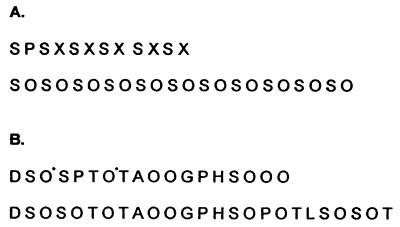Abstract
Design of hydroxyproline (Hyp)-rich glycoproteins (HRGPs) offers an approach for the structural and functional analysis of these wall components, which are broadly implicated in plant growth and development. HRGPs consist of multiple small repetitive “glycomodules” extensively O-glycosylated through the Hyp residues. The patterns of Hyp-O-glycosylation are putatively coded by the primary sequence as described by the Hyp contiguity hypothesis, which predicts contiguous Hyp residues to be attachment sites of small arabinooligosaccharides (1–5 Ara residues/Hyp); while clustered, noncontiguous Hyp residues are sites of arabinogalactan polysaccharide attachment. As a test, we designed two simple HRGPs as fusion proteins with green fluorescent protein. The first was a repetitive Ser-Hyp motif that encoded only clustered noncontiguous Hyp residues, predicted polysaccharide addition sites. The resulting glycoprotein had arabinogalactan polysaccharide O-linked to all Hyp residues. The second construct, based on the consensus sequence of a gum arabic HRGP, contained both arabinogalactan and arabinooligosaccharide addition sites and, as predicted, gave a product that contained both saccharide types. These results identify an O-glycosylation code of plants.
From green algae to flowering plants, hydroxyproline (Hyp)-O-glycosylation uniquely characterizes an ancient and diverse group of structural glycoproteins associated with the cell wall (1). These Hyp-rich glycoproteins (HRGPs) are broadly implicated in all aspects of growth and development, including fertilization (2, 3), differentiation and tissue organization (4), control of cell expansion growth (5), and responses to stress and pathogenesis (6). However, our level of understanding their role is largely superficial and conjectural. The problem remains to be explained as to why plants need so many HRGPs, what physiological roles they fulfill, and precisely how they fulfill them at the molecular level (7).
HRGPs are generally extended, repetitive glycoproteins (8). The repeats are usually small, ≈5- to 16-residue motifs that are frequently highly glycosylated. Most HRGPs consist of more than one type of repetitive motif. Thus, peptide sequence periodicity and glycosylation distinguish the three major HRGP families: arabinogalactan proteins (AGPs), extensins, and proline-rich proteins (PRPs). AGPs [>90% (wt/wt) sugar] have repetitive variants of (Xaa-Hyp)n motifs (7) with O-linked arabinogalactan polysaccharides involving an O-galactosyl-Hyp glycosidic bond (9, 10). Extensins [≈50% (wt/wt) sugar] have a diagnostic Ser-Hyp4 repeat that contains short oligosaccharides of arabinose (Hyp arabinosides) involving an O-l-arabinosyl-Hyp linkage (1, 11). Finally, the lightly arabinosylated PRPs [2–27% (wt/wt) sugar; ref. 12] are the most highly periodic, consisting largely of pentapeptide repeats, typically variants of Pro-Hyp-Val-Tyr-Lys (13, 14).
Because repetitive HRGP glycopeptide motifs are evolutionarily conserved, we consider them small functional units and refer to them as “glycomodules.” HRGP glycosylation is significant, because it defines the interactive molecular surface and hence should determine HRGP function as it does for other extensively glycosylated glycoproteins (15). If the molecular properties of these glycomodules depend on their glycosyl substituents precisely arranged along an extended Hyp-rich polypeptide template, then an O-glycosylation code is likely. The Hyp contiguity hypothesis (8, 16, 17) correlates Hyp arabinosylation with blocks of contiguous Hyp residues and predicts that Hyp galactosylation occurs on clustered noncontiguous Hyp residues (8, 16). For example, contiguous Hyp4 blocks of the Ser-Hyp4 glycomodules are extensively glycosylated with short chains comprised entirely of l-arabinose (1, 12). Similarly, dipeptidyl Hyp is the major arabinosylation site of a PRP in which noncontiguous Hyp was only rarely monoarabinosylated (17). On the other hand, we predict that Hyp galactosylation of clustered noncontiguous Hyp residues, such as the Xaa-Hyp-Xaa-Hyp repeats of AGPs, results in the addition of a galactan core with side chains of arabinose and other sugars to form characteristic Hyp-arabinogalactan polysaccharides. Hitherto, these sites of arabinogalactan polysaccharide attachment were poorly defined. AGPs resist proteases, and degradation by partial alkaline hydrolysis yields arabinogalactan glycopeptides that are difficult to purify. Therefore, as an approach to HRGP glycosylation site mapping and as a test of the code that directs Hyp-arabinogalactan polysaccharide addition, we designed a set of two synthetic genes that encode putative AGP glycomodules. Herein, we report that, when expressed and targeted for secretion, these modules behaved as simple endogenous substrates for HRGP glycosyl transferases. The construct expressing noncontiguous Hyp showed exclusive polysaccharide addition, whereas another construct containing noncontiguous Hyp and additional contiguous Hyp showed both polysaccharide and arabinooligosaccharide addition consistent with the predictions of the Hyp contiguity hypothesis.
Materials and Methods
Synthetic Gene and Plasmid Construction.
The signal sequence (Fig. 1) was modeled after an extensin signal sequence from Nicotiana plumbaginifolia (18); mutually priming oligonucleotides were extended by T7 DNA polymerase, and the duplex was placed in pUC18 as a BamHI–SstI fragment. Construction of a given synthetic gene involved the polymerization of three sets of partially overlapping, complementary oligonucleotide pairs as described (ref. 19; Fig. 1). The following subclonings were required to create DNA fragments/restriction sites, which allowed facile transfer of the signal sequence-synthetic gene-enhanced green fluorescent protein (EGFP) unit to the plant transformation vector pBI121 (ref. 20; CLONTECH); we placed the synthetic genes in pBluescript II SK(+) (Stratagene) as BamHI–EcoRI fragments and then subcloned the genes into pEGFP (CLONTECH) as BamHI–AgeI fragments preceding the EGFP gene (21, 22). The synthetic gene–EGFP fragments were then subcloned into pBluescript II KS(+) (Stratagene) as XmaI–NotI fragments, removed as XmaI–SstI fragments, and subcloned into pUC18 behind the signal sequence. DNA sequences were confirmed by sequence analysis before insertion into pBI121 as BamHI–SstI fragments, replacing the β-glucuronidase reporter gene. All constructs were under the control of the 35S cauliflower mosaic virus promoter. The oligonucleotides were synthesized by Life Technologies (Grand Island, NY). An Ala for Pro/Hyp substitution at residue 8 of the GAGP internal repeat module (Ser-Pro-Ser-Pro-Thr-Pro-Thr-Pro-Pro-Pro-Gly-Pro-His-Ser-Pro-Pro-Pro-Thr-Leu) was inadvertently introduced during synthesis by a G-for-C base substitution in the sense strand.
Figure 1.
Oligonucleotide sets used to build the synthetic genes. Internal repeat oligonucleotide sets encoding Ser-Pro repeats or the gum arabic glycoprotein (GAGP) sequence were polymerized head-to-tail in the presence of the 5′ linker set. After ligation, the 3′ linker was added, and the genes were then restricted with BamHI and EcoRI and inserted into pBluescript II SK(+). The signal sequence was built by primer extension of the overlapping oligonucleotides featured here. The overlap is underlined.
Tobacco Cell Transformation and Selection of Cell Lines.
Suspension cultured tobacco cells (Nicotiana tabacum, BY2) were transformed (23) with Agrobacterium tumifaciens strain LBA4404 containing the pBI121-derived plant transformation vector. Transformed cell lines were selected on solid Murashige–Skoog medium (Sigma, no. 5524) containing 100 μg/ml kanamycin. Timentin was initially included at 400 μg/ml to kill Agrobacterium. Cells were later grown in 1-liter flasks containing 500 ml of Shenck–Hildebrand medium (Sigma, no. 6765) and 100 μg/ml kanamycin, rotated at 100 rpm on an Innova 2000 New Brunswick Scientific gyrotary shaker.
Isolation of (Ser-Hyp)32-EGFP, (GAGP)3-EGFP, Native GAGP, and Endogenous Tobacco AGPs.
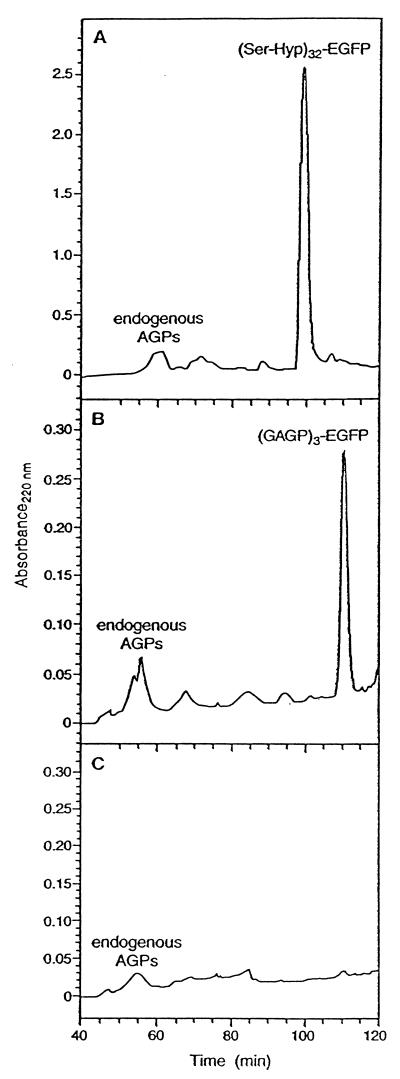
Culture medium was harvested 7–21 days after subculture, concentrated 10-fold via rotoevaporation, then injected onto a Superose-12 gel filtration column (Amersham Pharmacia) equilibrated in 200 mM sodium phosphate buffer (pH 7), and eluted at a flow rate of 1 ml/min. EGFP fluorescence was monitored by a Hewlett–Packard 1100 Series flow-through fluorometer (488-nm excitation; 520-nm emission). We calibrated the Superose-12 column with molecular mass standards (BSA, insulin, catalase, and sodium azide). Fluorescent Superose-12 fractions were injected directly onto a Hamilton PRP-1 reverse phase column, and gradient eluted at a flow rate of 0.5 ml/min. Start buffer consisted of 0.1% trifluoroacetic acid (aqueous), and elution buffer was 0.1% trifluoroacetic acid/80% (vol/vol) acetonitrile (aqueous). The sample was repeatedly injected (0.5 ml/min) onto the column over 35 min and then eluted with a gradient of elution buffer (0–70%/135 min). Native GAGP was isolated from gum arabic nodules as described by Qi et al. (10). Endogenous tobacco AGPs were isolated as described (see Fig. 4).
Figure 4.
PRP-1 reverse-phase fractionation of the Superose-12 peaks containing (Ser-Hyp)32-EGFP (A), (GAGP)3-EGFP (B), and (glyco)proteins in the medium of nontransformed tobacco cells (C). Endogenous tobacco AGPs eluted between 47 and 63 min; extensins eluted at ≈67 min. (C) Control medium collected from nontransformed tobacco cells was first fractionated on Superose-12, and the fractions eluting between 47 and 63 min were collected for further separation on PRP-1 to determine whether any endogenous AGPs/HRGPs cochromatographed with (Ser-Hyp)32-EGFP or with (GAGP)3-EGFP, which they did not.
Coprecipitation with Yariv Reagent.
We coprecipitated (Ser-Hyp)32-EGFP, (GAGP)3-EGFP, tobacco AGPs, and native GAGP with the Yariv reagent as described (24).
Monosaccharide and Glycosyl Linkage Analysis.
Monosaccharide compositions and linkage analyses were determined at the Complex Carbohydrate Research Center, University of Georgia, as described (25, 26).
Hyp-Glycoside Profiles.
Hyp-glycoside profiles were determined as described by Lamport and Miller (11). We hydrolyzed 5.8–12.2 mg of (Ser-Hyp)32-EGFP or (GAGP)3-EGFP in 0.44 M NaOH and neutralized the hydrolysate with 0.3 M HCl before injection onto a C2 cation exchange column.
Anhydrous Hydrogen Fluoride Deglycosylation.
We deglycosylated 4.5 mg each of (Ser-Hyp)32-EGFP and (GAGP)3-EGFP in anhydrous hydrogen fluoride containing 10% (vol/vol) dry methanol for 1 h at 0°C, and then quenched the reactions in double-distilled H2O (27).
Hyp Assay of Secreted EGFP.
Secreted EGFP, the product of the Sig-EGFP gene, was isolated by the Superose-12 fractionation. We removed EGFP from the fusion glycoproteins by overnight pronase digestion (1% ammonium bicarbonate/5 mM CaCl2; 27°C; 1:100 enzyme:substrate ratio), followed by isolation of EGFP by gel permeation chromatography as described above. After dialysis and freeze drying, we assayed Hyp on 0.5 mg of EGFP as described (12).
Protein and DNA Sequence Analysis.
Protein sequence analysis was performed at the Michigan State University Macromolecular Facility on a 477-A Applied Biosystems gas phase sequencer. DNA sequencing was performed at the Guelph Molecular Supercentre, University of Guelph, Ontario, Canada.
Results
Synthetic Gene and Plasmid Construction.
We built three plasmids, each encoding a tobacco signal sequence and EGFP. Two of the plasmids also contained a synthetic gene encoding either six (Ser-Pro) internal repeat units or three (GAGP) internal repeat units (Fig. 1) sandwiched between the signal sequence and EGFP.
Tobacco Cell Transformation and Selection of Cell Lines.
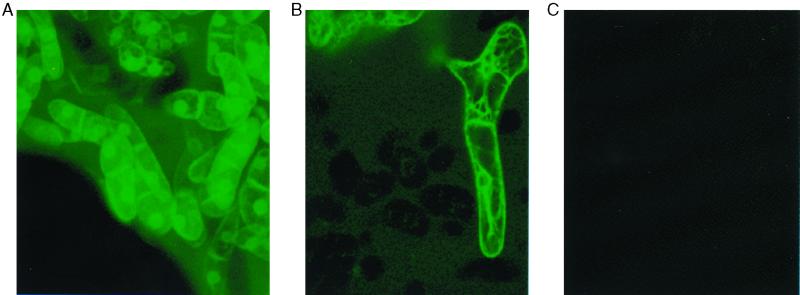
After transformation of tobacco cells with Agrobacterium harboring the plant transformation plasmid pBI121 outfitted with Sig-(GAGP)3-EGFP, Sig-(Ser-Pro)32-EGFP, or Sig-EGFP, selection on solid medium and subsequent growth in liquid culture yielded cells bathed in a green fluorescent medium (Fig. 2).
Figure 2.
Fluorescence micrographs of tobacco callus cells transformed with Sig-(Ser-Pro)32-EGFP (A) or Sig-(GAGP)3-EGFP (B); (C) nontransformed tobacco callus cells. The synthetic genes encoded a signal sequence to direct the products through the endoplasmic reticulum and Golgi, then out to the extracellular matrix (51). Not shown are cells transformed with Sig-EGFP, which looked like those in A and B; however, the medium fluorescence was much less intense. The fluorescence in these highly vacuolated, cultured cells surrounds the nuclei but is not inside of them, judging by optical sections (not shown). The microscope was a Molecular Dynamics Sarastro 2000 confocal laser scanning microscope with a 488-nm laser wave length filter, a 510-nm primary beam splitter, and a 510-nm barrier filter.
Isolation of (Ser-Hyp)32-EGFP and (GAGP)3-EGFP.
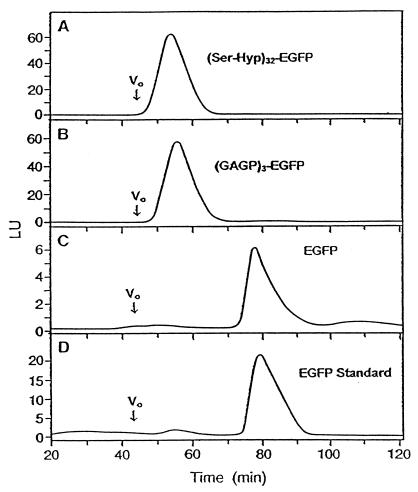
We harvested the culture medium and purified the gene products by gel permeation and reverse-phase chromatography (Figs. 3 and 4). Six cell lines examined [three each of (Ser-Hyp)32-EGFP and (GAGP)3-EGFP] synthesized fluorescent glycoproteins of comparable sizes, although product yields between lines differed by as much as 10-fold. For product characterization, we chose high-yielding lines (shown in Fig. 2), which typically produced 23 mg/liter (Ser-Hyp)32-EGFP and 8 mg/liter (GAGP)3-EGFP after isolation. Superose-12 fractionation of the two fusion glycoproteins (Fig. 3) compared with molecular mass standards (not shown) indicated mass ranges of ≈95–115 kDa for (Ser-Hyp)32-EGFP and ≈70–100 kDa for (GAGP)3-EGFP.
Figure 3.
Superose-12 gel permeation chromatography with fluorescence detection of culture medium containing (Ser-Hyp)32-EGFP (A), (GAGP)3-EGFP medium concentrated 4-fold (B), medium of EGFP targeted to the extracellular matrix concentrated 10-fold (C), or 10 μg of standard EGFP from CLONTECH (D). Not shown is the fractionation of medium from nontransformed tobacco cells, which gave no fluorescent peaks, consistent with the results presented in Fig. 2C. LU, luminescence units.
Coprecipitation with Yariv Reagent.
Both (Ser-Hyp)32-EGFP and (GAGP)3-EGFP precipitated with Yariv reagent (Table 1).
Table 1.
Yariv assay of (Ser-Hyp)32-EGFP and (GAGP)3-EGFP
| Sample weight, μg |
A at 420 nm
|
|||
|---|---|---|---|---|
| (Ser-Hyp)32-EGFP | (GAGP)3-EGFP | Standards
|
||
| GAGP | Tobacco AGP | |||
| 20 | 0.16 | 0.27 | 0.51 | 0.16 |
| 50 | 0.45 | 0.56 | 1.22 | 0.38 |
| 100 | 1.00 | 1.21 | 2.69 | 0.85 |
Hyp Glycoside Profiles.
Each Hyp residue in (Ser-Hyp)32-EGFP contained an arabinogalactan-polysaccharide substituent; (GAGP)3-EGFP Hyp residues contained arabinooligosaccharide substituents in addition to arabinogalactan polysaccharides (Table 2).
Table 2.
Hyp-glycoside profiles of (Ser-Hyp)32-EGFP (GAGP)3-EGFP, and native GAGP
| Hyp-glycoside | Percentage of total Hyp
|
||
|---|---|---|---|
| (Ser-Hyp)32-EGFP | (GAGP)3-EGFP | Native GAGP (ref. 10) | |
| Hyp-polysaccharide | 100 | 62 | 25 |
| Hyp-Ara | 0 | 4 | 10 |
| Hyp-Ara2 | 0 | 12 | 17 |
| Hyp-Ara3 | 0 | 7 | 31 |
| Hyp-Ara4 | 0 | 4 | 5 |
| Nonglycosylated Hyp | 0 | 11 | 12 |
Monosaccharide and Glycosyl Linkage Analysis.
Gal and Ara accounted for the bulk of the saccharides in both fusion proteins, with lesser amounts of Rha and GlcUA (Table 3); saccharide accounted for 58% (dry weight) of (Ser-Hyp)32-EGFP and 48% (dry weight) of (GAGP)3-EGFP. Methylation analyses indicated that 3- and 3,6-linked galactose species accounted for 50 mol % of the sugars in (Ser-Hyp)32-EGFP and 46 mol % of (GAGP)3-EGFP; 2-linked arabinofuranose accounted for 1.6 and 3.1 mol %, respectively; terminal arabinofuranose accounted for 20 and 21 mol %, respectively; 4-arabinopyranose or 5-arabinofuranose accounted for 6 and 8%, respectively; all rhamnose was terminal; all GlcUA was 4-linked.
Table 3.
Glycosyl compositions of (Ser-Hyp)32-EGFP, (GAGP)3-EGFP, native GAGP, and crude gum arabic
| Glycosyl residue | Mol percent
|
|||
|---|---|---|---|---|
| (Ser-Hyp)32-EGFP | (GAGP)3-EGFP* | Native GAGP (ref. 10) | Crude gum arabic (ref. 32) | |
| Ara | 28 | 23 | 36 | 28 |
| Gal | 45 | 49 | 46 | 37 |
| Rha | 8 | 8 | 10 | 13 |
| Xyl | 0 | 2 | 0 | 0 |
| GlcUA | 19 | 16 | 9 | 17 |
| Mann | 1 | 1 | 0 | 0 |
Values are corrected for a small amount of glucose contamination.
Hyp Assay of Secreted EGFP.
There was no Hyp in secreted EGFP or in EGFP removed from the fusion glycoproteins by pronase.
Anhydrous Hydrogen Fluoride Deglycosylation.
After deglycosylation of 4.5 mg of each fusion glycoprotein, we recovered 1 mg of deglycosylated (Ser-Hyp)32-EGFP (i.e., ≈23% weight recovery) and 2.2 mg of deglycosylated (GAGP)3-EGFP (i.e., ≈50% recovery).
Protein Sequence Analysis by Edman Degradation.
Edman degradation (Fig. 5) confirmed the gene sequences and identified which Pro residues had been hydroxylated to Hyp.
Figure 5.
Polypeptide sequences of (Ser-Hyp)32-EGFP and (GAGP)3-EGFP before and after deglycosylation. (A) N-terminal amino acid sequence of the glycoprotein (Ser-Hyp)32-EGFP. We obtained partial sequence of both the glycoprotein (Upper) and its polypeptide after deglycosylation (Lower). X denotes blank cycles that correspond to glycosylated Hyp; glycoamino acids tend to produce blank cycles during Edman degradation, an exception being arabinosyl Hyp (17). (B) Polypeptide sequence of glycosylated (GAGP)3-EGFP (Upper) and deglycosylated (GAGP)3-EGFP (Lower). Residues marked with an asterisk denote low molar yields of Hyp and likely sites of arabinogalactan polysaccharide attachment in glycosylated (GAGP)3-EGFP. For example, yields were 480 pM Asp in the first cycle, 331 pM Ser in the second, 194 pM Hyp in the third, and 508 pM Ser in the fourth.
Discussion
The general (Xaa-Pro)n motif is widespread in AGPs (7), where Pro is usually posttranslationally hydroxylated to form Hyp and Xaa is usually Ala, Thr, or Ser. Thus, the simple putative polysaccharide addition site (Xaa-Hyp)n is probably a major glycomodule of AGPs, including gum glycoproteins (16). To test that hypothesis, we chose the conserved (Ser-Hyp)n motif that occurs both in green algae (Chlamydomonas, ref. 28; and in higher plant AGPs, refs. 29 and 30). This noncontiguous Hyp motif is of particular interest, because it also occurs together with a contiguous Hyp motif in the consensus sequence of GAGP, which contains both oligoarabinoside and polysaccharide addition sites (10, 16). We designed three synthetic genes to test these ideas.
The first synthetic gene, dubbed Sig-(Ser-Pro)32-EGFP, encoded a signal sequence (Sig; ref. 18) at the N terminus followed by (Ser-Pro)32 and then EGFP at the C terminus. The predicted polysaccharide addition to noncontiguous Hyp should yield an expression product containing Hyp polysaccharide exclusively.
The second synthetic gene, dubbed Sig-(GAGP)3-EGFP, encoded three repeats of a slightly modified 19-residue GAGP consensus sequence (ref. 16; Figs. 1 and 5) and should yield an expression product that contains Hyp arabinosides as well as Hyp polysaccharide.
The third synthetic gene was a control construct, Sig-EGFP, that encoded only the signal sequence and EGFP. The expression product was a control to test whether any Hyp glycosylation could be attributed to EGFP modification.
Inclusion of the EGFP reporter protein facilitated the selection of transformed cells (Fig. 2) and subsequent detection of the expression products during isolation (Fig. 1). EGFP fluorescence in the growth medium was also a visual demonstration of Sig efficacy in directing secretion. The absence of any obvious cell lysis in the cultures and excellent product yields of the glycosylated expression products confirmed that the green fluorescence represented bona fide secretory products. Interestingly, EGFP without a glycomodule was secreted at very low levels, perhaps because of lower solubility.
The following experiments characterized the purified fusion proteins and showed that they are indeed new AGPs.
Coprecipitation with the β-galactosyl Yariv reagent.
Both fusion glycoproteins coprecipitated with the β-galactosyl Yariv reagent (Table 1), a specific property of β-1,3-linked AGPs (7, 24).
Protein sequence analysis.
N-terminal sequencing of both (Ser-Hyp)32-EGFP and (GAGP)3-EGFP (Fig. 5) verified the synthetic gene sequences and identified Hyp residues. Occasional incomplete proline hydroxylation has been observed elsewhere (12, 31) and may simply signify a prolyl hydroxylase with less than 100% fidelity.
Hyp-glycoside profiles.
A Hyp-glycoside profile of (Ser-Hyp)32-EGFP (Table 2) gave a single peak of Hyp corresponding to Hyp polysaccharide. Significantly, peaks corresponding to Hyp arabinosides and nonglycosylated Hyp were absent. This absence indicates that all of the Hyp residues in the glycomodule were linked to a polysaccharide.
In contrast, (GAGP)3-EGFP yielded peaks corresponding to Hyp arabinosides, nonglycosylated Hyp, and Hyp polysaccharide. However, (GAGP)3-EGFP (Figs. 1 and 5) was designed with fewer contiguous Hyp residues than the consensus sequence of native GAGP and yielded fewer Hyp arabinosides, consistent with fewer contiguous Hyp arabinosylation sites (8, 12, 16, 17). In addition, occasional incomplete hydroxylation of the middle proline residue in the Pro-Pro-Pro block (Fig. 5B) converted a region of contiguous Hyp (putative arabinosylation site) to noncontiguous Hyp (polysaccharide addition sites). Control EGFP targeted to the extracellular matrix contained no Hyp, and hence no glycosylated Hyp, judging by manual Hyp assays.
Sugar analyses.
Both fusion glycoproteins had sugar compositions typical of AGPs (Table 3)—a galactose:arabinose molar ratio of ≈2:1, with lesser amounts of glucuronic acid and rhamnose. The predominantly 3- and 3,6-linked galactose and terminal arabinofuranose determined by methylation analysis was in keeping with a β-1,3-linked galactan backbone having side chains of arabinose, glucuronic acid, and rhamnose (7). The very low amount of 1,2-linked arabinose in (Ser-Hyp)32-EGFP agreed with the absence of Hyp arabinosides; however, the presence of 1,2-linked arabinose in (GAGP)3-EGFP agreed with the presence of Hyp arabinosides in its Hyp glycoside profile, because they are known to be largely 1,2-linked (32). Thus, (GAGP)3-EGFP contained both types of Hyp glycosylation, consistent with the presence of a polypeptide having contiguous and noncontiguous Hyp as putative arabinosylation and polysaccharide addition sites, respectively.
Size of attached polysaccharide.
Hyp glycoside profiles showed the molar ratio of Hyp polysaccharides in each fusion glycoprotein (Table 2). This ratio gives the number of (polysaccharide)-Hyp residues in each glycoprotein molecule (e.g., Hyp polysaccharide accounted for 100% of the Hyp glycosides in (Ser-Hyp)32, i.e., 31–32 Hyp polysaccharides). Glycoprotein size before and after deglycosylation gave an approximate size for the attached polysaccharide. The size of each fusion protein before and after deglycosylation was ≈95–115 kDa and 34 kDa, respectively, for (Ser-Hyp)32-EGFP (≈71-kDa carbohydrate), and ≈70–100 kDa and 34 kDa, respectively, for (GAGP)3-EGFP (≈51-kDa carbohydrate). Judging by the gene sequence (not shown) and Fig. 5, (Ser-Hyp)32-EGFP contains ≈31–32 Hyp residues, all noncontiguous. Thus, the average polysaccharide size is 2.2–2.3 kDa (71 kDa/31), which corresponds to 14–15 sugar residues (average sugar residue weight of 155 calculated from the sugar composition in Table 3). This is consistent with the empirical formula Gal6 Ara3 GlcUA2 Rha, based on compositional data in Table 3. Similarly, (GAGP)3-EGFP contains ≈23–25 Hyp residues, of which 62% (Table 2) or ≈15 occur with polysaccharide attached. Hence, the polysaccharide approximates 51 kDa/15 = 3.4 kDa, corresponding to about 22 sugar residues; this value is a modest overestimate, because it includes arabinose from the Hyp arabinooligosaccharides.
The similarity of these fusion glycoproteins to native GAGP (Table 3) suggests a model for the Hyp polysaccharide based on the general arabinogalactan structure (33–35) of a galactan core, with small side chains containing rhamnose, arabinose, and glucuronic acid. Possibly larger arabinogalactan polysaccharides can be built up by repeated addition (36) of small ≈12-residue blocks represented by the empirical formula above.
Expression of the two repetitive glycomodules corroborates the Hyp contiguity hypothesis and has the following implications.
The Hyp contiguity hypothesis postulates that a glycosylation code based on contiguous versus noncontiguous Hyp motifs directs the addition of arabinosides or arabinogalactan polysaccharide, respectively. The repetitive Ser-Hyp motif directed the exclusive addition of arabinogalactan polysaccharide to Hyp in (Ser-Hyp)32-EGFP, whereas Hyp arabinosylation was correlated with the presence of contiguous Hyp blocks in (GAGP)3-EGFP. Thus, the O-Hyp glycosyltransferases of plants seem to resemble the O-Ser and O-Thr glycosyltransferases of animals in their multiplicity and ability to discriminate based on primary sequence and site clustering (37). Conformational requirements may also account for the sequence specificity of animal O-glycosyl transferases (38, 39). Possibly similar rules hold in plants. Thus, we should not rule out the possibility that a conformational switch determines the addition of arabinose or galactose to Hyp residues, because the polyproline II helix of contiguous Pro (or Hyp) residues may not propagate through regions of noncontiguity (40). However, in a PRP from Douglas fir, the sequence Ile-Pro-Pro-Hyp-Val was never arabinosylated, whereas the sequence Lys-Pro-Hyp-Hyp-Val was consistently arabinosylated, arguing for primary sequence rather than conformation as a determinant of Hyp arabinosylation (17). The ability to generate specific substrates for O-Hyp arabinosyltransferases and galactosyltransferases, as demonstrated here, should facilitate their isolation, their unambiguous identification, and the determination of their substrate preferences.
Transfer of an Acacia GAGP analog to Nicotiana gave a product containing both oligoarabinoside and arabinogalactan polysaccharide resembling the product of native GAGP. Therefore, the Hyp glycosylation code seems to be shared by Leguminosae and Solanaceae, two widely separated dicotyledonous families. Indeed, a Hyp oligosaccharide containing both arabinose and galactose was first reported for the green alga Chlamydomonas (41). Thus, the Hyp glycosylation code, together with conserved glycosyltransferases (42), may be global. A global code could help to identify potential sites of oligoarabinoside and polysaccharide addition in HRGPs inferred from their genomic sequences. Furthermore, it would permit the transfer of useful products, like exudate gum glycoproteins (43) such as GAGP from thorny desert scrub like Acacia, to more amenable crop plants.
Several strategies identify protein modules as functional units by mapping the regions involved (44, 45). We can now recognize two HRGP glycomodules, the arabinosylated Ser-Hyp4 (46), and the (Xaa-Hyp)n arabinogalactan polysaccharide glycomodule identified here. Fluorescently tagged, unimodular (or bimodular) HRGP analogs enable further functional analysis of HRGPs, because they allow sensitive fluorimetric assays of single module properties such as binding and cross-linking both in muro and in vitro. As possible endogenous competitive inhibitors of normal HRGP interactions, these glycomodules may show how HRGP scaffolds interact with other matrix components during wall self-assembly, for example in pollen tubes (47), protoplasts (48), and Chlamydomonas (49, 50).
Finally, a simple Hyp-glycosylation code facilitates the design of HRGPs and their manipulation module-by-module to enhance desirable properties. Thus, design of glycoproteins may have utilitarian value. A wide range of products is possible, such as exudate gum analogs and other useful hydrocolloids, including HRGP-enzyme hybrids stabilized as an insoluble cross-linked AGP-gel matrix. These environmentally benign products produced in plant factories may find application in the agricultural, food, pharmaceutical, and nanotechnology industries.
Acknowledgments
We thank Drs. Ron Sederoff and Alan Wenck for the tobacco cultures, for Agrobacterium strain LBA4404, and for tutoring Ms. Shpak in the art of plant transformation; Dr. Derk Lamport for comments on the manuscript; Ms. Carol Hahn for the figures and AEP colloids; and Gary Wine for the gum arabic nodules. We also thank the United States Department of Agriculture (National Research Initiative Competitive Research Program Grant 9701299) and the National Science Foundation (Grant MB-9805960) for supporting this work. This work was also supported in part by National Institutes of Health Grant 2-P41-RR05351-06 to the Complex Carbohydrate Research Center.
Abbreviations
- Hyp
hydroxyproline
- HRGP
Hyp-rich glycoprotein
- AGP
arabinogalactan protein
- PRP
proline-rich protein
- GAGP
gum arabic glycoprotein
- EGFP
enhanced green fluorescent protein
References
- 1.Lamport D T A. In: Structure, Biosynthesis and Significance of Cell Wall Glycoproteins. Loewus F A, Runeckles V C, editors. New York: Plenum; 1977. pp. 79–115. [Google Scholar]
- 2.Knox R B, Clarke A, Harrison S, Smith P, Marchalonis J J. Proc Natl Acad Sci USA. 1976;73:2788–2792. doi: 10.1073/pnas.73.8.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Wu H, Cheung A Y. Plant Cell. 1993;5:1639–1650. doi: 10.1105/tpc.5.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller B, Lamb C J. Genes Dev. 1989;3:1639–1646. doi: 10.1101/gad.3.10.1639. [DOI] [PubMed] [Google Scholar]
- 5.Sadava D, Chrispeels M J. Biochim Biophys Acta. 1973;227:278–287. doi: 10.1016/0005-2744(71)90060-x. [DOI] [PubMed] [Google Scholar]
- 6.Esquerre-Tugaye M T, Lamport D T A. Plant Physiol. 1979;64:314–319. doi: 10.1104/pp.64.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nothnagel E A. Int Rev Cytol. 1997;174:195–291. doi: 10.1016/s0074-7696(08)62118-x. [DOI] [PubMed] [Google Scholar]
- 8.Kieliszewski M J, Lamport D T A. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- 9.Pope D G. Plant Physiol. 1977;59:894–900. doi: 10.1104/pp.59.5.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qi W, Fong C, Lamport D T A. Plant Physiol. 1991;96:848–855. doi: 10.1104/pp.96.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamport D T A, Miller D H. Plant Physiol. 1971;48:454–456. doi: 10.1104/pp.48.4.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieliszewski M, de Zacks R, Leykam J F, Lamport D T A. Plant Physiol. 1992;98:919–926. doi: 10.1104/pp.98.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Varner J E. Proc Natl Acad Sci USA. 1985;82:4399–4403. doi: 10.1073/pnas.82.13.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcus A, Greenberg J, Averyhart-Fullard V. Physiol Plant. 1991;81:273–279. [Google Scholar]
- 15.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kielszewski M J. Int. Patent Appl. PIXXD2 and Publ. WO 9903978. 1999. [Google Scholar]
- 17.Kieliszewski M J, O'Neill M, Leykam J, Orlando R. J Biol Chem. 1995;270:2541–2549. doi: 10.1074/jbc.270.6.2541. [DOI] [PubMed] [Google Scholar]
- 18.De Loose M, Gheysen G, Tire C, Gielen J, Villarroel R, Genetello C, Van Montagu M, Depicker A, Inze D. Gene. 1991;99:95–100. doi: 10.1016/0378-1119(91)90038-d. [DOI] [PubMed] [Google Scholar]
- 19.McGrath K P, Tirrell D A, Kawai M, Mason T L, Fournier M J. Biotechnol Prog. 1990;6:186–192. doi: 10.1021/bp00003a004. [DOI] [PubMed] [Google Scholar]
- 20.Bevan M W. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 22.Haseloff J, Siemering K R, Prasher D C, Hodge S. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- 24.Serpe M D, Nothnagel E A. Planta. 1994;193:542–550. [Google Scholar]
- 25.Merkle R K, Poppe I. Methods Enzymol. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- 26.York W S, Darvill A G, McNeil M, Stevenson T T, Albersheim P. Methods Enzymol. 1985;118:3–40. [Google Scholar]
- 27.Mort A J, Lamport D T A. Anal Biochem. 1977;82:289–309. doi: 10.1016/0003-2697(77)90165-8. [DOI] [PubMed] [Google Scholar]
- 28.Woessner J P, Goodenough U W. Plant Sci. 1992;83:65–76. [Google Scholar]
- 29.Vijn I, Yang W-C, Pallisgaard N, Jensen E O, van Kammen A, Bisseling T. Plant Mol Biol. 1995;28:1111–1119. doi: 10.1007/BF00032671. [DOI] [PubMed] [Google Scholar]
- 30.de Blank C, Mylona P, Yang W C, Katinakis P, Bisseling T, Franssen H. Plant Mol Biol. 1993;22:1167–1171. doi: 10.1007/BF00028987. [DOI] [PubMed] [Google Scholar]
- 31.Sticher L, Hofsteenge J, Neuhaus J-M, Boller T, Meins F. Plant Physiol. 1993;101:1239–1247. doi: 10.1104/pp.101.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akiyama Y, Mori M, Kato K. Agric Biol Chem. 1980;44:2487–2489. [Google Scholar]
- 33.Aspinall G O, Knebl M C. Carbohydr Res. 1986;157:257–260. [Google Scholar]
- 34.Defaye J, Wong E. Carbohydr Res. 1986;150:221–231. [Google Scholar]
- 35.Clarke A E, Anderson R L, Stone B A. Phytochemistry. 1979;18:521–540. [Google Scholar]
- 36.Bacic A, Churms S C, Stephen A M, Cohen P B, Fincher G B. Carbohydr Res. 1987;162:85–93. [Google Scholar]
- 37.Gerken T A, Owens C L, Pasumarthy M. J Biol Chem. 1997;272:9709–9719. doi: 10.1074/jbc.272.15.9709. [DOI] [PubMed] [Google Scholar]
- 38.De Haan C A M, Roestenberg P, De Wit M, De Vries A A F, Nilsson T, Vennema H, Rottier P J M. J Biol Chem. 1998;273:29905–29914. doi: 10.1074/jbc.273.45.29905. [DOI] [PubMed] [Google Scholar]
- 39.Pisano A, Redmond J W, Williams K L, Gooley A A. Glycobiology. 1993;3:429–435. doi: 10.1093/glycob/3.5.429. [DOI] [PubMed] [Google Scholar]
- 40.Creamer T P. Proteins. 1998;33:218–226. [PubMed] [Google Scholar]
- 41.Miller D H, Lamport D T A, Miller M. Science. 1972;176:918–920. doi: 10.1126/science.176.4037.918. [DOI] [PubMed] [Google Scholar]
- 42.Breton C, Bettler E, Joziasse D H, Geremia R A, Imberty A. J Biochem (Tokyo) 1998;123:1000–1009. doi: 10.1093/oxfordjournals.jbchem.a022035. [DOI] [PubMed] [Google Scholar]
- 43.Islam A M, Phillips G O, Sljivo A, Snowden M J, Williams P A. Food Hydrocolloids. 1997;11:493–505. [Google Scholar]
- 44.Joseph G, Pick E. J Biol Chem. 1995;270:29079–29082. doi: 10.1074/jbc.270.49.29079. [DOI] [PubMed] [Google Scholar]
- 45.Hegyi H, Bork P. J Protein Chem. 1997;16:545–551. doi: 10.1023/a:1026382032119. [DOI] [PubMed] [Google Scholar]
- 46.Lamport D T A, Katona L, Roerig S. Biochem J. 1973;133:125–131. doi: 10.1042/bj1330125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy S, Jauh G Y, Hepler P K, Lord E M. Planta. 1998;204:450–458. doi: 10.1007/s004250050279. [DOI] [PubMed] [Google Scholar]
- 48.Cooper J B, Heuser J E, Varner J E. Plant Physiol. 1994;104:747–752. doi: 10.1104/pp.104.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hills G J, Phillips J M, Gay M R, Roberts K. J Mol Biol. 1975;96:431–444. doi: 10.1016/0022-2836(75)90170-9. [DOI] [PubMed] [Google Scholar]
- 50.Goodenough U W, Gebhart B, Mecham R P, Heuser J E. J Cell Biol. 1986;103:403. doi: 10.1083/jcb.103.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardiner M, Chrispeels M. Plant Physiol. 1975;55:536–541. doi: 10.1104/pp.55.3.536. [DOI] [PMC free article] [PubMed] [Google Scholar]