acyl chains are present in minor amounts (55). In the present study, the ends of the acyl chains were not resolved in the electron density, presumably because they were mobile and therefore disordered. The maximum chain lengths that could be supported by the density were 15 carbon atoms for chains 1 and 4, 14 carbon atoms for chain 2, and 9 carbon atoms for chain 3, and only these were included in the model. Double bonds were not included in the models of the acyl tails, because their exact position could not be determined from the electron density, and approximately 20% of the acyl tails in cardiolipin derived from Rb. sphaeroides membranes are fully saturated.
Fig. 2B shows a stereo view of the model of the cardiolipin and the surrounding protein, with each atom colored according to the crystallographic temperature factor. For the cardiolipin, the temperature factors were lowest for the head group and generally increased with progression down each acyl tail.
Cardiolipin–Protein Interactions.
The head group of the cardiolipin molecule was located on the cytoplasmic side of the membrane and came into close contact with residues from all three of the reaction center subunits (Fig. 3A). The contacts between atoms of the modeled cardiolipin and the surrounding protein were calculated by using the program contact (56) with a 4-Å cutoff, and the most significant hydrogen bond or salt bridge interactions are shown in Fig. 3A. A full list of contacts is available as supplementary material to this manuscript (see www.pnas.org). Several possible interactions were observed between the lipid headgroup and the surrounding protein, in particular between the phosphate oxygens of PA and His M145/Arg M267. In addition, the cardiolipin made contacts with the backbone amide of Lys M144 and with waters S76, S129, S148, and S149. These waters in turn made contacts with residues Lys M144, Trp M148, Arg M267, Trp M271, and Tyr H30.
Figure 3.
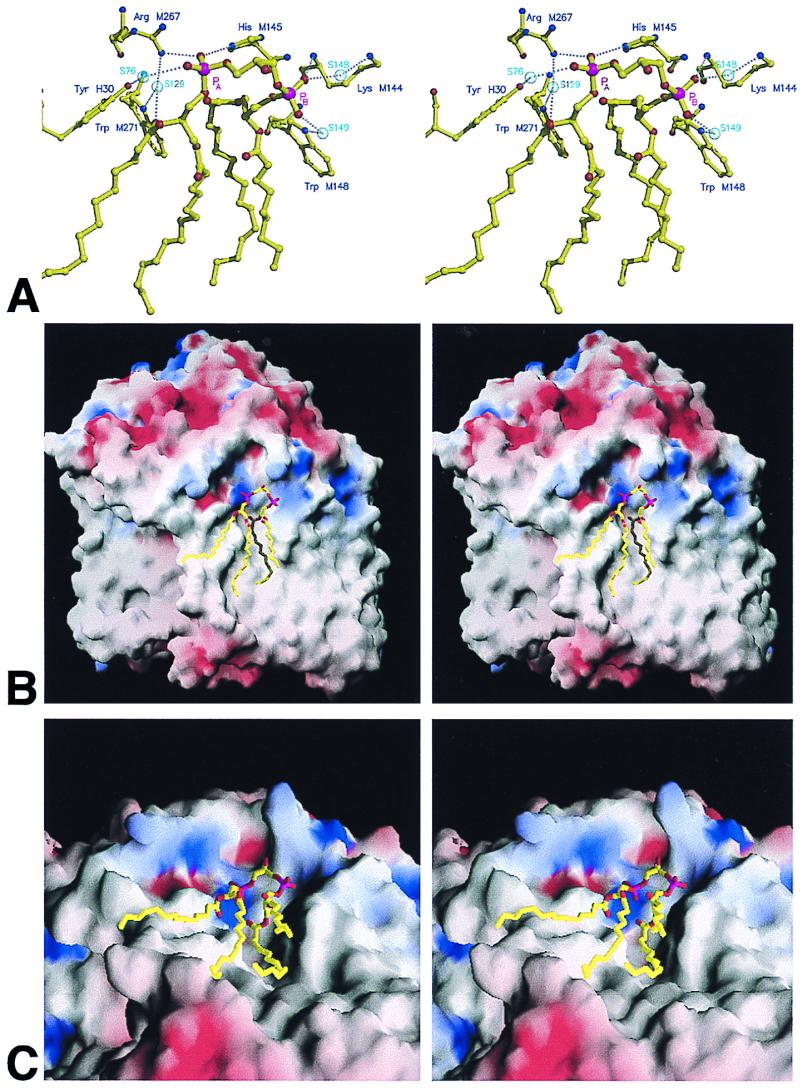
Stereo views of the interaction of cardiolipin with the surrounding protein. (A) Main bonding interactions between the cardiolipin headgroup and surrounding protein. (B) Solid model of the reaction center, colored according to surface potential with the program grasp (57) (blue positive, red negative, grey neutral), with cardiolipin shown in stick format. (C) Representation as in B viewed from the periplasmic side of the membrane.
The tail region interacted over a large surface area within the transmembrane region of the protein, with the closest contacts occurring at the top of chains 3 and 4 and toward the middle of chains 1, 2, and 4. Figs. 3 B and C show different stereo views of the highly irregular surface of the reaction center, colored according to surface potential (blue, positive; red, negative; grey, neutral) with the program grasp (57). The head group of the cardiolipin (stick format) was located on the cytoplasmic side of the membrane, close to the point where the transmembrane helix of the H subunit traverses the membrane, in a region of positive potential. The tails of the cardiolipin lie along grooves in the intramembrane surface of the protein, in regions that are largely electroneutral. Therefore, the strength of the lipid/protein interaction is contributed to by both ionic interactions with the cardiolipin headgroup and van der Waals interactions in the tail region.
Comparisons with Previous Studies.
A number of groups have determined structures for the wild-type Rb. sphaeroides reaction center and for several mutant complexes at resolutions ranging from 3.1 Å to 2.2 Å (12–14, 58–62). In some of these structures, density was seen on the surface of the M subunit, in a region adjacent to the transmembrane helix of the H subunit, that was modeled variously as a molecule of phosphate and/or one or more molecules of the detergent LDAO (12–14). In all cases where it was modeled, the phosphate molecule was located close to residues Arg M267 and His M145, which are exposed on the surface of the reaction center at the interface between the intramembrane and cytoplasmic regions of the protein (12, 13).
In addition to structures for the Rb. sphaeroides reaction center, a 2.3-Å structure has been determined for the reaction center from Rps. viridis (11, 63). The structural model for this complex has a sulfate molecule in close proximity to residues His M143 and Arg M265, the counterparts of residues M145 and M267 in Rb. sphaeroides. In addition, four more sulfate molecules were modeled in the structure of the Rps. viridis reaction center, at the interface between the intramembrane region and the cytoplasmic surface of the reaction center. Deisenhofer and coworkers (11) also commented on the fact that a number of mostly elongated features in the electron density map could not be satisfactorily modeled and suggested that these may have arisen from detergent, clusters of water molecules, or heptane-1,2,3-triol, the amphiphile used in crystallization of the complex.
In considering why the cardiolipin–protein interaction was resolved in the present study, we have discounted the possibility that this was because of the use of trisodium citrate as the precipitant for crystallization of the AM260W reaction center, rather than the more commonly used phosphate or sulfate (which could compete for binding with basic residues such as His M145 and Arg M267). Preliminary analysis of an electron density map at 2.5-Å resolution for a YM210W mutant reaction center has also clearly revealed the bound cardiolipin (K.E.M., F.P.K., M.R.J., R.J.C., and N.W.I., unpublished data). This reaction center was crystallized by using the conditions described in a recent publication (43), with potassium phosphate as the precipitant rather than trisodium citrate. In addition, in a 2.3-Å resolution structure for a WM115F/FM197R mutant reaction center reported recently (54), which was also crystallized by using potassium phosphate, electron density was observed at the same position on the protein surface as that occupied by cardiolipin in the structure described in this report. This density was not continuous and was modeled as two molecules of phosphate, 6.2 Å apart and at the same height in the membrane, adjacent to three molecules of LDAO also located on the protein surface toward the center of the membrane. In retrospect, it seems highly likely that this density was actually part of the electron density observed in the present report, corresponding to the two phosphoryl groups of the cardiolipin and part of acyl chains 1, 2, and 4.
Possible Function of Bound Cardiolipin.
To our knowledge, no study has been made of the relevance of cardiolipin to the structure and function of the bacterial reaction center. The findings described in this report provide a solid framework for such a study, which is currently under way. However, it is clear that cardiolipin is a key component of energy-transducing membranes and can affect a wide range of cellular functions. As indicated in the Introduction, cardiolipin is important for the maintenance of optimal activity of a number of major integral membrane proteins, including NADH dehydrogenase, the cytochrome bc1 complex, ATP synthase, cytochrome c oxidase, and a range of carrier proteins (1, 2, 10). The influence of cardiolipin on the activity of essential membrane-bound protein complexes provides a potential means of controlling the processes catalyzed by these proteins. It has been suggested, for example, that the control of mitochondrial respiration by thyroid hormones is exerted at the level of cardiolipin synthase (64), with changes in the cardiolipin content of the membrane leading to changes in the activity of major proteins such as cytochrome c oxidase (65).
In addition to their specific interactions with integral membrane proteins, anionic phospholipids such as cardiolipin and phosphatidyl glycerol play a crucial role in the adsorption of a range of peptides and soluble proteins onto the membrane surface, some proteins becoming embedded in the membrane bilayer (3). This process constitutes a second means by which cardiolipin can affect a diverse range of cellular processes. In general terms, the proteins involved may be agonists that interact with membrane-bound receptors, soluble electron transfer proteins that shuttle electrons between membrane-bound electron transfer complexes, or proteins that are to be translocated across the membrane by systems such as the Sec protein translocation apparatus of Escherichia coli (2, 3, 66, 67). Adsorption by anionic lipids concentrates the protein at the membrane surface, and the interaction with the membrane may orient the protein for correct binding to a receptor. Anionic lipids have also been found to interact with DNA- and RNA-binding proteins, leading to the suggestion that they may play a role in the control of chromosomal replication (3).
It has been reported that bacteriochlorophyll-containing proteins in Rb. sphaeroides preferentially associate with negatively charged lipids (68). When reaction center/light harvesting I core complexes in intracytoplasmic membranes of Rps. viridis were depleted of lipid by extraction with Triton X-100, cardiolipin was found to be the most abundant of the remaining lipids, perhaps reflecting a high affinity of cardiolipin for the proteins of the membrane (69). It has also been observed that the presence of anionic lipids in an artificial membrane bilayer increases the rate of electron transfer from soluble cytochrome c to photooxidized reaction centers, compared with that seen with neutral membranes (70).
It is intriguing that the residues that interact with phosphate headgroup A (Fig. 3A) are conserved among the available full and partial sequences for the M subunit of the bacterial reaction center (16 sequences for His M145 and 8 sequences for Arg M267). Residues Trp M148 and Tyr H30 are also conserved in the eight and four available sequences, respectively, and residue M271 is Phe in Rps. viridis but Trp in the seven remaining sequences that encompass this residue. These residues may therefore constitute a conserved site for binding of cardiolipin on the surface of the bacterial reaction center. Given the important role that cardiolipin plays in the stability and function of integral proteins in energy-transducing membranes, it will be of great interest to examine the consequences of disrupting cardiolipin binding at a molecular level. We are currently using site-directed mutagenesis to investigate whether the molecule of cardiolipin seen attached to the surface of the reaction center affects the function and/or assembly of the complex.
Supplementary Material
Acknowledgments
We thank the staff at beamline 9.6 of the Daresbury Synchrotron Radiation Source for their assistance with data collection. This work was funded by the Biotechnology and Biological Sciences Research Council of the United Kingdom.
Abbreviation
- LDAO
lauryldimethylamine oxide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1QOV).
References
- 1.Ioannou P V, Golding B T. Prog Lipid Res. 1979;17:279–318. doi: 10.1016/0079-6832(79)90010-7. [DOI] [PubMed] [Google Scholar]
- 2.Hoch F L. Biochim Biophys Acta. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 3.Dowhan W. Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 4.Awasthi Y C, Chuang T F, Keenan T W, Crane F L. Biochim Biophys Acta. 1971;226:42–52. doi: 10.1016/0005-2728(71)90176-9. [DOI] [PubMed] [Google Scholar]
- 5.Robinson N C. Biochemistry. 1982;21:184–188. doi: 10.1021/bi00530a031. [DOI] [PubMed] [Google Scholar]
- 6.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 7.Ostermeier C, Harrenga A, Ermler U, Michel H. Proc Natl Acad Sci USA. 1997;94:10547–10553. doi: 10.1073/pnas.94.20.10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 9.Fleming G R, van Grondelle R. Phys Today. 1994;47:48–55. [Google Scholar]
- 10.Hoff A J, Deisenhofer J. Phys Rep. 1997;287:1–247. [Google Scholar]
- 11.Deisenhofer J, Epp O, Sinning I, Michel H. J Mol Biol. 1995;246:429–457. doi: 10.1006/jmbi.1994.0097. [DOI] [PubMed] [Google Scholar]
- 12.Ermler U, Fritzsch G, Buchanan S K, Michel H. Structure (London) 1994;2:925–936. doi: 10.1016/s0969-2126(94)00094-8. [DOI] [PubMed] [Google Scholar]
- 13.Ermler U, Michel H, Schiffer M. J Bioenerg Biomembr. 1994;26:5–15. doi: 10.1007/BF00763216. [DOI] [PubMed] [Google Scholar]
- 14.Stowell M H B, McPhillips T M, Rees D C, Soltis S M, Abresch E, Feher G. Science. 1997;276:812–816. doi: 10.1126/science.276.5313.812. [DOI] [PubMed] [Google Scholar]
- 15.Doyle D A, Cabral J M, Pfuetzner R A, Kuo A, Gulbis J M, Cohen S L, Chait B T, MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 16.Pautsch A, Schulz G E. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 17.Forst D, Welte W, Wacker T, Diederichs K. Nat Struct Biol. 1998;5:37–46. doi: 10.1038/nsb0198-37. [DOI] [PubMed] [Google Scholar]
- 18.Schirmer T, Keller T A, Wang Y F, Rosenbusch J P. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 19.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Nature (London) 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 20.Kreusch A, Neubüser A, Schiltz E, Weckesser J, Schulz G E. Protein Sci. 1994;3:58–63. doi: 10.1002/pro.5560030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch A, Breed J, Saxena K, Richter O M H, Ludwig B, Diederichs K, Welte W. FEBS Lett. 1997;404:208–210. doi: 10.1016/s0014-5793(97)00131-2. [DOI] [PubMed] [Google Scholar]
- 23.Meyer J E W, Hofnung M, Schulz G E. J Mol Biol. 1997;266:761–775. doi: 10.1006/jmbi.1996.0823. [DOI] [PubMed] [Google Scholar]
- 24.Locher K P, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch J P, Moras D. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 25.Buchanan S K, Smith B S, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 26.Song L, Hobaugh M R, Shustak C, Cheley S, Bayley H, Gouaux J E. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 27.Xia D, Yu C-A, Kim H, Xia J-Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Huang L, Shulmeister V M, Chi Y-I, Kim K K, Hung L-W, Crofts A R, Berry E A, Kim S-H. Nature (London) 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 29.Iwata S, Lee J W, Okada K, Lee J K, Iwata M, Rasmussen B, Link T A, Ramaswamy S, Jap B K. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 30.McDermott G, Prince S M, Freer A A, Hawthornthwaite-Lawless A M, Papiz M Z, Cogdell R J, Isaacs N W. Nature (London) 1995;374:517–521. doi: 10.1016/s0969-2126(96)00050-0. [DOI] [PubMed] [Google Scholar]
- 31.Koepke J, Hu X, Muenke C, Schulten K, Michel H. Structure (London) 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 32.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 33.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 34.Kimura Y, Vassylyev D G, Miyazawa M, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y. Nature (London) 1997;389:206–211. doi: 10.1038/38323. [DOI] [PubMed] [Google Scholar]
- 35.Pebay-Peyroula E, Rummel G, Rosenbusch J P, Landau E M. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 36.Luecke H, Richter H-T, Lanyi J K. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 37.Kreusch A, Schulz G E. J Mol Biol. 1994;243:891–905. doi: 10.1006/jmbi.1994.1690. [DOI] [PubMed] [Google Scholar]
- 38.Weiss M S, Schulz G E. J Mol Biol. 1992;227:493–509. doi: 10.1016/0022-2836(92)90903-w. [DOI] [PubMed] [Google Scholar]
- 39.Ridge J P. Ph.D. thesis. Sheffield, U.K.: University of Sheffield; 1998. [Google Scholar]
- 40.Jones M R, Fowler G J S, Gibson L C D, Grief G G, Olsen J D, Crielaard W, Hunter C N. Mol Microbiol. 1992;6:1173–1184. doi: 10.1111/j.1365-2958.1992.tb01556.x. [DOI] [PubMed] [Google Scholar]
- 41.Jones M R, Visschers R W, van Grondelle R, Hunter C N. Biochemistry. 1992;31:4458–4465. doi: 10.1021/bi00133a011. [DOI] [PubMed] [Google Scholar]
- 42.Jones M R, Heer-Dawson M, Mattioli T A, Hunter C N, Robert B. FEBS Lett. 1994;339:18–24. doi: 10.1016/0014-5793(94)80376-5. [DOI] [PubMed] [Google Scholar]
- 43.McAuley-Hecht K E, Fyfe P K, Ridge J P, Prince S M, Hunter C N, Isaacs N W, Cogdell R J, Jones M R. Biochemistry. 1998;37:4740–4750. doi: 10.1021/bi971717a. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Brünger A T, Kuriyan J, Karplus M. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Lamzin V S, Wilson K S. Acta Crystallogr D. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 48.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 49.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 50.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 51.McRee D E. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 52.Ridge J P, van Brederode M E, Goodwin M G, van Grondelle R, Jones M R. Photosynth Res. 1999;59:9–26. [Google Scholar]
- 53.Fyfe P K. Ph.D. thesis. Glasgow, United Kingdom: University of Glasgow; 1997. [Google Scholar]
- 54.Fyfe P K, McAuley-Hecht K E, Ridge J P, Prince S M, Fritzsch G, Isaacs N W, Cogdell R J, Jones M R. Photosynth Res. 1998;55:133–140. [Google Scholar]
- 55.Russell N J, Harwood J L. Biochem J. 1979;181:339–345. doi: 10.1042/bj1810339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 57.Nicholls A, Sharp K A, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 58.Allen J P, Feher G, Yeates T O, Rees D C, Deisenhofer J, Michel H, Huber R. Proc Natl Acad Sci USA. 1986;83:8589–8593. doi: 10.1073/pnas.83.22.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen J P, Feher G, Yeates T O, Komiya H, Rees D C. Proc Natl Acad Sci USA. 1987;84:5730–5734. doi: 10.1073/pnas.84.16.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Komiya H, Yeates T O, Rees D C, Allen J P, Feher G. Proc Natl Acad Sci USA. 1988;85:9012–9016. doi: 10.1073/pnas.85.23.9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang C-H, El-Kabbani O, Tiede D, Norris J, Schiffer M. Biochemistry. 1991;30:5352–5360. doi: 10.1021/bi00236a005. [DOI] [PubMed] [Google Scholar]
- 62.Chirino A J, Lous E J, Huber M, Allen J P, Schenck C C, Paddock M L, Feher G, Rees D C. Biochemistry. 1994;33:4584–4593. doi: 10.1021/bi00181a020. [DOI] [PubMed] [Google Scholar]
- 63.Lancaster C R D, Michel H. Structure (London) 1997;5:1339–1359. doi: 10.1016/s0969-2126(97)00285-2. [DOI] [PubMed] [Google Scholar]
- 64.Hostetler K Y. Biochim Biophys Acta. 1991;1086:139–140. doi: 10.1016/0005-2760(91)90165-e. [DOI] [PubMed] [Google Scholar]
- 65.Paradies G, Ruggiero F M, Dinoi P, Petrosillo G, Quagliariello E. Arch Biochem Biophys. 1993;397:91–95. doi: 10.1006/abbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- 66.Oliver D B. Mol Microbiol. 1993;7:159–165. doi: 10.1111/j.1365-2958.1993.tb01107.x. [DOI] [PubMed] [Google Scholar]
- 67.den Blaauwen T, Driessen A J M. Arch Microbiol. 1996;165:1–8. doi: 10.1007/s002030050289. [DOI] [PubMed] [Google Scholar]
- 68.Birrell G B, Sistrom W R, Griffith O H. Biochemistry. 1978;17:3768–3773. doi: 10.1021/bi00611a015. [DOI] [PubMed] [Google Scholar]
- 69.Welte W, Kreutz W. Biochim Biophys Acta. 1982;692:479–488. [Google Scholar]
- 70.Overfield R E, Wraight C A. Biochemistry. 1980;19:3328–3334. doi: 10.1021/bi00555a035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.