Abstract
Like most proteins, complex RNA molecules often are modular objects made up of distinct structural and functional domains. The component domains of a protein can associate in alternative combinations to form molecules with different functions. These observations raise the possibility that complex RNAs also can be assembled from preexisting structural and functional domains. To test this hypothesis, an in vitro evolution procedure was used to isolate a previously undescribed class of complex ligase ribozymes, starting from a pool of 1016 different RNA molecules that contained a constant region derived from a large structural domain that occurs within self-splicing group I ribozymes. Attached to this constant region were three hypervariable regions, totaling 85 nucleotides, that gave rise to the catalytic motif within the evolved catalysts. The ligase ribozymes catalyze formation of a 3′,5′-phosphodiester linkage between adjacent template-bound oligonucleotides, one bearing a 3′ hydroxyl and the other a 5′ triphosphate. Ligation occurs in the context of a Watson–Crick duplex, with a catalytic rate of 0.26 min−1 under optimal conditions. The constant region is essential for catalytic activity and appears to retain the tertiary structure of the group I ribozyme. This work demonstrates that complex RNA molecules, like their protein counterparts, can share common structural domains while exhibiting distinct catalytic functions.
Keywords: in vitro selection, in vitro evolution, RNA enzyme, RNA structure
Large naturally occurring RNAs are modular structures (1, 2). The group I intron and the RNA component of RNase P, for example, can be dissected into independent folding domains that associate through tertiary interactions to form an active structure (3, 4). The 23S rRNA of Salmonella species occurs in multiple fragments that assemble noncovalently to form a functional rRNA (5). This modular view of large structured RNAs suggests that they may have arisen in nature through the assembly of smaller pieces, and that RNAs with enhanced functional complexity could be obtained in the laboratory by similar means (2).
The concept of combining small molecular units to create larger molecules with new or enhanced functionality is well known in organic chemistry. It also has precedent in the biological evolution of proteins, in which the modular architecture of proteins derives from a common set of structural domains that can be assembled in various combinations (6). It has been proposed that new proteins may arise in nature by combining the exons of unrelated genes in a process known as “exon shuffling” (7). Recently, several protein enzymes have been designed on the basis of this concept (8–10). However, there are few data to suggest whether complex RNA molecules with novel function could be obtained in a similar manner.
Structurally complex and highly active ribozymes have been developed starting from a limited number of random-sequence RNAs through in vitro selection and evolution (11, 12). In some cases the complexity of the evolved ribozymes greatly exceeded what would have been expected to occur with reasonable probability among the RNAs that were examined. This finding suggests that many different complex functional RNAs are possible and that only a small fraction of these are represented in any limited sampling (13–15).
One of the most remarkable in vitro evolved ribozymes is the class I RNA ligase, which was obtained from a starting pool of 1015 RNAs, each containing 220 random nucleotides (13). This ribozyme catalyzes formation of a 3′,5′-phosphodiester linkage between two template-bound RNAs, one bearing a 3′ hydroxyl and the other a 5′ triphosphate, analogous to the reaction catalyzed by modern polymerase proteins. Could a molecule with this activity be obtained by starting from a preexisting RNA molecule rather than random-sequence RNAs? If so, the evolution of a complex functional RNA need not involve the chance acquisition of a complete RNA structure, but rather the elaboration of an existing structure to attain the new function.
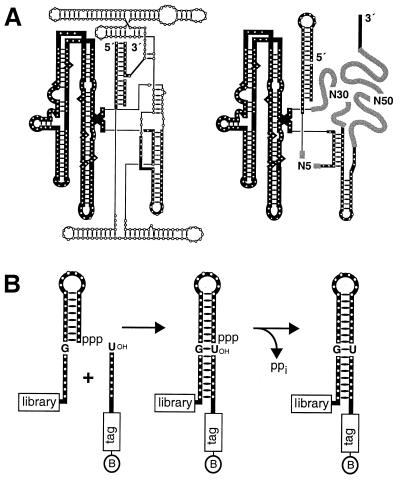
A combinatorial library of RNA molecules was constructed on the basis of a constant scaffold region derived from the self-splicing group I intron of Tetrahymena, with attached hypervariable regions that had the potential to form a novel catalytic domain (Fig. 1). The constant and random regions of the RNAs can be viewed as analogous to the constant and hypervariable regions of an antibody, respectively. The constant portion was based on the P4–P6 domain of the group I ribozyme, which is a stable, independent folding domain whose structure has been determined by x-ray crystallography (16). During the hierarchical folding of the Tetrahymena ribozyme, the P4–P6 domain folds rapidly and acts as a scaffold for the subsequent folding steps (17, 18). This process suggests that the P4–P6 domain within each member of the combinatorial library would promote folding and provide structural stabilization of catalytic molecules that might emerge from the in vitro evolution process.
Figure 1.
Secondary structure of the starting pool of RNAs. (A) Comparison of the Tetrahymena group I ribozyme (Left) and the RNA pool (Right), with conserved elements shown as thick lines. N5, N30, and N50 are regions of random-sequence nucleotides. (B) Schematic representation of the RNA ligation reaction that was the basis for in vitro selection.
The library of RNA molecules was subjected to 10 rounds of selective amplification based on the ability to catalyze the template-directed ligation of RNA. This procedure led to the isolation of a class of complex ribozymes that have the same activity as the class I RNA ligase but are based on the three-dimensional structure of the P4–P6 domain of the group I ribozyme. This work demonstrates that evolutionary pathways can exist between RNA molecules with similar structures but distinct functions.
Materials and Methods
Construction of the RNA Library.
A pool of 1.4 × 1016 different RNAs (13 copies each, 200 nmol total) was constructed, starting with three fragments of synthetic DNA. These fragments were amplified separately by PCR, digested with restriction enzymes, and ligated to form a DNA template that was transcribed to produce the RNA pool. Nonpalindromic restriction enzymes, BanI and StyI, were employed so that the subsequent ligation occurred in a directional manner. The ligated DNA was PCR amplified on a 400-ml scale, then transcribed on a 40-ml scale to maintain a sequence diversity of >1016.
In Vitro Evolution.
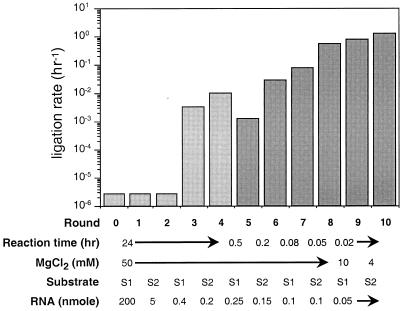
The scheme for each round of selective amplification was adapted from Bartel and Szostak (13). After in vitro transcription with T7 RNA polymerase, the DNA templates were digested with DNase I and the RNA was purified by denaturing polyacrylamide gel electrophoresis and precipitation with ethanol. The purified RNA was dissolved in water, denatured by incubating at 85°C for 2 min and 50°C for 2 min, then renatured in 4-fold concentrated reaction buffer at 50°C for 10 min. This protocol folded the P4–P6 domain within 80% of the molecules in the initial RNA pool, as judged by chemical probing using dimethyl sulfate. The folded RNAs, bearing a 5′ triphosphate, were incubated at 30°C in the presence of a biotinylated synthetic oligonucleotide substrate. During the first four rounds, the incubation time was 24 hr and the reaction mixture contained 1 μM RNA, 2 μM substrate, 50 mM MgCl2, 400 mM KCl, and 30 mM Hepes (pH 7.5 at 25°C). During subsequent rounds, the incubation time was progressively decreased to 1 min, the concentration of substrate was increased to 5 μM, the concentration of MgCl2 was progressively decreased to 4 mM, and the concentration of KCl was decreased to 50 mM (Fig. 2).
Figure 2.
Improvement of RNA ligase activity over successive rounds of in vitro selection. Self-ligation rates were measured for the population as a whole at 30°C, as described in Materials and Methods. Calculated ligation rates are based on the first 2% of the reaction. Mutagenic PCR was performed after rounds 4 and 6. After round 5, the 5′ stem–loop of the ribozyme was changed and the concentration of KCl was decreased from 400 to 50 mM.
Two different chimeric DNA-RNA substrates were used during in vitro evolution, each containing a different DNA portion that served as a primer recognition sequence during selective amplification. Substrate S1 had the sequence 5′-AA(biotinT)AACTTGACGTCAGCCTGGAAAAUCUUACUU-3′ and substrate S2 had the sequence 5′-AA(biotinT)AAGCTGAGCCTGCGATTGGAAAAUCUUACUU-3′ (RNA portion underlined). The ligation reaction was quenched by precipitation with ethanol, and the products were isolated by denaturing polyacrylamide gel electrophoresis to separate the reacted and unreacted RNAs from the unreacted substrate. After elution from the gel and precipitation with ethanol, biotinylated molecules were captured on streptavidin paramagnetic particles (PMP; Promega), allowed to hybridize with a DNA primer having the sequence 5′-AGGTCCGACTGCAGTTC-3′, and reverse transcribed with Superscript II (GIBCO/BRL) as described previously (19). The resulting cDNAs were eluted from the particles by degrading the RNA with 150 mM KOH, then neutralizing with 150 mM HCl. The cDNAs were selectively amplified by PCR using the reverse transcription primer and a selective primer corresponding to the DNA portion of the substrate, excluding the AA(biotinT)AA sequence at the 5′ end and the AAAA sequence at the 3′ end. A nested PCR was carried out with the reverse transcription primer and an oligonucleotide containing the sequence of the T7 RNA polymerase promoter element. Two different versions of the latter primer were used: 5′-CTGCAGAATTCTAATACGACTCACTATAGGAATATATCGTGCCTGTG-3′ during rounds 1–4, and 5′-TTCTAATACGACTCACTATAGGTAGACTCGCAGGAAGTCTACCGAGTAAGAGAAANNNNNAAGACGGCCAAATTGCGGGAAAG-3′ during rounds 6–10 (promoter sequence underlined; N corresponds to an equal mixture of A, G, T, and C). The products of the nested PCR were used as templates to transcribe the pool of RNAs used to begin the next round of selective amplification.
To maintain diversity in the population, random mutations were introduced after rounds 4 and 6 by error-prone PCR (20). To prevent the ribozymes from recognizing a particular substrate sequence, the stem–loop at their 5′ end was changed at the end of round 5, and the 5 nucleotides located immediately downstream from the stem–loop were maintained as a random sequence during all subsequent rounds. To prevent the ribozymes from specifically recognizing the DNA portion of the substrate, two different substrates (S1 and S2) were employed in an alternating manner during successive rounds of selective amplification. After round 10, individuals were isolated from the population by cloning the PCR DNA (TA cloning kit; Invitrogen) and the sequences of 38 clones were determined.
Mutagenesis of Individual Ribozymes.
Variants of cloned ribozymes were generated by PCR using the primer containing the T7 promoter sequence to introduce mutations near the 5′ end of the RNA. The presence of these mutations was confirmed by DNA sequence analysis. The mutant RNAs were transcribed from PCR-generated DNA templates and purified by denaturing polyacrylamide gel electrophoresis.
Measurement of Ligation Activity.
Self-ligation assays were carried out in the presence of 50 mM MgCl2, 200 mM KCl, 5 mM spermidine, and 50 mM Hepes (pH 7.5) at either 30° or 50°C. RNA molecules were first denatured by incubating in H2O at 85°C for 2 min and 50°C for 2 min, then renatured in 4-fold concentrated reaction buffer at 50°C for 10 min. Reactions were initiated by mixing the ribozyme (typically 0.1 μM final concentration) and various concentrations of substrate, and were stopped by adding gel loading buffer that contained 60 mM Na2EDTA and 4 M urea. Reaction products were separated by electrophoresis in a denaturing polyacrylamide gel and quantitated by using a PhosphorImager (Molecular Dynamics). Catalytic rates were determined by measuring the fraction ligated as a function of time in the presence of a saturating concentration of substrate. The hc16 ligase reacted to a maximum extent of 65% in a reproducible manner at either 30° or 50°C, indicating that 35% of the molecules were either mistranscribed or misfolded.
Characterization of Ligation Products.
The identity of the nucleotides at the ligation junction was confirmed by sequencing individual clones isolated after the tenth round of selective amplification. The regiospecificity of ligation catalyzed by the hc16 ribozyme was determined as described previously (13). The 5′ stem–loop was changed to 5′-GGAGUCUACCGAGUAAGAGAAA-3′ to allow binding of two oligonucleotide substrates, 5′-AUCUUACUU-3′ and 5′-pppGGUAGACUU-3′. The latter substrate was produced by in vitro transcription in the presence of [α-32P]GTP to label the 5′ phosphate of each guanylate residue. The ribozyme (2 μM) and substrates (20 μM each) were incubated under standard ligation conditions at 30°C for 17 hr. The ligation product was purified by denaturing polyacrylamide gel electrophoresis and digested with ribonuclease T2, which cleaves 3′,5′- but not 2′,5′-phosphodiester linkages. The reaction mixture for ribonuclease digestion also contained an unlabeled 18-nucleotide carrier molecule having the same sequence as the ligation product but lacking a 5′ triphosphate and with a 2′,5′ linkage at the ligation junction. The carrier molecule was prepared synthetically from the corresponding nucleoside phosphoramidites (ChemGenes; Ashland, MA) and purified by denaturing polyacrylamide gel electrophoresis. The products of nuclease digestion were separated by two-dimensional thin-layer chromatography, as described previously (21, 22).
Results
Structure-Based Combinatorial Library.
To obtain novel ribozymes that take advantage of a preexisting RNA structural domain, a library of 1.4 × 1016 different RNAs was constructed, each molecule containing a constant scaffold region and three hypervariable regions. The hypervariable regions contained a total of 85 random nucleotides (N5 + N30 + N50) that had the potential to interact with the constant region to form a catalytic motif. The constant region of 255 nucleotides was derived from the Tetrahymena group I ribozyme (Fig. 1A) and included the P3 and P8 helices and the P4–P6 domain. Despite the presence of the three hypervariable regions, roughly 80% of the molecules in the library contained a properly folded P4–P6 domain, as judged by chemical probing with dimethyl sulfate. All molecules in the library contained a stem–loop structure at their 5′ end that was designed to bind the oligonucleotide substrate (Fig. 1B).
The structure-based library had two main advantages for the selection of ribozymes that can perform a reaction within the context of a Watson–Crick duplex. First, the constant scaffold region is preadapted for docking RNA helices. The P4–P6 domain contains internal loops that might participate in RNA helix packing interactions, as they do in the group I ribozyme (23). Second, the constant region provides a soluble support for the attached random sequences, which might otherwise form insoluble aggregates.
The Tetrahymena group I ribozyme was tested for its ability to perform an RNA ligation reaction in the context of the 5′ stem–loop structure shown in Fig. 1B. Surprisingly, the ribozyme was able to catalyze the template-directed joining of the substrate 5′-CCCUCU-3′ to its own 5′ end, albeit at a rate of only 3 × 10−5 min−1. This is about 300-fold greater than the uncatalyzed rate of reaction (24). Ligation was detected by monitoring the release of labeled inorganic pyrophosphate from ribozymes that had a guanosine 5′-[γ-32P]triphosphate at the first nucleotide position. The ribozyme also catalyzed a phosphoester transfer reaction involving joining of the substrate to the second nucleotide with concomitant release of GTP, at a rate of about 0.1 min−1. When the P2–P2.1 domain was deleted from the ribozyme, as was the case for the starting pool of RNAs, the ligation rate fell to 8 × 10−7 min−1, which is only about 8-fold greater than the uncatalyzed rate of reaction.
In Vitro Evolution.
Starting with a pool of an average of 13 copies each of 1016 different RNAs, the selection protocol was designed to enrich those RNAs that could ligate an oligonucleotide substrate to their own 5′ end (Fig. 1B). Each RNA had an 8-base-pair stem–loop structure at its 5′ end, located adjacent to 8 nucleotides that were complementary to nucleotides at the 3′ end of the substrate. This arrangement allowed the 5′ triphosphate of the pool RNA to be held in close proximity to the 3′ hydroxyl of the substrate within the context of a Watson–Crick duplex. The substrate bound at the 5′ end of the pool RNA formed a nicked helical duplex of 16 base pairs that can be regarded as the true substrate for ligation. This structure included a wobble pair at the ligation site, analogous to the wobble pair at the 5′ splice site of group I introns. The substrate was biotinylated so that members of the RNA pool that carried out ligation became biotinylated and thus could be captured on streptavidin beads and selectively reverse transcribed. The substrate oligonucleotide also had a tag sequence that allowed selective amplification of only those cDNAs that were generated from reacted molecules. Amplification was carried out by PCR, followed by a nested PCR to remove the tag sequence. The final PCR products were transcribed in vitro to produce a progeny population of RNAs that could be used to begin the next round of selective amplification.
The in vitro selection process was repeated for a total of 10 rounds. After the third round, the population exhibited a modest level of catalytic activity, with 5% of the molecules becoming ligated to the substrate over 24 hr (Fig. 2). After the fourth and sixth rounds, the sequence diversity of the population was increased by performing error-prone PCR (20). This was done to generate variants of the selected individuals that might have even higher catalytic activity. Beginning after the fourth round, the selection pressure was made more stringent by progressively decreasing the reaction time and lowering the concentration of Mg2+ in the reaction mixture (Fig. 2), thus favoring molecules that reacted faster and were less dependent on Mg2+ for structural stabilization and catalysis. Finally, to favor template-dependent ligases that were generalizable with respect to template sequence, the sequence of the stem–loop structure at the 5′ end of the ribozymes was changed at the end of the fifth round of selective amplification. This led to a 10-fold decrease in the activity of the population after the fifth round (Fig. 2), presumably because most of the sequence-dependent catalysts had been eliminated. By the end of the tenth round, the population of RNAs exhibited a ligation rate of about 1 hr−1, which is about 106-fold faster than the uncatalyzed rate of template-directed ligation of two oligonucleotides (13). This rate enhancement is likely to be an underestimate because the uncatalyzed ligation of adjacent template-bound oligonucleotides is 50-fold slower when the base pair on the 5′ side of the ligation junction is a wobble pair rather than a Watson–Crick pair (measured in the presence of 100 mM MgCl2 at pH 9.0 and 25°C; ref. 25).
A Previously Undescribed Class of RNA Ligases.
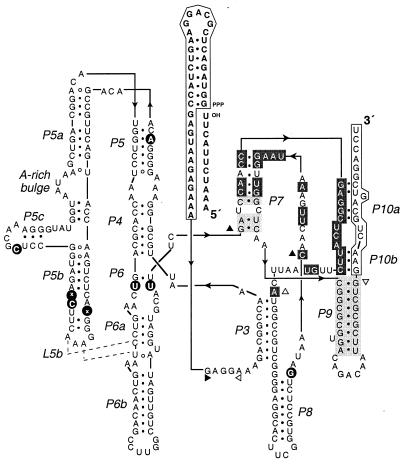
Examination of the nucleotide sequence of 38 cloned individuals obtained after the tenth round of selective amplification revealed two distinct families, one represented by 32 clones and the other by 6 clones. The two families shared a consensus sequence of 29 nucleotides within the N30 and N50 hypervariable regions, including 13 highly conserved residues within the N30 region. A putative secondary structural model of one of the most efficient ligases, clone hc16, is shown in Fig. 3. Comparative sequence analysis revealed that the ligase ribozymes contained a conserved stem of 9 base pairs within the N50 region (the P9 pairing) and a conserved stem of up to 8 base pairs between the N30 and N50 regions (the P7 pairing). The 13 nucleotides at the 3′ end of the ribozyme, which bind the primer used to initiate reverse transcription, could not be deleted without a complete loss of catalytic activity. Further structural analysis is required to determine how these 3′-terminal nucleotides interact with the remainder of the ribozyme (the putative Pl0a and Pl0b pairings) and whether other conserved nucleotides within the N30 and N50 regions are involved in secondary and tertiary interactions.
Figure 3.
Putative secondary structure of the hc16 ligase ribozyme. Nucleotides shaded in dark gray are invariant among the clones that were examined; nucleotides shaded in light gray varied but maintained Watson–Crick complementarity. Solid and open triangles indicate the first and last nucleotide, respectively, of each of the three hypervariable regions. Positions that were mutated within the constant region are indicated by black circles. Of these, only U67 → A occurred in all of the clones. Other mutations that occurred in the majority of clones were U104 → C, G105 → deleted, U115 → C, C163 → U, and G205 → U.
The self-ligation reaction catalyzed by the hc16 ribozyme proceeded with a rate of 0.054 ± 0.002 min−1 and 0.26 ± 0.012 min−1 at 30° and 50°C, respectively, measured in the presence of 50 mM MgCl2. There was no detectable activity in the absence of Mg2+. The identity of the ligated product was confirmed by sequencing across the ligation junction and by measuring the release of labeled inorganic pyrophosphate from ribozymes that had a guanosine 5′-[γ-32P]triphosphate residue at the first nucleotide position. The released inorganic pyrophosphate was visualized by two-dimensional thin-layer chromatography (21, 22). No labeled GTP was detected, indicating that the hc16 ribozyme does not have residual nucleotidyl transfer activity, as occurs with the group I ribozyme.
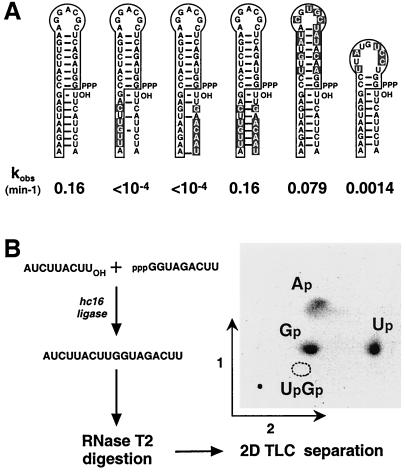
The N5 region of the class hc ligases, which separates the 5′ stem–loop from the constant region, does not appear to contribute significantly to catalysis. Changing these nucleotides from 5′-GAGGA-3′ to random sequence reduced the catalytic rate by only about 2-fold, to 0.16 min−1 at 50°C. Additional site-directed mutagenesis experiments were carried out to determine whether ligation takes place in a sequence-general manner within the context of a Watson–Crick duplex. Changing the sequence of either the RNA substrate or the substrate-binding portion of the ribozyme abolished catalytic activity, whereas combining these changes in a way that reestablished complementarity restored full activity (Fig. 4A). Replacing the 8-base-pair stem–loop at the 5′ end of the ribozyme by a hairpin of very different sequence but similar structure reduced ligase activity by only about 2-fold. However, when this stem–loop was shortened to 2 base pairs, activity decreased by 120-fold (Fig. 4A). The wobble pair on the 5′ side of the ligation junction contributed to optimal activity; when it was replaced by a G⋅C pair, activity decreased by about 7-fold. Taken together, these results strongly suggest that the evolved ligase ribozymes operate in the context of a Watson–Crick helical duplex. Accordingly, they have been designated “class hc” ligases (h, helical; c, context).
Figure 4.
Self-ligation occurs in a helical context. (A) Effect of changing the substrate-binding portion of the ribozyme and/or the sequence of the RNA substrate. Catalytic rates were measured in the presence of 50 mM MgCl2 at 50°C. (B) The regiospecificity of ligation was determined in a reaction with two oligonucleotide substrates, one containing 32P-labeled guanylate residues. The ligation products were purified, digested with RNase T2, mixed with unlabeled G(2′→5′)Up (dotted circle), and analyzed by two-dimensional thin-layer chromatography.
Ribozyme hc16 can be engineered to operate as a true enzyme that joins two separate RNA substrates, albeit very inefficiently. The ligation product that results from this reaction was purified and subjected to endoribonuclease digestion followed by thin-layer chromatography, demonstrating the presence of a 3′,5′-phosphodiester linkage at the ligation junction (Fig. 4B). The class hc ligases obtained in the present study and the previously described class I ligases (13, 14) are the only reported ribozymes that catalyze this 3′,5′-regiospecific reaction.
Folding of the P4–P6 Domain.
The ability of the class hc ligases to catalyze oligonucleotide ligation within the context of a helical duplex and in a largely sequence-independent manner is reminiscent of group I ribozymes, which catalyze nucleotidyl phosphoester transfer reactions in a similar context. These two ribozymes share the P4–P6 structural domain, although this domain need not be as important for the activity of the class hc ligase as it is for the group I ribozyme. Replacing the P4–P6 domain (residues U54–U206) by a two-nucleotide linker abolished the activity of the hc16 ligase (data not shown). However, portions of this domain could be deleted while retaining catalytic activity. For example, removing the P5abc subdomain (residues C74–G142) reduced ligase activity by about 30-fold, similar to what has been observed for the group I ribozyme (26). There is a U → A mutation within the P5 stem of all sequenced clones of the class hc ligase (Fig. 3). When this mutation was reverted to a U, ligase activity was reduced by 5-fold, demonstrating that detailed structural features of the P4–P6 domain influence the molecule's catalytic activity.
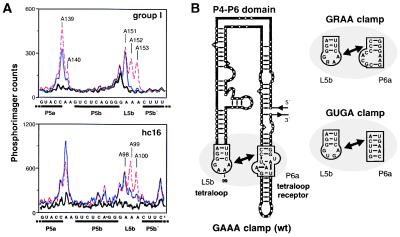
To assess the structural integrity of the P4–P6 domain, chemical probing experiments were carried out by using dimethyl sulfate to modify unpaired A and C residues within either the hcl6 ligase or Tetrahymena group I ribozyme, in either the presence or the absence of Mg2+. The two ribozymes exhibited very similar modification patterns (Fig. 5A). In both cases, the A residues within the L5b tetraloop and the A-rich bulge of P5a were protected from methylation in the presence, but not the absence, of Mg2+. These residues are known to be involved in long-range Watson–Crick interactions that are essential for proper folding of the P4–P6 domain within the group I ribozyme.
Figure 5.
Folding of the P4–P6 domain. (A) Dimethyl sulfate (DMS) modification patterns for the L5b tetraloop region within either the Tetrahymena group I or hc16 ligase ribozyme, based on primer extension analysis using reverse transcriptase (27). Thick line, no DMS and 50 mM MgCl2; thin line, DMS and 50 mM MgCl2; dashed line, DMS and 1 mM Na2EDTA. Nucleotide numbering follows the sequence of each ribozyme. After renaturation, the RNA (0.3 μM) was preincubated at 30°C for 5 min in 40 μl containing 200 mM KCl, 5 mM spermidine, 50 mM cacodylate (pH 7.5), and either MgCl2 or Na2EDTA, followed by addition of 1 μl of DMS in ethanol to a final concentration of 17 mM and incubation at 30°C for 5 min. (B) Tertiary interaction between the GAAA tetraloop of L5b and the corresponding tetraloop receptor of P6a (Left). Analogous interactions occur with the GRAA and GUGA clamps (Right).
The overall shape of the P4–P6 domain is a bent helix that is stabilized by tertiary contacts between the GAAA tetraloop of L5b and an 11-nucleotide “tetraloop receptor” within P6a (Fig. 5B; refs. 28–30). If the properly folded P4–P6 domain is required for activity of the hc16 ligase, then disruption of the clamp between L5b and P6a would be expected to impair catalytic function (31, 32). Replacing the GAAA tetraloop by either GGAA or GUGA reduced activity by 25- or 50-fold, respectively. However, a compensatory change of the tetraloop receptor improved activity in both cases (data not shown). This observation suggests that the integrity of the L5b–P6a clamp and the consequent folding of the P4–P6 domain are essential for optimal activity of the class hc ligase.
Modification interference experiments were carried out, using dimethyl sulfate to modify the hc16 ligase in a statistical manner and then comparing the modification pattern of reacted and unreacted ribozymes. Details of these experiments will be published elsewhere (L.J., unpublished results). However, it is worth noting that several A residues within the P4–P6 domain were shown to be intolerant of modification, most significantly within L5b and the region joining P4 and P5.
Discussion
A sophisticated RNA-based catalytic function, 3′,5′-regiospecific oligonucleotide ligation driven by pyrophosphorolysis, which was isolated previously from random-sequence RNAs (13), can also be obtained from a structural scaffold derived from a group I ribozyme. The two resulting ligases have very different nucleotide composition, although their catalytic sites may have similar organization. It is not surprising to discover that at least two distinct structural solutions exist for the catalysis of a complex reaction. Several small catalytic RNA motifs have been described that catalyze the relatively facile RNA-cleavage reaction involving attack by a 2′ hydroxyl on an adjacent phosphate. Furthermore, the selection pressures that were imposed during development of the class I and class hc ligases were somewhat different and could have led to different catalytic motifs even if the two procedures had been applied to the same starting pool of RNAs. The class I ligase specifically recognizes the stem–loop structure at its own 5′ end (14, 19), and thus would have been excluded during the evolution of the class hc ligase. It was a requirement for selection of the latter ribozyme that ligation occur within an extended Watson–Crick duplex of variable sequence, similar to the context for nucleotidyl transfer catalyzed by the group I ribozyme.
The class hc ligase contains structural elements that do not occur in the group I ribozyme. However, the overall architecture of the two ribozymes is similar (33). Approximately 55% of the nucleotides in the hc16 ligase are identical to their counterparts in the Tetrahymena group I ribozyme. This includes the P4–P6 domain, which is essential for the activity of both ribozymes. The Tetrahymena ribozyme was found to have a very slight amount of ligase activity, although with the P2–P2.1 domain deleted it was only about 8-fold greater than the uncatalyzed rate of reaction. The hc16 ligase, on the other hand, exhibited a catalytic rate enhancement of more than 106-fold in the ligation reaction and had no detectable nucleotidyl transferase activity. Thus the two ribozymes have distinct functions despite having common structural features.
The complex function performed by the class hc ligase requires a complex structure. It is likely that the constant scaffold region facilitated selection of this ribozyme, which might otherwise have been absent in a population of RNAs that contained only 85 random nucleotides. It has been estimated that the probability of finding a catalytic motif of 92 nucleotides within a pool of RNAs containing 220 random nucleotides is about 360,000 times greater than the probability of finding that same motif within a pool containing only 92 random nucleotides (14, 15). The ease with which structurally complex ligase ribozymes were isolated from a pool of RNAs containing only 85 random nucleotides suggests that the degree of structural preorganization provided by the P4–P6 domain conferred a similar advantage. An in vitro evolution experiment that was carried out in parallel beginning with a pool of RNAs containing 85 random nucleotides failed to produce any ligase ribozymes that could operate within a helical context (data not shown). However, this negative result should not be taken as proof that such ligases could not exist. Instead, the limited size of the initial pool or technical aspects of the selection procedure may have prevented them from being obtained.
The observation that a ribozyme with novel catalytic function can arise from the structural foundation of a different ribozyme supports the concept that preexisting RNA structural domains in nature may have facilitated the evolution of more complex functional RNAs. This principle has been used to obtain ribozymes with polynucleotide kinase or ligase activity, starting from randomized pools of RNA molecules that contained a preexisting ATP-binding domain (34, 35), although in neither case was it clear whether the evolved catalysts continued to rely on the original ATP-binding domain. The results of the present study suggest that RNAs, like their protein counterparts, can be assembled from smaller domains to attain a higher level of structural and functional organization.
It has been proposed that in an “RNA world,” prior to the emergence of DNA and proteins, RNA replicase function was carried out by ribozymes analogous, and perhaps evolutionarily related, to the group I ribozyme (36). There is no direct evidence to support this conjecture. However, the results of the present study demonstrate that nucleotidyl phosphoester transferase activity, as carried out by a group I ribozyme, may be linked via evolutionary pathways to an activity that is more closely related to the template-directed polymerization of RNA. Thus it is not difficult to imagine that ribozymes with RNA splicing and RNA polymerization activity could have arisen from a common ancestor.
Acknowledgments
This work was initiated by L.J. and M.C.W. in the laboratory of G.F.J. when both were recipients of a postdoctoral fellowship from the National Aeronautics and Space Administration Specialized Center for Research and Training in Exobiology. Since November 1995, the work has been pursued by L.J. in Strasbourg with support from the Centre National de la Recherche Scientifique and the Centre National d'Etudes Spatiales (Grant 96/CNES/0247). L.J. gratefully acknowledges Eric Westhof and Bernard Ehresmann for their support and encouragement. This paper is dedicated to the memory of Dr. Robert D. Tschirgi.
References
- 1.Westhof E, Masquida B, Jaeger L. Folding Design. 1996;1:78–88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 2.Jaeger L. Curr Opin Struct Biol. 1997;7:324–335. doi: 10.1016/s0959-440x(97)80047-4. [DOI] [PubMed] [Google Scholar]
- 3.Murphy F L, Cech T R. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 4.Loria A, Pan T. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 5.Burgin A B, Parodos K, Lane D J, Pace N R. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 6.Bork P. Curr Opin Struct Biol. 1992;2:413–421. [Google Scholar]
- 7.Gilbert W. Nature (London) 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 8.Nixon A E, Warren M S, Benkovic S J. Proc Natl Acad Sci USA. 1997;94:1069–1073. doi: 10.1073/pnas.94.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J-H, Dawes G, Stemmer W P C. Proc Natl Acad Sci USA. 1997;94:4504–4509. doi: 10.1073/pnas.94.9.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skandalis A, Encell L P, Loeb L A. Chem Biol. 1997;4:889–898. doi: 10.1016/s1074-5521(97)90297-0. [DOI] [PubMed] [Google Scholar]
- 11.Lorsch J R, Szostak J W. Acc Chem Res. 1996;29:103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 12.Tsang J, Joyce G F. Methods Enzymol. 1996;267:410–426. doi: 10.1016/s0076-6879(96)67025-6. [DOI] [PubMed] [Google Scholar]
- 13.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 14.Ekland E H, Szostak J W, Bartel D P. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 15.Sabeti P C, Unrau P J, Bartel D P. Chem Biol. 1997;4:767–774. doi: 10.1016/s1074-5521(97)90315-x. [DOI] [PubMed] [Google Scholar]
- 16.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1684. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 17.Treiber D K, Rook M S, Zarrinkar P P, Williamson J R. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 18.Sclavi B, Sullivan M, Chance M R, Brenowitz M, Woodson S A. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 19.Ekland E H, Bartel D P. Nucleic Acids Res. 1995;23:3231–3238. doi: 10.1093/nar/23.16.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 21.Keith G. In: Chromatography and Modifications of Nucleosides, Chromatography Library Series. Gehrke C W, Kuo K C, editors. 45A. Amsterdam: Elsevier; 1990. pp. 103–141. [Google Scholar]
- 22.Keith G. Biochimie. 1995;77:142–144. doi: 10.1016/0300-9084(96)88118-1. [DOI] [PubMed] [Google Scholar]
- 23.Strobel S A, Ortoleva-Donnelly L, Ryder S P, Cate J H, Moncoeur E. Nat Struct Biol. 1998;5:60–66. doi: 10.1038/nsb0198-60. [DOI] [PubMed] [Google Scholar]
- 24.Rohatgi R, Bartel D P, Szostak J W. J Am Chem Soc. 1996;118:3332–3339. doi: 10.1021/ja953712b. [DOI] [PubMed] [Google Scholar]
- 25.Rohatgi R, Bartel D P, Szostak J W. J Am Chem Soc. 1996;118:3340–3344. doi: 10.1021/ja9537134. [DOI] [PubMed] [Google Scholar]
- 26.van der Horst G, Christian A, Inoue T. Proc Natl Acad Sci USA. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaeger L, Westhof E, Michel F. J Mol Biol. 1993;234:331–346. doi: 10.1006/jmbi.1993.1590. [DOI] [PubMed] [Google Scholar]
- 28.Murphy F L, Cech T R. J Mol Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 29.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Szewczak A A, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 30.Costa M, Michel F. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaeger L, Michel F, Westhof E. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 32.Costa M, Michel F. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 34.Lorsch J R, Szostak J W. Nature (London) 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- 35.Hager A J, Szostak J W. Chem Biol. 1997;4:607–617. doi: 10.1016/s1074-5521(97)90246-5. [DOI] [PubMed] [Google Scholar]
- 36.Cech T R. Proc Natl Acad Sci USA. 1986;83:4360–4363. doi: 10.1073/pnas.83.12.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]