Abstract
Yeast Sec18p and its mammalian orthologue N-ethylmaleimide-sensitive fusion protein (NSF) are hexameric ATPases with a central role in vesicle trafficking. Aided by soluble adapter factors (SNAPs), Sec18p/NSF induces ATP-dependent disassembly of a complex of integral membrane proteins from the vesicle and target membranes (SNAP receptors). During the ATP hydrolysis cycle, the Sec18p/NSF homohexamer undergoes a large-scale conformational change involving repositioning of the most N terminal of the three domains of each protomer, a domain that is required for SNAP-mediated interaction with SNAP receptors. Whether an internal conformational change in the N-terminal domains accompanies their reorientation with respect to the rest of the hexamer remains to be addressed. We have determined the structure of the N-terminal domain from Sec18p by x-ray crystallography. The Sec18p N-terminal domain consists of two β-sheet-rich subdomains connected by a short linker. A conserved basic cleft opposite the linker may constitute a SNAP-binding site. Despite structural variability in the linker region and in an adjacent loop, all three independent molecules in the crystal asymmetric unit have the identical subdomain interface, supporting the notion that this interface is a preferred packing arrangement. However, the linker flexibility allows for the possibility that other subdomain orientations may be sampled.
Sec18p is an essential protein for transport through the yeast secretory pathway. The gene coding for this protein (SEC18) originally was identified on the basis of a temperature-conditional mutant that blocks the progress of proteins toward the yeast cell surface (1). The cloning and sequencing of yeast SEC18 and its mammalian orthologue N-ethylmaleimide-sensitive fusion protein (NSF), in conjunction with biochemical studies on their gene products, led to the remarkable realization that this key player in vesicle trafficking is conserved structurally and functionally from yeast to the mammalian brain (2–4). Sec18p/NSF is an ATPase that primes membranes for fusion by chaperoning disassembly of a 20S complex that it forms with soluble NSF-attachment proteins (SNAPs) and the integral membrane SNAP receptors (5–8). In addition, Sec18p/NSF has been proposed to play a direct role in lipid bilayer fusion (9).
Sec18p/NSF is a three-domain protein that self-assembles in the presence of ATP into ring-shaped homohexamers (10–12). The central and carboxyl-terminal domains (D1 and D2, respectively) are homologous to one another (3) and contain sequences common to the AAA+ protein family (13–15). Nucleotide binding by D2 is required for hexamerization (12), and ATP hydrolysis by D1 is needed for catalytic activity (16). The structure of hexameric NSF D2 has been determined crystallographically (17, 18), providing molecular details about ATP-dependent hexamerization and also a model for the fold of D1. The remaining N-terminal domain is required for coupling Sec18p/NSF ATPase activity with 20S complex disassembly, as deletion mutants lacking this domain cannot interact with SNAPs and SNAP receptors (19).
Quaternary structural changes involving the N-terminal domain may underlie the Sec18p/NSF catalytic mechanism. As visualized by electron microscopy, ATP hydrolysis by the D1 domains appears to affect the orientation and/or conformation of the adjacent N-terminal domains. In the presence of nonhydrolyzable ATP analogs, the N-terminal domains splay from the hexameric assembly like petals of a flower, and the enzyme is competent to bind the complex of SNAPs and SNAP receptors (12). Under conditions in which nucleotide hydrolysis occurs, the N-terminal domains pack back against the D1 domains, and the enzyme cannot bind its protein substrates (12).
Although stylistic interpretations of the electron micrographs illustrate the N-terminal domain as undergoing an internal conformational change rather than simply a rigid-body motion within the hexamer (12), the resolution of the micrographs is not sufficient to rule out either model. Also unknown are the regions of the Sec18p/NSF N-terminal domain that interact with the SNAP/SNAP receptor complex or that contact the other domains in the Sec18p/NSF hexamer. As a result, the biophysical mechanism by which Sec18p/NSF disassembles the 20s complex has not yet been detailed.
To contribute toward a structure-based description of the Sec18p/NSF catalytic mechanism, we have determined the structure of the N-terminal domain of Saccharomyces cerevisiae Sec18p (Sec18p-N) by x-ray crystallography. Sec18p-N consists of two subdomains: a double-psi β-barrel and a four-stranded β-sheet backed by a helix. Although Sec18p-N has the same overall fold as the N-terminal domain of NSF (20, 21), one face of the structure is conformationally divergent from its mammalian orthologue. Opposite this face is a basic groove that is conserved between the Sec18p and NSF N-terminal domain structures and that may constitute a SNAP-binding site.
Materials and Methods
Plasmid Construction, Protein Expression, and Protein Purification.
The region coding for the N-terminal domain of Sec18p (residues 22–210) was obtained by PCR from S. cerevisiae genomic DNA (gift of Richard Young, Massachusetts Institute of Technology). A lysine AAA second codon was inserted to potentially increase expression (22). This PCR fragment was cloned into the pAED4 vector (23). The resulting expression construct was transformed into BL21 (DE3) plysS cells, which were grown at 37°C in LB containing 100 mg/liter ampicillin and 30 mg/liter chloramphenicol. When the cell culture reached an optical density of A600 = 0.5, it was shifted to 32°C and isopropyl β-d-thiogalactoside was added to 1 mM to induce protein expression. Four hours later, cells were harvested by centrifugation and lysed by sonication in 20 mM Tris, pH 6.8, 50 mM NaCl, 1 mM EDTA. The homogenate was centrifuged for 10 min at 10,000 g, and the supernatant was applied at 4°C to a Fast S Sepharose column (Sigma). Protein was eluted with an NaCl gradient in 20 mM Tris, pH 6.8, 1 mM EDTA. Coomassie-stained gels of the eluted fractions revealed two closely spaced bands apparently corresponding to the presence and absence of an internal disulfide bond between the two cysteines in this domain. In addition, a small fraction of disulfide-bonded dimers could be seen. DTT was added to 50 mM, and incubation proceeded overnight at 4°C. DTT then was removed and the protein was exchanged into 10 mM sodium phosphate buffer, pH 6.8, by desalting over PD-10 columns (Amersham Pharmacia). Immediately after buffer exchange, N-ethylmaleimide (NEM) was added to a concentration of 10 mM from a 1 M NEM stock in acetonitrile. NEM-modified Sec18p-N then was applied to a type I hydroxyapatite column (Bio-Rad) and eluted with increasing sodium phosphate, pH 6.8. Peak fractions were pooled, dialyzed against 10 mM sodium phosphate, pH 6.8, and concentrated in a Centricon 10 (Amicon) to 10 mg/ml as determined spectrophotometrically in 6 M GuHCl (24), assuming an extinction coefficient of 18,880 at 280 nm.
Crystallization and Heavy Atom Derivatization.
Crystals of NEM-modified Sec18p-N were grown by hanging-drop vapor diffusion at 4°C. Protein stock solution was mixed 1:1 with a well solution containing 50 mM sodium phosphate, pH 5.6, 5–10% DMSO, 5% PEG 8000, 5 mM ATP. Crystals grew to their maximum size (0.05 × 0.05 × 0.5 mm3) in 2–3 days. Native crystals were transferred directly to Exxon Paraton oil before freezing and storage in liquid nitrogen. For derivatization, crystals were transferred to 300 μl of well solution containing the heavy atom (1 mM PCMBS or 1 mM AgNO3) and soaked at 4°C for 24 hr before transfer to Paraton oil for freezing.
Data Collection and Structure Determination.
Diffraction data from a native Sec18p-N crystal were collected at 100 K on an UltraX generator (Rigaku, Tokyo) equipped with an RaxisIV detector (Rigaku) and Osmic mirrors. Crystals were of space group P21 and unit cell dimensions a = 93.7 Å, b = 32.2 Å, c = 94.8 Å, α = 90°, β = 118.7°, γ = 90°. A self-rotation function calculated to 4.0-Å resolution by using polarrfn (25) revealed 6σ peaks at ϕ = 90°, φ = 90°, κ = 60° and at ϕ = 90°, φ = 90°, κ = 120°. Three molecules per asymmetric unit results in a calculated solvent content of approximately 39%. Heavy atom derivative data were collected on an RU-300 generator (Rigaku) equipped with an RaxisIV detector (Rigaku). All data were processed and scaled by using denzo and scalepack (26). Multiple isomorphous replacement statistics are reported from mlphare output (25), and phasing for electron density map calculation was performed with sharp (27). Approximate noncrystallographic symmetry (ncs) operators were calculated by using the two heavy atom sites per molecule and the information that the self-rotation axis is parallel to the crystallographic two-fold axis. Ncs operators were improved with imp (28). Phases were modified by solvent-flattening and ncs averaging using dm (29). The Sec18p-N model was built by using o (30) and refined with the Crystallography and NMR System (31). Ncs restraints were applied to residues 27–102, 114–122, 136–174, and 187–206 from each molecule during refinement.
Structure Analysis.
Global rms deviation calculations between Sec18p-N and NSF-N were carried out over Sec18p-N residues 26–61, 64–105, 111–125, 133–178, and 194–210 and NSF-N residues 3–38, 45–86, 89–103, 111–156, and 173–189. To compare the relative orientations of the subdomains in Sec18p-N and NSF-N, the structures were aligned over residues 26–61 and 64–100 from Sec18p-NA and residues 3–38 and 45–81 from NSF-NA. A rotation matrix relating the resulting positions of the NB subdomains (Sec18p-NB residues 114–123, 135–178, and 196–208; NSF-NB residues 92–101, 113–156, and 175–187) then was calculated by using lsqkab (32). This matrix was used to approximate a single angle rotation θ using the equation: θ = cos−1((m11 + m22 + m33 −1)/2) (http://www.ecse.rpi.edu/Homepages/wrf/geom/rotation.html.).
Results
Structure Determination.
The N-terminal domain of S. cerevisiae Sec18p (Sec18p-N, residues 22–210) was expressed in Escherichia coli. The expression construct was designed to begin at Met-22 to increase expression over that observed from Met-1. Moreover, the region upstream of Met-22 in S. cerevisiae Sec18p is largely absent from mammalian NSF. The truncated construct gave high protein yields of which approximately half could be obtained in soluble form upon cell lysis. Sec18p-N migrated as a closely spaced doublet by nonreducing SDS/PAGE. After incubation with DTT, only the upper band was observed (data not shown). Because Sec18p-N contains two cysteine residues, and mutation of one of these cysteines resulted in a single band by SDS/PAGE (data not shown), the doublet was interpreted as representing a mixture of internally disulfide-bonded and nonbonded species.
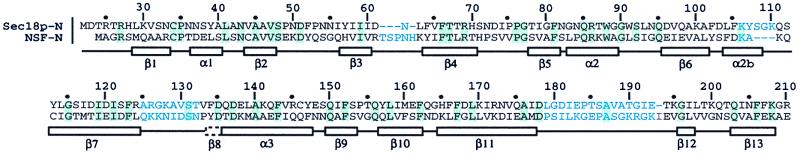
To obtain a single protein species for crystallization, Sec18p-N was fully reduced, and the free cysteine thiols were blocked with the alkylating agent NEM. NEM-modified Sec18p-N was crystallized in the space group P21 with three molecules per asymmetric unit, and its structure was determined to 2.3-Å resolution by multiple isomorphous replacement (Table 1). The Sec18p-N structure was refined to a working R factor of 23.0% and a free R factor of 27.7% (Table 2). While refinement of Sec18p-N was in progress, the structure of the N-terminal domain from mammalian NSF (NSF-N) was reported (20, 21). Although the N-terminal domain is the least conserved of the three Sec18p/NSF domains, the sequence identity between S. cerevisiae Sec18p and hamster NSF is still 27% in this region (Fig. 1), and the structures have similar folds.
Table 1.
Data collection statistics
| Data set | Res., Å | Reflections unique/total | Rmerge (%)* | Aver I/σ | Completeness (%) | Riso (%)† | Phasing power‡ | Rcullis§ |
|---|---|---|---|---|---|---|---|---|
| Native | 2.3 | 22,632/176,811 | 4.0 (11.5) | 23.9 | 99.0 (99.8) | — | — | — |
| PCMBS | 2.8 | 12,696/199,928 | 6.3 (14.1) | 24.0 | 100 (100) | 20.3 (23.6) | 1.43 | 0.75 (0.86) |
| AgNO3 | 3.0 | 10,368/69,049 | 10.8 (21.8) | 12.2 | 99.0 (97.6) | 24.3 (8.8) | 0.68 | 0.91 |
PCMBS, p-chloromercuribenzenesulfonate.
*Rmerge = ΣΣj|Ij − 〈I〉|/Σ〈I〉.
†Riso = Σ∥Fph| − |Fp∥/Σ|Fp|. Values in parentheses are weighted R factors calculated with the program scaleit (38).
‡Phasing power = FH/lack of closure. Phasing statistics are calculated with the program mlphare (25) to 3.0-Å resolution. The overall figure of merit is 0.51.
§Rcullis = ∑||Fph ± Fp| − |Fh,c||/∑|Fph ± Fp|. Value in parentheses is for anomalous signal.
Table 2.
Refinement statistics
| Resolution, Å | 20.0–2.3 |
| Rcryst, % | 23.0 |
| Rfree, % | 27.7 |
| Number of reflections, free/work (%) | 1,547/20,640 (7%) |
| Number of protein atoms | 4,233 |
| Number of NEM atoms | 54 |
| Number of water molecules | 270 |
| rmsd bond lengths, Å | 0.007 |
| rmsd bond angles, degrees | 1.34 |
rmsd, rms deviation.
Figure 1.
Sequence alignment of S. cerevisiae Sec18p-N and hamster NSF-N based on a visual comparison of the two structures. Sequence alignments performed before knowledge of the Sec18p-N structure introduced gaps between strands β2 and β3 in subdomain NA. Such gaps were hypothesized to be inconsistent with a double-psi β-barrel (33). In fact, the only gap in the proper alignment of NA occurs in the loop between strands β3 and β4, which is elongated in NSF vs. Sec18p by an additional four residues. Insertions in this loop are proposed to be tolerated in the double-psi β-barrel (33). The Sec18p-N sequence is labeled every 10 residues according to the full-length Sec18p numbering. Secondary structural elements are boxed below the sequences. The box with a dashed border indicates a short strand (β8) present in NSF-N but not observed in Sec18p-N. Sequence identities are highlighted in light green. Sequences shown in light blue indicate regions with structures that diverge significantly between NSF-N and Sec18p-N, or that were not built in one or the other structural models.
Comparison of the Sec18p and NSF N-Terminal Domain Folds.
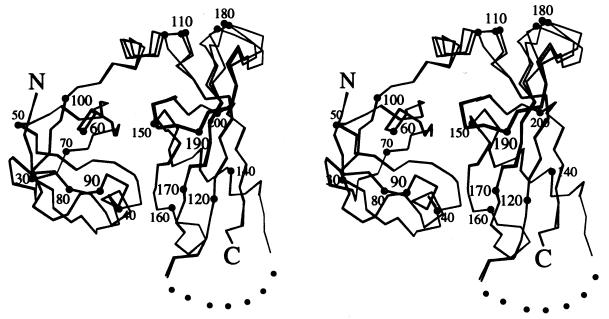
Like NSF-N, Sec18p-N forms a compact, globular structure consisting of two subdomains rich in β-sheet (Fig. 2). The first subdomain (Sec18p-NA) is a double-psi β-barrel (33) and the second (Sec18p-NB) contains a four-stranded β-sheet with an α-helix packed at an angle of approximately 70° to the strands. The importance of the NA fold in Sec18p function is illustrated by the temperature-sensitive sec18–1 mutation, which replaces Gly-89 by aspartic acid (20); the aspartate side chain would be expected to insert into the hydrophobic cluster formed by residues Phe-64, Phe-66, Trp-88, and Trp-91 at the core of the NA subdomain. The three copies of Sec18p-N in the crystal asymmetric unit are identical in the NA subdomain and through most of the NB subdomain. However, α-carbons in a region of the linker connecting the two subdomains, as well as in a loop (residues 175–185) from NB, have shifted by as much as 4 Å (Fig. 2).
Figure 2.
Stereo Cα trace of the three Sec18p-N molecules in the crystal asymmetric unit. The N and C termini, and every 10th Cα, are labeled. Dots are a visual aid to bridge the gap between residues 125 and 133, through which the chain could not be traced. Figure was generated with molscript (36).
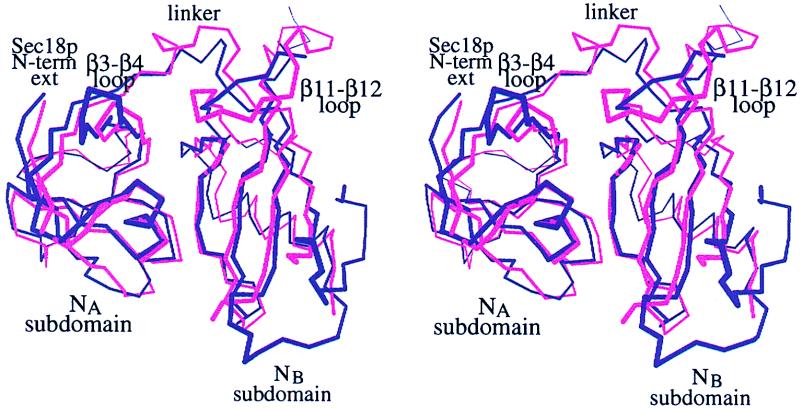
The most significant conformational differences between the Sec18p-N and NSF-N structures are localized to the surface loops, and these differences are concentrated on one face of the structure (Fig. 3). The loop between strands β3 and β4 in Sec18p is shorter than that in NSF, and the structure of the linker between the two Sec18p-N subdomains diverges from NSF between residues Phe-105 and Gln-111. The loop between strand β7 and helix α3 could not be traced in Sec18p-N, and the region between strands β11 and β12 is structurally dissimilar between the mammalian and yeast proteins. The rms deviation over the remaining, comparable regions of Sec18p and NSF (see Materials and Methods) is 1.43 Å.
Figure 3.
Superposition of Sec18p-N (pink) and NSF-N (purple). Sec18p-N (residues 26–61, 64–105, 111–125, 133–178, and 194–210) was aligned on NSF-N (residues 3–38, 46–86, 89–103, 111–156, and 173–189). The NA and NB subdomains are indicated, and structurally divergent regions are labeled. “Sec18p N-term ext” refers to a 19-aa extension at the N terminus of Sec18p that is lacking in mammalian NSF and was deleted from the Sec18p-N protein expression construct. Figure was generated with molscript (36).
Opposite the structurally divergent face is a groove at the subdomain interface. The backbone conformation in this region is similar between Sec18p-N and NSF-N, as is the basic character of the groove surface. This region of Sec18p/NSF recently was proposed as a SNAP binding site (21), and the putative site of interaction on SNAPs is highly acidic (34). In addition, SNAPs contain a conserved leucine that is required for stimulating ATPase activity by NSF (35). Sec18p-N and NSF-N both have partially exposed hydrophobic residues (Leu-41 and Phe-166 in Sec18p; Leu-18 and Leu-144 in NSF) in the basic groove.
Comparison of Sec18p-N and NSF-N Subdomain Interfaces.
Superposition of the NA subdomain of Sec18p-N onto NA of NSF-N results in a rotation of approximately 11° between NB subdomains (Fig. 4). The overall character of the subdomain interface, a mixture of hydrophobic, polar, and water-mediated contacts, is preserved between NSF-N and Sec18p-N, but the specific contacts are different in each case to accommodate the variation in subdomain orientation (Fig. 5). For example, in Sec18p-N, the side chains of residues Ile-192 from subdomain NB and Leu-63, Phe-64, and Trp-88 from subdomain NA form a single hydrophobic cluster that extends through Phe-66 into the core of subdomain NA. In NSF-N, the contribution from NB is absent because the loop between strands β11 and β12 does not approach the subdomain interface in NSF-N as it does in Sec18p-N. In addition, the side chain of Asn-84 in Sec18p-NA participates in a hydrogen-bonding network with the side chains of Arg-124 and Asp-168 from NB and, through a water molecule, with the backbone amide of Leu-169. Asn-84 is replaced by proline (Pro-65) in NSF-N, Arg-124 by leucine (Leu-102), and Asp-168 by glycine (Gly-146). The only inter-subdomain interaction in this region of NSF-N is a hydrophobic interaction between Pro-64 and Val-137. In Sec18p-N, a hydrophobic cluster is formed by the side chains of residues Tyr-57, Ile-151, and Leu-104. In NSF-N, the comparable residues in the cluster are His-34, Ala-129, and Phe-85, and the cluster is more compact (Fig. 5A). At other positions in the interface, direct hydrogen-bonding contacts are observed in the N-terminal domain from one species, whereas a water-mediated bond is seen in the other to compensate for backbone shifts. For example, the side chain of Thr-67 in Sec18p-NA forms a water-mediated hydrogen bond with the backbone amide of Ile-151 in NB, whereas the comparable residues in NSF-N are directly hydrogen-bonded (Fig. 5B). In summary, the Sec18p-N and NSF-N subdomain interfaces contain a sampling of conserved and nonconserved residues, but even conserved residues may exhibit distinct interaction modes.
Figure 4.
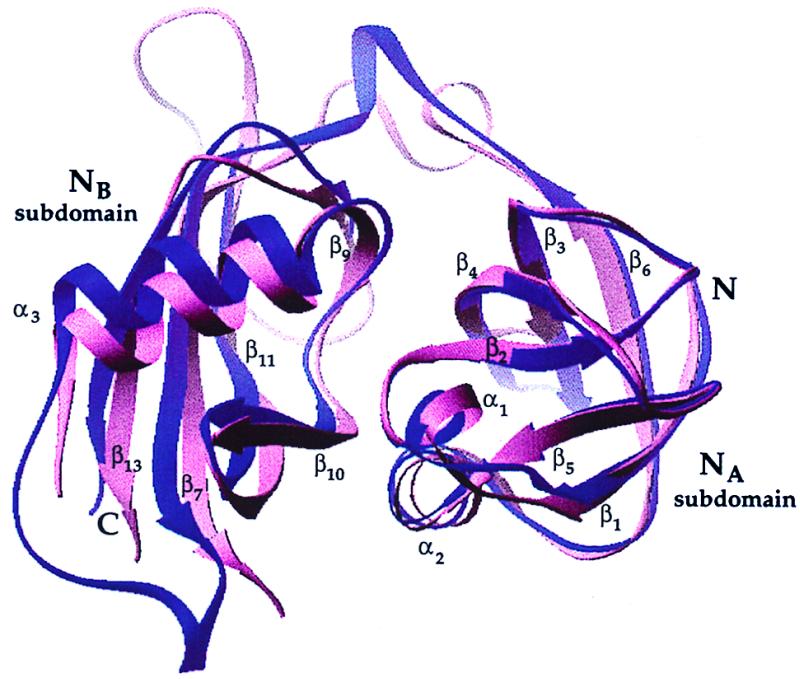
Comparison of the subdomain orientations in Sec18p-N and NSF-N. The NA subdomain of Sec18p (residues 26–61 and 64–100) was structurally aligned with NSF-NA (residues 3–38 and 45–81). A ribbon diagram (37) of the resulting superposition is displayed, with Sec18p-N in pink and NSF-N in purple. The region of NSF-N between strands β11 and β12 (residues 154 and 174) was removed for clarity. Although the NA subdomains align well, the NB subdomains are rotated with respect to one another by approximately 11°.
Figure 5.
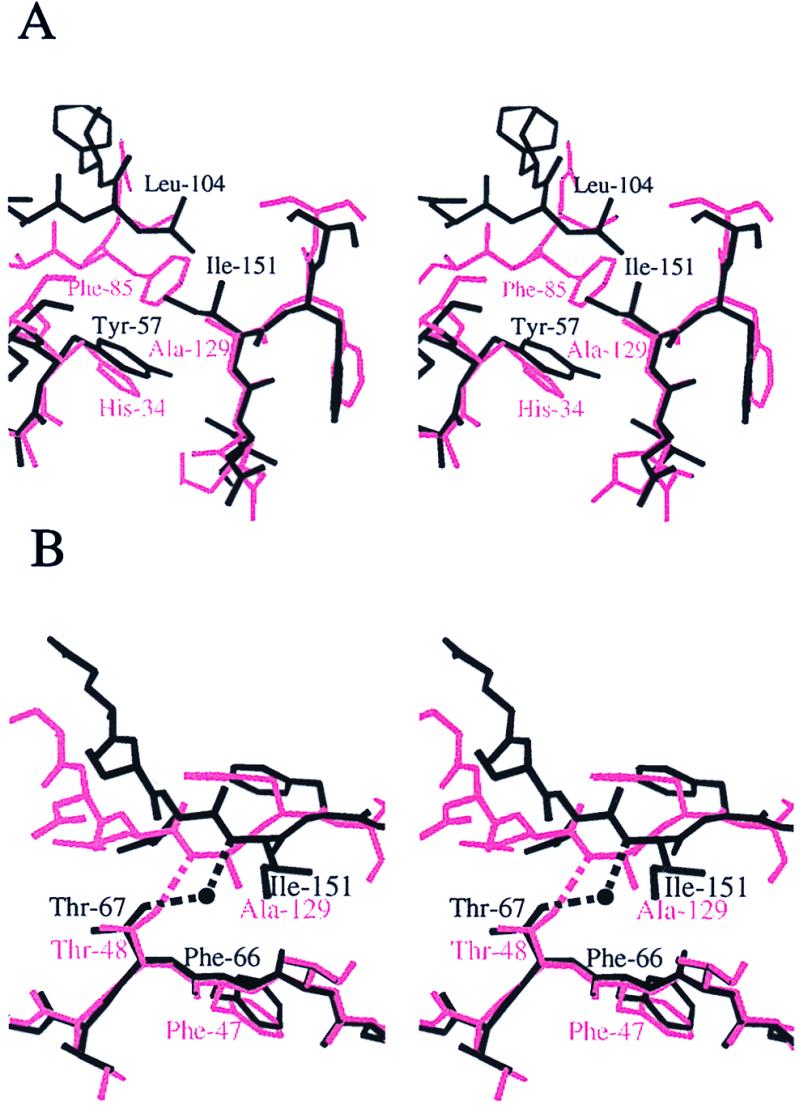
Stereo diagram comparing the subdomain interfaces of Sec18p-N (black) and NSF-N (pink). (A) In NSF-N, the small side chain of Ala-129 allows Phe-85 to pack against His-34. In Sec18p-N, the larger Ile-151 prevents the close approach of Leu-104 to Tyr-57. A hydrophobic cluster is preserved in this region, but the backbone in the vicinity of the linker must shift to accommodate the particular amino acids in the cluster. (B) In NSF-N, the side chain of Thr-48 is in position to form a hydrogen bond directly with the backbone amide of Ala-129. In Sec18p-N, the distance between these atoms is increased, and the hydrogen bond is water-mediated. Figure was generated with molscript (36).
Cysteine Residues in Sec18p-N.
One of the two NEM-modified cysteine residues (Cys-34) in Sec18p-N is located at the end of the first β-strand in Sec18p-NA. The orientations of Cys-34 and Pro-35 are different from that seen for the comparable residues in NSF-N, Cys-11 and Pro-12, with the α-carbon of Sec18p-N Pro-35 located 5.2 Å from the α-carbon of NSF Pro-12 when Sec18p-NA is aligned with NSF-NA. It is not clear whether this difference is the result of the presence of the NEM or is rather an inherent difference between the yeast and hamster proteins. The backbone conformation around the other modified cysteine residue in Sec18p-N, Cys-146, is similar to that around the comparable residue, Gln-124, in NSF. Both cysteine residues in Sec18p-N are partially solvent exposed. The distance between the two sulfur atoms is approximately 15 Å, however, and the shortest path between them passes through helix α1. Therefore, the presence of a disulfide bond between these two cysteines would require a degree of unfolding or a different orientation of the two subdomains with respect to one another.
Discussion
Three molecules per asymmetric unit were observed in both the Sec18p-N crystals and in one report of NSF-N crystals (20). The arrangements of the three molecules differ, however, between the two cases. NSF-N protomers are related by a 3-fold noncrystallographic rotation axis, whereas the packing observed for Sec18p-N more closely resembles a 3-fold screw axis. Although a physiological interaction between N-terminal domains cannot be ruled out, there is to date no evidence that the Sec18p or NSF N-terminal domains self-associate during the catalytic cycle.
Comparison of the three independent copies of Sec18p in the crystal asymmetric unit reveals that the domain contains a region of structural heterogeneity. A concerted displacement of the linker region between the two subdomains (residues 104–113) and the loop between strands β11 and β12 (residues 175–186), at the top of Sec18p-NB as viewed in Fig. 2, suggests that this region is flexible. Electron density corresponding to the loop between strand β7 and helix α3 (residues 125–133) was poor or absent, and this region could not be traced in any of the three molecules. However, the flexibility of this second loop is most likely an artifact of the choice of C terminus of the expressed Sec18p-N domain. The NSF-N structures contain 12 additional residues at the C terminus (20, 21); these residues pack against the loop between strand β7 and helix α3 and would be expected to stabilize its structure.
The regions that are flexible in the Sec18p-N structure are part of a larger area on the protein surface that has diverged evolutionarily between S. cerevisiae Sec18p-N and hamster NSF (20, 21) (Fig. 3). This region includes the linker between the two subdomains, a small loop in the NA subdomain and a large loop in NB. In addition, the N-terminal extension of 19 residues that is present in S. cerevisiae Sec18p but absent in mammalian NSF would be expected to contribute at least partially to this face. Because the D1 and D2 domains are highly conserved between yeast and hamsters, it is likely that regions of the N-terminal domain aside from the intersubdomain linker and the other heterogeneous loops form the major interactions with the rest of the hexamer.
The remaining faces of Sec18p-N and NSF-N are structurally more similar to one another. The site of interaction between Sec18p/NSF and SNAPs has been proposed to lie far from the region of conformational variability, in a basic groove between the two subdomains lined by helices α1 and α2 from NA and strand β10 from NB (21). Interestingly, full-length Sec18p treated with the nonhydrolyzable ATP analog AMP-PNP binds to cation exchange columns, whereas protein under conditions in which nucleotide hydrolysis can occur does not (unpublished observations). In other words, the structural changes that occur in Sec18p/NSF upon ATP binding and result in exposure of SNAP-binding regions correlate with exposure of basic surface area. This observation is consistent with the basic groove on the structurally conserved face of Sec18p/NSF being a binding site for the acidic SNAP C-terminal helix (21, 34).
Upon ATP hydrolysis, Sec18p/NSF disassembles the 20s complexes and releases the SNAPs. A number of models can be proposed for the mechanism of SNAP release, including hiding of the basic groove by rigid-body motion of the N-terminal Sec18p/NSF domains or breaking of the interface between the NA and NB subdomains and eliminating the basic groove altogether. The likelihood of the second model can be discussed in terms of the Sec18p-N structure. On one hand, the flexibility of the intersubdomain linker, as evidenced by the differing conformations of the linker region between Sec18p-N molecules in the crystal asymmetric unit, would be expected to allow the subdomains to change relative orientation. On the other, the linker itself does not appear to constrain the subdomains in the observed orientation, suggesting that the subdomain interface present in Sec18p-N is favored because of the packing interactions. Although the interface residues are not among the most highly conserved in the domain, the overall character of the interface, a mixture of hydrophobic, hydrogen-bonding, and water-mediated contacts, is similar in both Sec18p-N and NSF-N. Furthermore, certain amino acid residues in the interface of these structures appear to have covaried during evolution (Fig. 5A). The slight difference in orientation of the subdomains with respect to one another in Sec18p-N as compared with NSF-N is likely to be the result of sequence differences near the interface and does not represent two alternate conformations that the domains from each species can populate. Whether, in the context of the intact hexamer, the Sec18p and NSF N-terminal domains can indeed assume different subdomain arrangements remains to be determined.
Acknowledgments
We thank M. Mevarech and J. Sussman for valuable suggestions and encouragement, J. M. Berger and F. Frolow for providing data collection facilities, H. Rozenberg for advice on crystal freezing, and H. Greenblatt for assistance with refinement. This research was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities. D.F. is incumbent of the Lilian and George Lyttle Career Development Chair.
Abbreviations
- NSF
N-ethylmaleimide-sensitive fusion protein
- SNAP
soluble NSF-attachment protein
- NEM
N-ethylmaleimide
Footnotes
Data deposition: The atomic coordinates and data have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1CR5).
References
- 1.Novick P, Ferro S, Schekman R. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 2.Eakle K A, Bernstein M, Emr S D. Mol Cell Biol. 1988;8:4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson D W, Wilcox C A, Flynn G C, Chen E, Kuang W-J, Henzel W J, Block M R, Ullrich A, Rothman J E. Nature (London) 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- 4.Graham T R, Emr S D. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hohl T M, Parlati F, Wimmer C, Rothman J E, Söllner T H, Engelhardt H. Mol Cell. 1998;2:539–548. doi: 10.1016/s1097-2765(00)80153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson D W, Whiteheart S W, Wiedmann M, Brunner M, Rothman J E. J Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söllner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 8.Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 9.Otter-Nilsson M, Hendriks R, Pecheur-Huet E-I, Hoekstra D, Nilsson T. EMBO J. 1999;18:2074–2083. doi: 10.1093/emboj/18.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tagaya M, Wilson D W, Brunner M, Arango N, Rothman J E. J Biol Chem. 1993;268:2662–2666. [PubMed] [Google Scholar]
- 11.Fleming K G, Hohl T M, Yu R C, Müller S A, Wolpensinger B, Engel A, Engelhardt H, Brünger A T, Söllner T H, Hanson P I. J Biol Chem. 1998;273:15675–15681. doi: 10.1074/jbc.273.25.15675. [DOI] [PubMed] [Google Scholar]
- 12.Hanson P I, Roth R, Morisaki H, Jahn R, Heuser J E. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 13.Beyer A. Protein Sci. 1997;6:2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel S, Latterich M. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- 15.Neuwald A F, Aravind L, Spouge J L, Koonin E V. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 16.Whiteheart S W, Rossnagel K, Buhrow S A, Brunner M, Jaenicke R, Rothman J E. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenzen C U, Steinmann D, Whiteheart S W, Weis W I. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- 18.Yu R C, Hanson P I, Jahn R, Brünger A T. Nat Struct Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 19.Nagiec E E, Bernstein A, Whiteheart S W. J Biol Chem. 1995;270:29182–29188. doi: 10.1074/jbc.270.49.29182. [DOI] [PubMed] [Google Scholar]
- 20.May A P, Misura K M S, Whiteheart S W, Weis W I. Nat Cell Biol. 1999;1:175–182. doi: 10.1038/11097. [DOI] [PubMed] [Google Scholar]
- 21.Yu R C, Jahn R, Brünger A T. Mol Cell. 1999;4:97–107. doi: 10.1016/s1097-2765(00)80191-4. [DOI] [PubMed] [Google Scholar]
- 22.Looman A C, Bodlaender J, Comstock L J, Eaton D, Jhurani P, de Boer H A, van Knippenberg P H. EMBO J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doering D S. Ph.D. thesis. Cambridge: Massachusetts Institute of Technology; 1992. [Google Scholar]
- 24.Edelhoch H. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computing Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 26.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 276. Boston: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Fortelle Ed-L, Bricogne G. In: Methods in Enzymology. Carter C W Jr, Sweet R M, editors. Vol. 276. Boston: Academic; 1997. pp. 472–494. [DOI] [PubMed] [Google Scholar]
- 28.Jones T A. In: Molecular Replacement. Dodson E J, Gover S, Wolf W, editors. Warrington, U.K.: SERC Daresbury Laboratory; 1992. pp. 91–105. [Google Scholar]
- 29.Cowtan K. Joint CCP 4 ESF-EACBM Newsl Protein Crystallogr. 1994;31:34–38. [Google Scholar]
- 30.Jones T A, Zou J Y, Cowan S, Kjeldgaard M. Acta Crystallogr A. 1991;42:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W. Acta Crystallogr A. 1976;32:922–923. [Google Scholar]
- 33.Castillo R M, Mizuguchi K, Dhanaraj V, Albert A, Blundell T L, Murzin A G. Structure (London) 1999;7:227–236. doi: 10.1016/s0969-2126(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 34.Rice L M, Brünger A T. Mol Cell. 1999;4:85–95. doi: 10.1016/s1097-2765(00)80190-2. [DOI] [PubMed] [Google Scholar]
- 35.Barnard R J O, Morgan A, Burgoyne R D. J Cell Biol. 1997;139:875–883. doi: 10.1083/jcb.139.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraulis P. J Appl Crystallogr. 1991;24:924. [Google Scholar]
- 37.Carson M. J Appl Crystallogr. 1991;24:958. [Google Scholar]
- 38.Howell L, Smith D. J Appl Crystallogr. 1992;25:81–86. [Google Scholar]