Abstract
We present evidence that the sporulation protein SpoIVFB of Bacillus subtilis is a member of a newly recognized family of metalloproteases that have catalytic centers adjacent to or within the membrane. SpoIVFB is required for converting the membrane-associated precursor protein, pro-σK, to the mature and active transcription factor σK by proteolytic removal of an N-terminal extension of 20 amino acids. SpoIVFB and other family members share the conserved sequence HEXXH, a hallmark of metalloproteases, as well as a second conserved motif NPDG, which is unique to the family. Both motifs, which are expected to form the catalytic center of the protease, overlap hydrophobic segments that are predicted to be separate transmembrane domains. The only other characterized member of this family of membrane-embedded metalloproteases is the mammalian Site-2 protease (S2P), which is required for the intramembrane cleavage of the eukaryotic transcription factor sterol regulatory element binding protein (SREBP). We report that amino acid substitutions in the two conserved motifs of SpoIVFB impair pro-σK processing and σK-directed gene expression during sporulation. These results and those from a similar analysis of S2P support the interpretation that both proteins are founding members of a family of metalloproteases involved in the activation of membrane-associated transcription factors. Thus, the pathways that govern the activation of the prokaryotic transcription factor pro-σK and the mammalian transcription factor SREBP not only are analogous but also use processing enzymes with strikingly homologous features.
Keywords: sporulation, σ factor, metalloprotease, membrane protein, HEXXH
Sequestration and regulated proteolysis of transcription factors is emerging as a theme in the control of gene expression in both prokaryotes and eukaryotes. Two well characterized examples are the prokaryotic transcription factor σK (1) and the mammalian sterol regulatory element binding protein (SREBP) (2). σK plays a central role in the regulation of gene expression during the process of sporulation in the Gram-positive bacterium, Bacillus subtilis. Sporulation takes place in a two-chamber sporangium, which consists of a large cell called the mother cell and a small cell called the forespore. The two cells initially lie side by side, but later in development the forespore becomes wholly engulfed by the mother cell to create a cell within a cell (1). σK is produced after engulfment and is responsible for directing gene expression in the mother cell. Its activity, however, is linked to transcription in the forespore by an intercellular signal transduction pathway, which ensures that gene expression in one compartment is coordinated with gene expression in the other (3, 4). This pathway operates at the level of the conversion of an inactive membrane-associated precursor protein, known as pro-σK, to the mature and active form of the transcription factor (3–5). The activation of pro-σK involves the proteolytic removal of a 20-residue-long N-terminal extension, which results in release of mature σK into the mother-cell cytoplasm (3, 4, 6). The N-terminal pro domain, which is rich in hydrophobic residues, is necessary for membrane association (5). Pro-σK processing is triggered by a putative signaling protein SpoIVB, which is produced in and presumably secreted from the forespore under the control of the late-appearing forespore transcription factor σG (7). The signal transduction pathway resulting in the proteolytic activation of pro-σK is a timing device that delays the appearance of mature σK in the mother cell for a period of about 1 hr (3).
SREBP, on the other hand, is a eukaryotic transcription factor that is involved in the activation of genes involved in cholesterol synthesis and uptake (2). SREBP contains an N-terminal transcription activation domain and a C-terminal regulatory domain separated by two transmembrane helices (8, 9). When sterol levels are high, full-length SREBP is sequestered to the membranes of the endoplasmic reticulum and nuclear envelope with the N- and C-terminal domains facing the cytosol (10, 11). When sterols are depleted, SREBP is activated by a two-step proteolytic cleavage process (12). In the first step, SREBP is cleaved in a sterol-dependent manner at Site-1 in its luminal loop. The N-terminal transcription activation domain, however, remains tethered to the membrane by the first transmembrane helix. In the second step, the transcription activation domain is released from the membrane by cleavage at Site-2, which is located within the first transmembrane segment of SREBP (12, 13). The soluble transcription factor domain then translocates to the nucleus and activates the expression of genes that cause an increase in cellular pools of sterols (2). Thus, the pathways controlling σK and SREBP are analogous in that both transcription factors are derived from membrane-tethered precursors and are activated by regulated proteolysis.
Here we are concerned with the enzyme responsible for converting pro-σK to σK, the integral membrane protein, SpoIVFB. SpoIVFB is produced in the mother cell during the engulfment process and is located solely in the mother-cell membrane that surrounds the forespore (14, 15). Three lines of evidence suggest that SpoIVFB is the pro-σK processing enzyme. First, SpoIVFB is required for processing of pro-σK and for σK-directed gene transcription during sporulation, and no other sporulation protein has been identified that acts downstream of SpoIVFB in the processing pathway (1). Second, the requirement for SpoIVFB in spore formation and in σK-directed transcription can be bypassed by use of a mutated form of the gene for pro-σK from which the pro amino acid coding sequence has been removed (3). This shows that SpoIVFB has no role in sporulation other than in processing. Third, cells that have been engineered to produce pro-σK and a modified form of SpoIVFB during growth are capable of converting the proprotein to mature σK (16). This indicates that SpoIVFB is the only sporulation-specific protein necessary for processing.
As part of an effort to further characterize SpoIVFB, we have discovered that it is a member of a newly recognized family of putative metalloproteases. All members of this family contain the canonical metalloprotease motif, HEXXH, and a second conserved motif unique to this family. These two motifs are predicted to form the catalytic center of the protease. Unlike in other metalloproteases, both motifs overlap hydrophobic regions that are predicted to be separate transmembrane domains, positioning the catalytic center adjacent to or within the membrane. Interestingly, the only other characterized member of this new family of putative membrane-embedded metalloproteases is the Site-2 protease (S2P), which is required for the intramembrane cleavage of SREBP at Site-2 (17). Both SpoIVFB and S2P are predicted to cleave membrane-associated substrates, consistent with the unusual location of the catalytic center and with the interpretation that this is a family of proteases. We have analyzed the effects of amino acid substitutions in many of the conserved residues in SpoIVFB on pro-σK processing, σK activation, and sporulation efficiency. Our molecular genetic analysis, together with those of Rawson et al. (17) and Zelenski et al. (18), supports the interpretation that SpoIVFB and S2P are founding members of a newly recognized family of metalloproteases. Our findings reinforce the view that SpoIVFB is the pro-σK processing enzyme and suggest that the analogous pathways governing the activation of the prokaryotic transcription factor, σK, and the mammalian regulatory protein, SREBP, use processing enzymes that share homologous features.
Materials and Methods
Database Analysis.
The complete amino acid sequence of SpoIVFB was used to search for signature sequences by using the blocks 10.1 database (19). Proteins with conserved motifs were identified by blast alignments generated by the Genequiz website (http://columba.ebi.ac.uk:8765/gqsrv/submit) (20). These sequence data were used to hand construct a hidden Markov model of the conserved regions by using the hmmer 2.1 package (http://hmmer.wustl.edu) (21). This model was used to search release 113 of the nonredundant GenPept database. An annotated list that includes hydrophobicity plots (22) and topological predictions (23) of all the family members can be found on our website (http://mcb.harvard.edu/losick).
General Methods.
B. subtilis strains used were PY79 (wild type) (24), BSL51 (spoIVFΔ∷cat) (25), and RL13 (SPβ∷gerE-lacZ) (26). Escherichia coli strains used were DH5α and CJ236. Routine growth of B. subtilis and E. coli was as described (16). β-Galactosidase activity was determined during sporulation by resuspension (27, 28). Sporulation efficiency was determined in 36-hr cultures induced to sporulate by exhaustion in DS medium (27).
Plasmid Construction.
Point mutations in spoIVFB were generated by site-directed mutagenesis by using uracil-substituted single-stranded DNA (29) or PCR. Fragments containing the point mutations were sequenced completely and subcloned into the intact spoIVF operon in pLD30 (30). For a complete description of the oligonucleotides used to generate the spoIVFB point mutations and the intermediate and final cloning steps, see http://mcb.harvard.edu/losick.
Strain Construction.
amyE insertion plasmids containing wild-type or spoIVFB mutations were linearized and transformed into PY79 selecting for spectinomycin resistance (SpR). Chromosomal DNAs from SpR amylase negative (Amy−) transformants were used to transform BSL51. Specialized transduction was performed on each strain by using a fusion-bearing derivative of the prophage SPβ (SPβ∷gerE-lacZ) (26). For a complete list of the SpoIVFB mutant strains, see http://mcb.harvard.edu/losick.
Immunoblot Analysis.
At indicated times after the initiation of sporulation by resuspension, 1.0-ml samples were collected. Whole-cell extracts were prepared by resuspension of cell pellets in 50 μl lysis buffer (20 mM Tris, pH7.5/10 mM EDTA/1 mg/ml lysozyme/10 μg/ml DNaseI/100 μg/ml RNaseA/1 mM PMSF/10 μg/ml leupeptin/10 μg/ml pepstatin) and incubation at 37°C for 10 min, followed by addition of 50 μl SDS sample buffer. Equivalent amounts of extract (15–20 μl) based on OD600 were separated by SDS/PAGE on 12% polyacrylamide gels, electroblotted onto Immobilon-P membranes (Millipore), and blocked in 5% nonfat milk in PBS 0.5% Tween-20. The blocked membranes were probed with anti-σK polyclonal antiserum affinity-purified anti-SpoIVFB antibodies or anti-SpoIVFA polyclonal antiserum, as previously described (14, 16).
Results
SpoIVFB Is a Member of a Family of Putative Metalloproteases.
To investigate further the nature of SpoIVFB, we sought to identify conserved motifs that would guide us in determining its function. We searched the blocks 10.1 database (19) for signature motifs in SpoIVFB and identified the metalloprotease signature sequence, HEXXH. Fifteen of thirty different families of metalloproteases contain the HEXXH motif (31). The two histidines are required for metal binding, and the glutamic acid is thought to serve as the catalytic residue by activating a water molecule that is used to cleave the peptide bond.
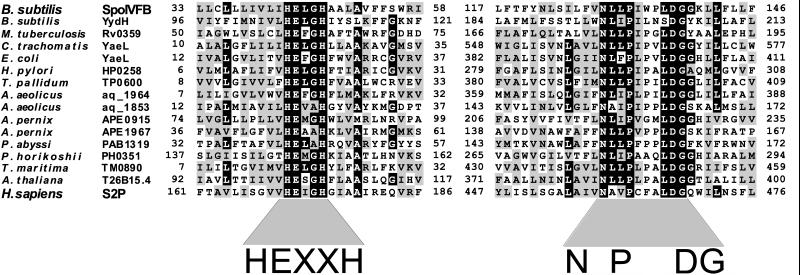
Interestingly, unlike other characterized metalloprotease families, the HEXXH motif in SpoIVFB lies within a large hydrophobic segment that is predicted to be a transmembrane domain (15). The presence of this metalloprotease motif and its location within a putative transmembrane domain were intriguing in light of the predicted role of SpoIVFB in processing the membrane-associated pro-σK. A Genequiz blast search (20) using the complete SpoIVFB sequence indicated that this configuration was not unique to SpoIVFB. We identified a family of proteins similar to SpoIVFB, which all have HEXXH motifs adjacent to or overlapping with putative transmembrane domains. All family members were predicted to be polytopic membrane proteins (see Materials and Methods and http://mcb.harvard.edu/losick) and contained a second conserved motif at varying distances downstream of the HEXXH motif (Fig. 1). We have named this second conserved motif the NPDG motif, because these four residues are virtually invariant among all family members. Like the HEXXH motif, the NPDG motif is adjacent to or overlaps with a stretch of hydrophobic residues predicted to be a separate transmembrane domain. To identify all members of this family, we constructed a hidden Markov model (21) of the two conserved regions and used it to search the GenPept database (see Materials and Methods). Members of this family were identified from the three branches of life, but the overwhelming majority are bacterial. No family members were identified in the completely sequenced genome of Saccharomyces cerevisiae and only one, a homolog of Site-2 protease (see below), was found in the completely sequenced genome of Caenorhabditis elegans. Unknown to us at the time of our analysis, this novel family was also identified by Lewis and Thomas (32) using similar database search programs. These researchers also identified an additional conserved sequence located between the HEXXH and NPDG motifs. This third motif contains a conserved glycine and several hydrophobic residues (32).
Figure 1.
Amino acid sequence comparisons of conserved motifs in some members of the newly recognized family of putative metalloproteases. A black box was assigned if 50% of the family members contained the identical residue at that position. A gray box was assigned if 50% of the family members had a similar residue at that position. Similar residues were I, L, V, and M; F, Y, and W; D and E; N and Q; S and T; A and G; and R and K. Amino acid positions are indicated. Gene name and species of origin are shown (Left). An alignment containing the entire 46-member family can be found on our website (http://mcb.harvard.edu/losick).
The only other characterized member of this newly recognized family of putative metalloproteases with active sites abutting hydrophobic regions is the mammalian S2P (Fig. 1). S2P and SpoIVFB have been implicated in strikingly similar regulatory processes: S2P is required for the proteolytic activation of the sterol-responsive transcription factor SREBP. Mutations in the gene encoding S2P block the intramembrane cleavage of SREBP at Site-2, suggesting that S2P is the Site-2 processing enzyme (17). The identification of this family of putative metalloproteases raises the possibility that the pathways governing the proteolytic activation of pro-σK and SREBP might be based on processing enzymes that share homologous features.
The HEXXH Motif in SpoIVFB Is Required for Pro-σK Processing.
To determine whether the HEXXH motif is required for pro-σK processing, we analyzed the effect of amino acid substitutions in the conserved residues on pro-σK processing in vivo. The amino acid substitution mutants were created by site-directed mutagenesis of the spoIVFB gene (contained within a copy of the two-cistron spoIVF operon). The mutant operon was introduced into the chromosome at the nonessential amyE locus and analyzed in a strain containing a deletion of the endogenous spoIVF operon. The SpoIVFB mutant strains and a matched control strain (harboring a wild-type copy of the spoIVF operon at amyE and the spoIVF deletion mutation) were induced to sporulate, and processing of pro-σK was assessed by immunoblot analysis. In the control strain, mature σK could be detected by 3 hr after the initiation of sporulation and continued to accumulate over the next 2 hr (data not shown). Processing was examined at 5 hr after the start of sporulation in anticipation that some of the mutants would be partially impaired in processing and might not have accumulated detectable levels of mature σK at earlier times. By hour 5, >80% of pro-σK had been converted to mature σK in the control strain harboring wild-type spoIVFB, whereas no processing was detected in a spoIVF null mutant (Fig. 2A). Replacement of the conserved glutamate residue in the HEXXH motif in SpoIVFB with alanine (E44A) or glutamine (E44Q) completely blocked pro-σK processing (Fig. 2A). However, pro-σK processing was unaffected by substitution of Glu-44 with aspartate (E44D) (Fig. 2A). In three metalloproteases (33–35) in which glutamic acid was changed to aspartic acid in the HEXXH motif, protease activity was abolished. Interestingly, when the corresponding amino acid substitution (E172D) was introduced into S2P, as in the case of SpoIVFB, activation of SREBP (and presumably Site-2 processing) was virtually unaffected (17).
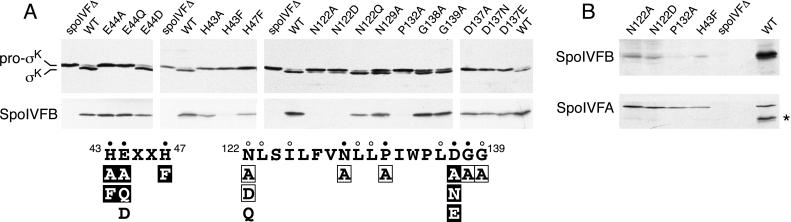
Figure 2.
Pro-σK processing in SpoIVFB mutants. (A) Immunoblots of whole-cell extracts from sporulating cells. All strains analyzed contained a deletion of spoIVF at its normal locus (spoIVFΔ) and a copy of the spoIVF operon at the amyE locus containing either wild-type spoIVFB (WT) or the indicated spoIVFB point mutation. (Upper) Extracts from hour 5 sporulating cells were analyzed by using antibodies that recognize both pro-σK and σK. (Lower) Extracts from hour 3 sporulating cells were analyzed by using antibodies that recognize the C terminus of SpoIVFB (a region that does not contain the two motifs). The results of the analysis are shown schematically below the immunoblots. The amino acid sequences of the two conserved motifs in SpoIVFB are shown in black. Amino acid positions are indicated. Closed and open dots indicate amino acids that are virtually invariant and highly conserved, respectively. Amino acid substitutions that completely abolished pro-σK processing are in black boxes. Mutations that reduced processing efficiency are in black and are surrounded by a box, and those that had no detectable effect on processing are in black. (B) An immunoblot of the four point mutants that had no detectable SpoIVFB protein in A. Twenty-five percent more extract (from hour 3 sporulating cells) was used in this experiment than in those in A. The immunoblot was analyzed by using anti-SpoIVFB antibodies (Upper) and was overexposed to detect the low level of SpoIVFB mutant protein. The immunoblot was reprobed with antibodies that recognize SpoIVFA to control for loading (Lower). The asterisk (*) indicates the position of residual SpoIVFB signal on the reprobed blot.
We also examined the effect of substitutions of the putative metal-coordinating residues in the HEXXH motif. Replacement of the histidines with alanine (H43A) or phenylalanine (H43F, H47F) in the HEXXH motif of SpoIVFB abolished pro-σK processing (Fig. 2A). SpoIVFB levels were examined to determine whether the effect on pro-σK processing could be explained by a reduction in the level of the mutant proteins. SpoIVFB is detectable by 2 hr after the initiation of sporulation and reaches its maximal level at around hour 3 (data not shown). SpoIVFB levels were determined by immunoblot analysis at hour 3 from the same sporulating cultures that were used for analysis of pro-σK processing. Other than the H43F mutant (discussed below), all HEXXH mutants had protein levels that were similar to the level of wild-type SpoIVFB (Fig. 2A). We conclude that the HEXXH motif is important for pro-σK processing, consistent with the idea that SpoIVFB is a metalloprotease.
The NPDG Motif Is Important for Pro-σK Processing.
We then analyzed the requirement for the second conserved motif in SpoIVFB for pro-σK processing. Single amino acid changes were created in many of the highly conserved residues in the NPDG motif (Fig. 2A). Five hours after the initiation of sporulation, the efficiency of pro-σK processing was examined by immunoblot analysis. Almost all the NPDG motif mutants were less efficient than the wild-type control strain in processing pro-σK (Fig. 2A). Only amino acid substitution mutants of D137 were completely impaired in processing. All substitutions analyzed at this amino acid position completely blocked pro-σK processing (Fig. 2A). Importantly, when the corresponding aspartic acid (D467) in S2P was replaced with asparagine, activation of SREBP (and presumably Site-2 cleavage) was also completely abolished (18). HEXXH-containing metalloproteases possess a third residue that acts together with the two histidines to coordinate the active-site metal ion (31, 36). This third coordinating residue can be histidine, glutamate, tyrosine, or aspartate. Frequently, the third coordinating residue is present in the context of a second conserved motif, which is unique to individual families (31, 36). Our mutagenesis studies of SpoIVFB and those of Zelenski et al. (18) on S2P are consistent with the conserved aspartic acid in the NPDG motif being the third metal ligand.
The protein levels in the NPDG motif mutants were examined to determine whether the effect on pro-σK processing could be explained by a reduction in SpoIVFB levels. All but three mutants had protein levels comparable to wild-type SpoIVFB (Fig. 2A). N122A, N122D, and P132A had virtually undetectable SpoIVFB in this assay. Overexposure of an immunoblot containing 25% more whole-cell extract revealed that all three mutants as well as the H43F mutant described above had low but detectable levels of mutant SpoIVFB protein (Fig. 2B). As a control for loading, we reprobed the immunoblot with anti-SpoIVFA antibodies. The SpoIVFA levels were comparable to wild type (Fig. 2B). It is possible that the impaired processing by the N122A, N122D, and P132A mutants is a secondary consequence of their reduced protein levels. However, it is likely that the H43F protein is intrinsically defective in pro-σK processing because the other three mutants, which were present at comparable levels to H43F, were able to process pro-σK, albeit inefficiently. We conclude that the second conserved motif is important for pro-σK processing, and that D137 is essential for converting pro-σK to mature σK.
Analysis of σK Activity in SpoIVFB Mutants.
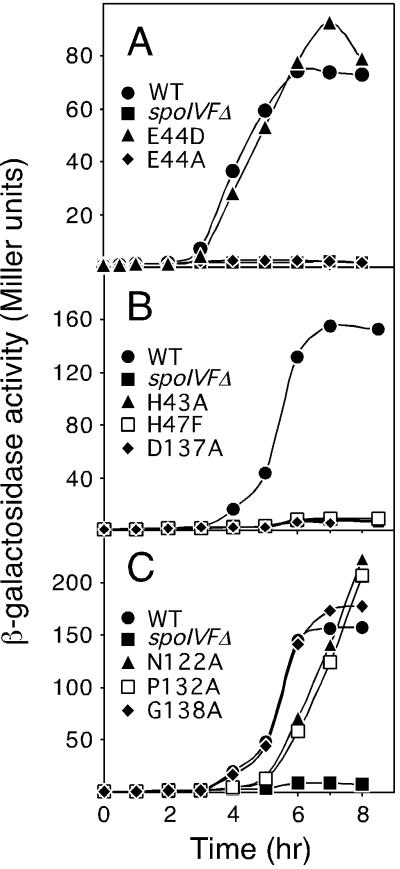
As an independent measure of pro-σK processing, we analyzed σK activity in the SpoIVFB mutants using a σK-responsive promoter (gerE) fused to lacZ (26). This assay allowed us to monitor the effect of the mutations throughout sporulation rather than at a particular time point. Consistent with the immunoblot analysis, changes in the glutamic acid residue in the HEXXH motif to alanine (E44A) or glutamine (E44Q) completely abolished σK activity, whereas σK activity in the E44D mutant was similar to wild type (Fig. 3A and data not shown). All of the amino acid substitutions in the putative metal coordinating ions [the conserved histidines (H43 and H47) in the HEXXH motif and the conserved aspartic acid (D137) in the NPDG motif] completely abolished σK activity (Fig. 3B and data not shown). Interestingly, all of the NPDG mutants that were partially impaired in pro-σK processing had high σK activity (Fig. 3C and data not shown). All the mutants that had protein levels similar to wild-type SpoIVFB (N122Q, N129A, G138A, and G139A) displayed similar kinetics and extent of activation of σK (Fig. 3C and data not shown). The three mutants that had low SpoIVFB protein levels (N122A, N122D, and P132A) were delayed in σK activation by about 1 hr but reached similar levels of β-galactosidase activity compared with wild type (Fig. 3C and data not shown). We conclude that the mutants that were completely blocked in pro-σK processing were also completely blocked in σK activity.
Figure 3.
Analysis of σK-directed β-galactosidase synthesis in SpoIVFB mutant strains. Samples were collected at the indicated times after the initiation of sporulation and assayed for β-galactosidase activity. Each strain contained the gerE-lacZ fusion in the prophage SPβ, a deletion of the spoIVF operon at its normal locus (spoIVFΔ), and a copy of the spoIVF operon at amyE with either wild-type spoIVFB (WT) or a spoIVFB point mutation, as indicated.
Sporulation Efficiency of SpoIVFB Mutants.
Finally, to determine the phenotypic consequences of amino acid substitutions in the two conserved motifs, we analyzed the effect of these mutations on sporulation (see http://mcb.harvard.edu/losick for a table of sporulation efficiencies). Consistent with the immunoblot analysis and the β-galactosidase assays, the HEXXH motif mutants that were blocked in pro-σK processing and σK activity were also severely impaired in sporulation (efficiencies of 0.001%–0.5%). The E44D mutant sporulated as efficiently as wild-type SpoIVFB (85% sporulation efficiency). Interestingly, all of the NPDG mutants that were partially impaired in pro-σK processing, including the mutants N122A, N122D, and P132A, which had very low SpoIVFB protein levels and processed pro-σK inefficiently, sporulated as well as wild type (efficiencies of 79%–100%). Importantly, amino acid substitution mutants of D137 were severely blocked in sporulation (efficiencies of 0.007%–0.03%). On the basis of these findings and those of Fig. 3C, we infer that mature σK is present in excess, and that a low level of processing suffices for a high level of σK-directed gene expression and efficient spore formation.
Discussion
A Family of Membrane-Embedded Metalloproteases.
SpoIVFB is a member of a newly identified family of putative metalloproteases with catalytic pocket adjacent to or within the membrane. All family members contain the canonical HEXXH motif and a second conserved motif that is unique to this family. We have named this second motif the NPDG motif, because these four residues are virtually invariant among family members. The aspartic acid in the NPDG motif is likely to participate in metal coordination at the active site with the two histidines in the HEXXH motif. Unlike any other metalloprotease family, both motifs lie adjacent to, or overlap with, hydrophobic regions that are predicted to be separate transmembrane segments. The only two characterized members of this family are SpoIVFB and S2P. Both proteins are predicted to cleave membrane-associated substrates, consistent with the unusual location of the catalytic pocket and with the interpretation that this is a family of proteases. The mutational analysis of SpoIVFB presented in this study strengthens this interpretation and reinforces the view that SpoIVFB is the pro-σK processing enzyme. Amino acid substitutions in the HEXXH motif of SpoIVFB (except E44D; see below) abolished processing of pro-σK. In the NPDG motif, only mutants of the conserved aspartic acid were completely blocked in processing, a finding consistent with the idea that D137 is part of the active site. Substitutions at other conserved residues in the NPDG motif caused impaired processing, suggesting that these amino acids are important for efficient conversion of pro-σK to mature σK. Our mutational analysis of SpoIVFB is in perfect agreement with the analysis of S2P: SREBP processing was abolished when amino acid substitutions were made in the HEXXH motif of S2P (17). When the aspartic acid in the NPDG motif was changed to asparagine, SREBP activity was lost (18). Taken together, these studies provide strong support to the interpretation that S2P and SpoIVFB are founding members of a new family of metalloproteases. Yu and Kroos, who have independently noted the HEXXH and NPDG motifs in SpoIVFB, have come to similar conclusions (Y. N. Yu and L. Kroos, personal communication).
The only result possibly at variance with the interpretation that SpoIVFB is a metalloprotease is the observation that substitution of the glutamic acid for aspartic acid in the HEXXH motif (E44D) had no effect on pro-σK processing. Although aspartic acid could perform the same water-activating function as glutamic acid in the active site, in the three other studies in which this substitution was analyzed, protease activity was lost (33–35). The observation that the analogous S2P mutant (E172D) retained significant activity (17), and the relatively small number of studies that analyzed Glu to Asp mutants in the HEXXH motif, allay our concern that the activity of the E44D mutant is contradictory to the view that SpoIVFB is a metalloprotease.
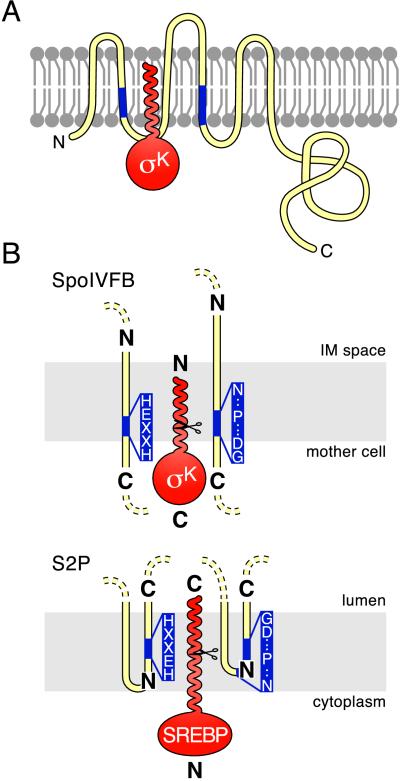
If the aspartic acid in the NPDG motif is involved in metal coordination with the histidines in the HEXXH motif, both conserved motifs should be adjacent to the same face of the membrane. The membrane topology of SpoIVFB has recently been determined in E. coli by using phoA and lacZ fusions (37). Both conserved motifs were predicted to overlap the mother-cell membrane that surrounds the forespore (Fig. 4A). This orientation of the two conserved motifs is consistent with the idea that the HEXXH and NPDG motifs are near each other, as is required if both participate in metal ion coordination. Also, the presumed catalytic center would lie close to the surface of the membrane upon which pro-σK processing is thought to occur (Fig. 4A). If this model is correct, the catalytic center in SpoIVFB must presumably be shielded within the lipid bilayer by other membrane segments in SpoIVFB or associated proteins. Extending the topological studies of Green and Cutting (37), we further hypothesize that the pro domain of σK is inserted into the mother-cell membrane, positioning its cleavage site in the catalytic pocket of SpoIVFB (Fig. 4A). In support of this hypothesis, biochemical and cytological studies indicate that the amino-terminal extension of pro-σK is required for membrane association (5). Also, the pro domain is very hydrophobic, and this hydrophobicity is a conserved feature of the pro sequence of pro-σK homologs from other endospore-forming bacteria (D.Z.R. and R.L., unpublished observations).
Figure 4.
Topological models for SpoIVFB, S2P and their substrates. Membranes are shown schematically in gray. Pro-σK and SREBP are in red. SpoIVFB and S2P are in yellow with their conserved motifs in blue (the invariant residues in the two motifs are shown in B). N and C termini are indicated. (A) Proposed insertion of the pro domain of pro-σK into the membrane. Pro-σK is shown with SpoIVFB. The membrane topology of SpoIVFB is based on the work of Green and Cutting (37). The N-terminal pro domain of pro-σK is drawn as a helix based on its predicted secondary structure (40). The mother-cell cytoplasm is below the membrane, and the space between the mother-cell and forespore membranes (the intermembrane space) is above it. (B) Proposed orientation of the catalytic centers in SpoIVFB and S2P. Relevant membrane segments of SpoIVFB and S2P are shown relative to the mother cell or cytoplasmic face of the membrane and to the intermembrane space (IM) or the lumen. Scissors indicate cleavage sites in pro-σK and SREBP. To accommodate the opposite orientation of the transcription factors, the catalytic centers in the two processing enzymes are shown in opposite orientations relative to the compartment into which the mature transcription factors are released (see text).
A second sporulation σ factor, pro-σE, is also regulated by membrane-sequestration and proteolysis (1). As is the case with pro-σK, membrane association of pro-σE is conferred by its N-terminal pro domain (38, 39). Interestingly, however, the putative processing enzyme of pro-σE, SpoIIGA, does not belong to this family of metalloproteases.
The Catalytic Centers of SpoIVFB and S2P Are Likely to Be in Opposite Orientation.
If our hypothesis (above) that the amino-terminal extension of pro-σK is inserted into the membrane is correct, then the orientation of pro-σK in the membrane must be opposite to that of SREBP relative to the compartment into which the two transcription factors will be released after cleavage (Fig. 4B). In the case of pro-σK, the transcription factor domain is in the C-terminal portion of the proprotein (6), whereas in the case of SREBP the transcription factor domain is in the N-terminal portion of the protein (9). If the two proteins, and hence their cleavage sites, are in opposite orientations, then it is likely that the catalytic centers of S2P and SpoIVFB are also in opposite orientations relative to each other.
The topological analysis of SpoIVFB strongly suggests that the HEXXH motif resides at or near the mother-cell face of the membrane at the C-terminal end of a transmembrane segment (Fig. 4B) (37). In the case of S2P, the topology of the 40-residue hydrophobic segment that contains the HEXXH motif could not be assigned definitively (18). However, it was suggested that the hydrophobic segment that contains the NPDG motif dipped into the membrane without actually crossing it (18). This assignment places the NPDG motif within the membrane and near the N-terminal end of the switch back to the lumen (Fig. 4B). To accommodate the topology data on S2P, given the opposite orientations of the substrates for the two proteases, we propose that this 40-residue hydrophobic segment comprises two transmembrane domains, with the HEXXH motif near the N-terminal end of the second transmembrane segment (Fig. 4B). This topology would position the catalytic center in the opposite orientation relative to that in SpoIVFB (Fig. 4B). As a result, the catalytic pockets of the two processing enzymes would lie in the same orientation relative to the cleavage site of their respective substrates.
The proposed orientation of the HEXXH motif in S2P is appealing for three additional reasons. First, it places the HEXXH motif in S2P entirely within the membrane, an expectation that is consistent with the idea that the predicted cleavage site of SREBP lies within a transmembrane helix and hence within the membrane (12, 13). Second, the HEXXH motif would be oriented in the same direction relative to the NPDG motif in S2P, as is the case in SpoIVFB. Third, it permits the conserved aspartic acid in the NPDG motif to lie in close proximity to the HEXXH motif, consistent with its predicted role in metal coordination. Confirmation of this hypothesis and verification of the functional similarity between S2P and SpoIVFB awaits biochemical reconstitution of processing with the membrane-embedded proteases using purified components.
Acknowledgments
The authors acknowledge P. Stragier and members of the Losick lab for critical reading of the manuscript, D. Green, S. Cutting, Y. Yu, and L. Kroos for communicating results before publication, U. Davé and M. Brown for useful discussions, and R. Hellmiss for help with digital art. This work was supported by National of Institutes of Health Grant GM18568 to R.L. D.Z.R. was supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship, DRG-1514.
Abbreviations
- SREBP
sterol regulatory element binding protein
- S2P
Site-2 protease
References
- 1.Stragier P, Losick R. Annu Rev Genet. 1996;30:297–241. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 2.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Cutting S, Oke V, Driks A, Losick R, Lu S, Kroos L. Cell. 1990;62:239–250. doi: 10.1016/0092-8674(90)90362-i. [DOI] [PubMed] [Google Scholar]
- 4.Lu S, Halberg R, Kroos L. Proc Natl Acad Sci USA. 1990;87:9722–9726. doi: 10.1073/pnas.87.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B, Hofmeister A, Kroos L. J Bacteriol. 1998;180:2434–2441. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kroos L, Kunkel B, Losick R. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 7.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 8.Sakai J, Nohturfft A, Cheng D, Ho Y K, Brown M S, Goldstein J L. J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- 9.Sato R, Yang J, Wang X, Evans M J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- 10.Hua X, Sakai J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1995;270:29422–29427. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 12.Sakai J, Duncan E A, Rawson R B, Hua X, Brown M S, Goldstein J L. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 13.Duncan E A, Dave U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 14.Resnekov O, Alper S, Losick R. Genes Cells. 1996;1:529–542. doi: 10.1046/j.1365-2443.1996.d01-262.x. [DOI] [PubMed] [Google Scholar]
- 15.Cutting S, Roels S, Losick R. J Mol Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 16.Resnekov O, Losick R. Proc Natl Acad Sci USA. 1998;95:3162–3167. doi: 10.1073/pnas.95.6.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M T, Chang T Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 18.Zelenski N G, Rawson R B, Brown M S, Goldstein J L. J Biol Chem. 1999;274:21973–21980. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff J G. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 20.Scharf M, Schneider R, Casari G, Bork P, Valencia A, Ouzounis C, Sander C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI; 1994. [PubMed] [Google Scholar]
- 21.Durbin R, Eddy R, Krogh A, Mitchison G. Biological Sequence Analysis: Probalistic Models of Proteins and Nucleic Acids. Cambridge, U.K.: Cambridge Univ. Press; 1998. [Google Scholar]
- 22.Engelman D M, Steitz T A, Goldman A. Annu Rev Biophys Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 23.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 24.Youngman P, Perkins J B, Losick R. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 25.Lu S, Kroos L. J Bacteriol. 1994;176:3936–3943. doi: 10.1128/jb.176.13.3936-3943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cutting S, Panzer S, Losick R. J Mol Biol. 1989;207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 27.Harwood C R, Cutting S M. Molecular Biological Methods for Bacillus. New York: Wiley; 1990. [Google Scholar]
- 28.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 29.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 30.Garsin D A, Paskowitz D M, Duncan L, Losick R. J Mol Biol. 1998;284:557–568. doi: 10.1006/jmbi.1998.2201. [DOI] [PubMed] [Google Scholar]
- 31.Rawlings N D, Barrett A J. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 32.Lewis A P, Thomas P J. Protein Sci. 1999;8:439–442. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGwire B S, Chang K P. J Biol Chem. 1996;271:7903–7909. doi: 10.1074/jbc.271.14.7903. [DOI] [PubMed] [Google Scholar]
- 34.Vazeux G, Wang J, Corvol P, Llorens C C. J Biol Chem. 1996;271:9069–9074. doi: 10.1074/jbc.271.15.9069. [DOI] [PubMed] [Google Scholar]
- 35.Fujimura K K, Nouvet F J, Michaelis S. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hooper N M. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 37.Green, D. H. & Cutting, S. M. J. Bacteriol., in press.
- 38.Hofmeister A. J Bacteriol. 1998;180:2426–2433. doi: 10.1128/jb.180.9.2426-2433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju J, Luo T, Haldenwang W G. J Bacteriol. 1997;179:4888–4893. doi: 10.1128/jb.179.15.4888-4893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kneller D G, Cohen F E, Langridge R. J Mol Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]