Abstract
Prophenoloxidase, a melanin-synthesizing enzyme, is considered to be an important arthropod immune protein. In mosquitoes, prophenoloxidase has been shown to be involved in refractory mechanisms against malaria parasites. In our study we used Anopheles gambiae, the most important human malaria vector, to characterize the first arthropod prophenoloxidase gene at the genomic level. The complete nucleotide sequence, including the immediate 5′ flanking sequence (−855 bp) of the prophenoloxidase 1 gene, was determined. The gene spans 10 kb and is composed of five exons and four introns coding for a 2.5-kb mRNA. In the 5′ flanking sequence, we found several putative regulatory motifs, two of which were identified as ecdysteroid regulatory elements. Electrophoretic mobility gel-shift assays and supershift assays demonstrated that the Aedes aegypti ecdysone receptor/Ultraspiracle nuclear receptor complex, and, seemingly, the endogenous Anopheles gambiae nuclear receptor complex, was able to bind one of the ecdysteroid response elements. Furthermore, 20-hydroxyecdysone stimulation was shown to up-regulate the transcription of the prophenoloxidase 1 gene in an A. gambiae cell line.
Keywords: insect immunity, gene regulation, development, melanization
The melanizing effects of phenoloxidases were identified nearly 60 years ago in grasshopper eggs (1). Since then, tyrosinase-type phenoloxidases (monophenol, l-dopa: oxidoreductase; EC 1.14.18.1), which are widely distributed in both prokaryotes and eukaryotes, have been characterized as copper-containing enzymes catalyzing two key reactions in the synthesis of melanin (2). Tyrosinase-type phenoloxidase (PO)-mediated melanin synthesis plays a major role in wound healing and the formation of melanotic capsules that sequestrate parasites, parasitoids, and pathogens as they breach the cuticular exoskeleton or midgut epithelium or enter into the hemocoel of the insect (3). The role of PO in melanotic encapsulation has bestowed upon this enzyme the status of “immune protein.” Because melanization/encapsulation can lead to refractoriness of certain Plasmodium species, the genes controlling melanization/encapsulation in Anopheles gambiae are considered to be prime candidates for genetic manipulation of mosquito vectors as a future means of malaria control (4, 5). At present, six prophenoloxidase cDNAs have been cloned and sequenced from A. gambiae (6–8).
Ashida (9) demonstrated that PO from Bombyx mori exists in a zymogen form and is activated via a serine proteinase cascade, which is set into motion by injury or microbial challenge (3, 10). To date, all insect prophenoloxidases (PPOs) seem to follow this activation pattern. However, apart from studies on the hormonal regulation of granular cuticular phenoloxidase synthesis (11) and a very recent study by Müller et al. (8), in which the authors show that 20-hydroxyecdysone (20E) is able to modulate PPO gene expression in A. gambiae cells, little is known about arthropod PPO gene regulation or direct hormonal regulation of insect immune genes.
It is well established that 20E, the principal steroid hormone in arthropods, regulates a wide variety of developmental processes such as molting, metamorphosis, and reproduction (12–14). The identification of ecdysteroid response elements (EcREs) and the ecdysteroid receptor–Ultraspiracle complex (EcR/USP) in Drosophila has greatly advanced our understanding of the molecular basis of 20E action on this insect (15–24). Recently, the characteristics of DNA binding and transactivation of 20E receptor–Ultraspiracle complex were established in the mosquito Aedes aegypti (25), which provided the foundation for further 20E studies in A. gambiae. Very recently in Manduca sexta, Lan et al. (26) also demonstrated selective 20E regulation of a retinoid orphan receptor homolog through the ecdysone receptor heterodimer EcR-B1-USP1 (but not EcR-B1-USP2).
Despite the accumulation of ever more striking evidence showing parallels between insect development and immune response, no immune genes have yet been found to contain EcREs in their promoter sequences. In this paper, we demonstrate that A. gambiae PPO1 (AgPPO1) responds to 20E and that the promoter region of AgPPO1 contains an EcRE that is able to functionally bind to the A. aegypti EcR/USP heterodimer and, seemingly, to its endogenous heterodimer EcR/USP in nuclear extracts of adult A. gambiae.
Materials and Methods
Mosquitoes Rearing.
A. gambiae G3 strain was maintained at 26°C with 75% relative humidity under a 12-hr photoperiod. Adult mosquitoes were provided a 10% honey solution, and females were blood-fed from anesthetized rabbits biweekly. Larvae were fed on Friskies Cat Chow (Rueil–Malmaison, France). A. gambiae GASUA strain was used for pWE15 cosmid construction (27).
Maintenance of A. gambiae Cell Line 4a-3B.
The 4a-3B cells characterized by Müller et al. (8) were maintained at 27°C in Schneider medium with 10% FCS (GIBCO/BRL) in Corning 25-cm2 cell-culture flasks.
Bacterial Injection of A. gambiae, Hormonal, Microbial, and Parasitic Stimulation of 4a-3B Cells.
A. gambiae fourth instar larvae, pupae, and newly emerged adults were pricked in the thorax with a tungsten needle dipped in a mixture (vol/vol) of heat-inactivated Escherichia coli 1106 and Micrococcus luteus A270 and then maintained for 4 or 12 hr at 27°C before mRNA extraction. Confluent 4a-3B cells were treated for 0, 6, 12, and 24 hr with the above bacterial suspension (100 μl/flask), curdlan beads (2.5 mg/flask), live Plasmodium gallinaceum ookinetes resuspended in Schneider medium (4 × 105 ookinetes per flask), and microfilaria of Wuchereria bancrofti (1.25 × 104 microfilaria per flask). 4a-3B cells also were treated with 20E (Sigma) in ethanol at a final concentration of 200 nM for 2, 8, 16, 24, and 48 hr. Wash-out experiments to remove 20E were conducted according to Müller et al. (8).
Screening and Sequencing of AgPPO1 Genomic Clone.
The GASUA cosmid library was constructed by Mathiopoulos et al. (27). The cDNA of AgPPO1 (7) was labeled by random priming and was used to screen the cosmid library. Seven positive clones were identified, and the longest clone was used for structural analysis. The fragments of AgPPO1 gene were subcloned into pBluescript SK and prepared for DNA sequencing by using a Sequenase Version 2.0 kit (Amersham Pharmacia/United States Biochemical) with specific internal oligonucleotide primers.
Reverse Transcription–PCR Analysis.
Except where otherwise noted, all DNA and RNA manipulations were carried out by using standard techniques (28). mRNA was extracted from the various mosquito stages after bacterial injection and from 4a-3B cells after microbial, parasitic, and 20E treatment by using the Oligotex Direct mRNA minikit (Qiagen). First-strand cDNA was synthesized in 20 μl of reaction mixture containing 100 ng of mRNA, 1 mM of each dNTP, 20 units of AMV reverse transcriptase, and 2 μl of oligo(dT) primer (A260 = 0.04). The PCR was performed in a 50-μl reaction mixture containing all cDNAs derived from the reverse transcription reaction, 1.25 units of Thermus aquaticus DNA polymerase, 200 μM of each dNTP, 200 nM of each primer and, 1 mM MgCl2. The specific AgPPO1 primer sequences were sense (5′-TTCGATGCCTCTAACCGGGCGA-3′), antisense (5′-GCGGGATGCGGTTACCGGATTCA-3′), and S7 ribosomal protein (29) sense (5′-GGCGATCATCATCTACGT-3′) and antisense (5′-GTAGCTGCTGCAAACTTCGG-3′). PCR cycle numbers were chosen empirically to obtain comparable band intensities for the different markers in each experiment while avoiding saturation. The number of cycles was constant for a particular sequence in the multiple samples analyzed in a given experiment. PCR was performed under the following conditions: 1 min at 94°C, 1 min at 54°C, and 1 min at 72°C for the amplification of S7 ribosomal protein; 1 min at 94°C, 1 min at 63°C, and 1 min at 72°C for AgPPO1 amplification.
In Vitro Transcription and Translation.
The A. aegypti AaEcR and AaUSP cDNAs were cloned in pGEM-3Z (Promega) under the control of the SP6 polymerase (30). The TNT system (Promega) was used for in vitro transcription and translation of the cDNAs utilizing the SP6 promoter.
Electrophoretic Mobility-Shift Assay (EMSA).
Nuclear cell extracts were prepared from 4-day-old adult A. gambiae, as described previously (31). DNA probes for EMSA were obtained by annealing complementary oligonucleotides and by gel purifying those with electroelution from a 15% nondenaturing PAGE. The oligonucleotides used to generate the A. gambiae PPO1 ecdysteroid response element AgEcRE-60 (only one strand of each probe is shown) were 5′-CTTAAACCGGGGGTGCGTACCATTGACCTTCCCA- ATGCGC-3′. The Drosophila melanogaster hsp27 probe was composed of 5′-AGAGACAAGGGTTCAATGCACTTGTCCAAT-3′. The DR-4 probe was composed of 5′-AAGCGAAAGGTCAAGGAAGGTCAAGGAAAAT-3′. The probes were labeled by backfilling with Klenow fragment by using [α-32P]dATP. EMSA was carried out in a 20-μl volume containing 1 μl of each TNT reaction or 2–4 μg of nuclear extracts, 10 mM Tris⋅HCl (pH 8.0), 50 mM NaCl, 0.5 mM DTT, 0.5 mM EDTA, 1 μg of poly(dI-dC), 4% glycerol, 1 μg of single-stranded DNA (5′-CTAACAAAGTTCGCCTGGACTAGAACGGCC-3′), 0.5 μM 20E (only with nuclear extracts), and, for competition experiments, the indicated amounts of unlabeled competitor oligonucleotides. After 15 min of incubating at 4°C, 1 ng of 32P-labeled DNA probe was added, and the solution was incubated 45 min further at the same temperature. The reaction mixture was resolved by using a 5% nondenaturing PAGE in 0.5× TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at a constant voltage of 150 V for 2 hr at 4°C. The gel was then dried and autoradiographed with an intensifying screen at −70°C. In the antibody-binding supershift experiments, 1 μl of anti-EcR or anti-USP was added to the binding reaction during the first incubation (26).
Results
Organization of the PPO1 Gene.
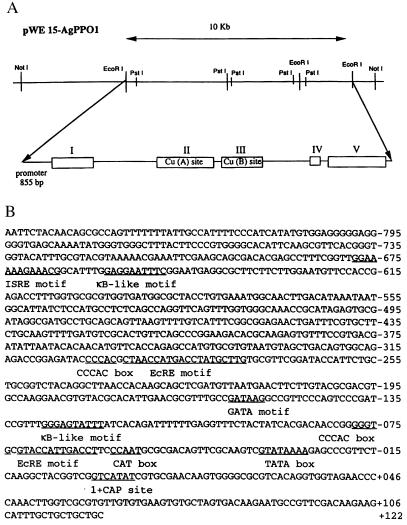
To obtain the complete A. gambiae PPO1 gene, we screened a cosmid library by using the AgPPO1 cDNA (7). Cosmid clone pWE15 was found to contain a 10-kb insert corresponding to the entire AgPPO 1 gene (Fig. 1A). The sequencing showed that the AgPPO 1 gene is composed of five exons separated by four introns (intron I, 2,831 bp; intron II, 74 bp; intron III, 2,175 bp; and intron IV, 101 bp). Exons also varied in sequence length (exon I, 447 bp; exon II, 683 bp; exon III, 440 bp; exon IV, 197 bp; and exon V, 670 bp) (Fig. 1A). The coding sequence of AgPPO 1 is consistent with the earlier-published cDNA sequence (7). Furthermore, the characteristic copper-binding sites, Cu(A) and Cu(B), found in all arthropod PPOs, were located in exon II and exon III, respectively. In addition, we sequenced the 855 bp of the 5′ flanking region (promoter region) (Fig. 1B). A potential insect cap site was situated 27 bp downstream from the TATA box. A polyadenylation signal was localized in the 3′ region of the gene (7).
Figure 1.
Structure of the AgPPO1 gene. (A) Schematic representation of the cosmid clone pWE15-AgPPO1. Complete nucleotide sequence of 10 kb spanning the entire AgPPO1 has been deposited in the GenBank database (accession no. AF031626). Exons are indicated by open boxes with roman numerals. Copper-binding sites [Cu (A) site and Cu (B) site] are situated in exons II and III, respectively. (B) The nucleotide sequence of the AgPPO1 promoter region. Various putative cis-regulatory elements are underlined with their names.
The upstream sequence of the AgPPO1 gene contained three putative immune regulatory motifs: GGAAAAAGAAACG, a putative motif conforming to the mammalian complete IFN-stimulated responsive element consensus (GGAAANNGA- AANN) located at −666; two putative κB-like motifs, GGGAGTATTT and GAGGAATTTC at −119 and at −649, respectively; and a GATA motif, GATAAG at position −153 (Fig. 1B). In addition, we detected two putative EcREs located at −60 and −276 immediately flanked by CCCAC boxes (Fig. 1B) known to work synergistically with nuclear receptors of glucocorticoids in mammals (32, 33).
PPO1 Expression.
To investigate the inducibility of AgPPO1 mRNA after microbial and parasitic challenge, AgPPO1 immunoresponsive 4a-3B cells were challenged with heat-treated Gram +/− bacteria, curdlan beads, live P. gallinaceum ookinetes, and W. bancrofti microfilaria. Furthermore, A. gambiae of various developmental stages were pricked with Gram +/− bacteria. Reverse transcription–PCR analysis showed that no up-regulation of the AgPPO1 mRNA could be detected in any of the microbial or parasitic treatments in the 4a-3B cells (data not shown). In fact, slight down-regulation was observed in bacterial- and curdlan-treated cells (data not shown). Similarly, bacteria-challenged larvae, pupae, or newly emerged adults at 4 and 12 hr postinjection did not manifest any AgPPO1 mRNA up-regulation (data not shown). However, AgPPO1 is constitutively expressed in larvae and pupae, and very low constitutive expression was detected in adults (7).
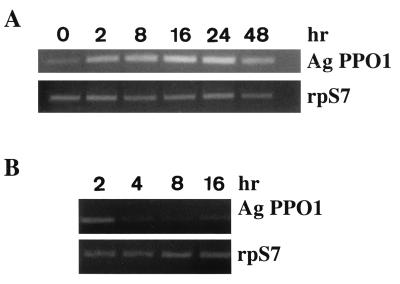
After identifying two putative EcREs in the promoter region, we examined whether the A. gambiae PPO-producing cell line 4a-3B could respond to physiological concentrations of 20E (200 nM). There was a noticeable increase in AgPPO1 expression 2 hr after 20E stimulation. AgPPO1 expression continued to increase for up to 24 hr and then decreased (Fig. 2A). When the 4a-3B cells were stimulated for 16 hr and 20E subsequently was removed by washing the cells twice with culture medium alone, AgPPO1 transcription decreased after 2 hr and returned to basal levels after 16 hr (Fig. 2B). These results demonstrate that AgPPO1 can be regulated by 20E in vitro.
Figure 2.
Differential effects of 20E on transcription of AgPPO1 gene in 4a-3B cells. (A) Kinetics of AgPPO1 expression after ecdysone stimulation. Cells were exposed to 200 nM 20E for various periods of time (0, 2, 8, 16, 24, and 48 hr). Induction of AgPPO1 mRNA was analyzed by reverse transcription–PCR (RT-PCR) (rpS7, 20 cycles; AgPPO1, 30 cycles). (B) Effect of ecdysone removal on AgPPO1 expression. After 16 hr of 20E treatment (200 nM), cells were washed twice with culture medium and incubated further in fresh medium for various times (2, 4, 8, and 16 hr). RT-PCR was performed as above.
The Promoter Region of AgPPO 1 Gene Contains One Functional Binding Site for EcR/USP.
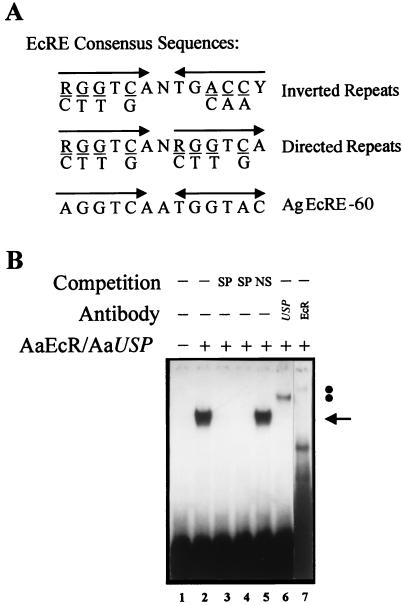
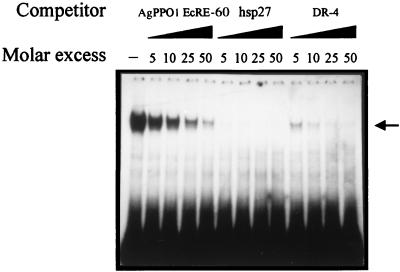
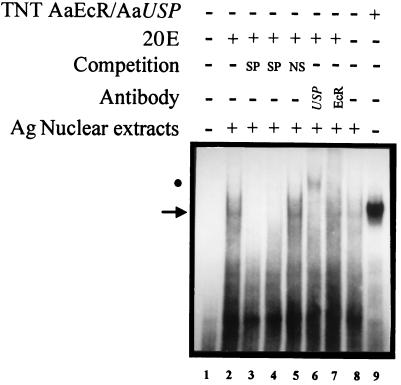
The above results prompted us to investigate whether these AgEcREs could serve as binding sites for the A. aegypti 20E receptor/Ultraspiracle nuclear receptor complex. Using a labeled EcRE from the D. melanogaster hsp27 gene (18) as a probe, the AgEcRE-60 was able to compete for the binding of the in vitro-translated functional AaEcR/AaUSP heterodimer, whereas the AgEcRE-276 did not bind well in preliminary experiments (data not shown). Hence, we focused our functional study on AgEcRE-60. A detailed inspection of the nucleotide sequence of AgEcRE-60 showed that its sequence matches 10 of 13 positions with the inverted (IR) EcRE consensus and 10 of 13 with the directed (DR) consensus sequence (Fig. 3A). To test whether this putative AgEcRE-60 was able to directly bind a 20E receptor, we performed EMSA experiments by using in vitro-produced AaEcR/AaUSP with the AgEcRE-60 as a probe. As shown in Fig. 3B (lane 2), incubation of the AgEcRE-60 with AaEcR/AaUSP resulted in a single retarded band. The specificity of the complex was confirmed by competition with oligonucleotides containing related (Fig. 3B, lanes 3 and 4) and unrelated (Fig. 3B, lane 5) sequences. Moreover, the identity of the complex was confirmed to be due to the AaEcR/AaUSP heterodimer by using either anti-EcR or anti-USP, both of which were able to supershift the DNA–protein complex (Fig. 3B, lanes 6 and 7). To characterize the affinity of the AgEcRE-60, we carried out competition analysis by using AaEcR/AaUSP with different concentrations of three unlabeled oligonucleotides containing different EcREs (Fig. 4). In previous studies, Wang et al. (25) showed that AaEcR/AaUSP bound to a broad spectrum of binding sequences including IR and DR repeats of the half-site consensus. In this context, we used the D. melanogaster hsp27 EcRE as a high-affinity element with an IR structure and a perfect DR-4 as the most efficient DR EcRE (25). As expected, both hsp27 and DR-4 exhibited higher binding affinity than the AgEcRE-60 (Fig. 4). Although more than 95% of the labeled AgEcRE-60 was competed in the presence of a 10-fold molar excess of the unlabeled hsp27 and DR-4, 25–30% still remained in the presence of the same molar excess of unlabeled AgEcRE-60.
Figure 3.
Binding of the heterodimer AaEcR/AaUSP to the AgEcRE-60. (A) Comparison of the AgEcRE-60 motif of A. gambiae with EcRE IR or DR half-site consensus sequences originating from D. melanogaster. R, purine residues; Y, pyrimidines; and N, any nucleotide. (B) EMSA was performed with an in vitro-translated AaEcR/AaUSP and AgEcRE-60 as a probe. The retarded complex, resulting from the specific interaction between AaEcR/AaUSP and AgEcRE-60, is indicated by an arrow (lane 2). One hundred-fold molar excess of unlabeled probe (lane 3) or hsp27 EcRE (lane 4) was included as a specific competitor (SP). The same excess of unlabeled double-stranded oligonucleotide of unrelated sequence (NS) was included in lane 5. The complex was supershifted by anti-Drosophila USP (lane 6) and anti-A. aegypti EcR (lane 7). The positions of each supershifted complex are indicated by solid circles.
Figure 4.
Relative affinity of the AgEcRE-60 for the AaEcR/AaUSP complex. Binding of the heterodimer AaEcR/AaUSP to the AgEcRE-60 in the presence of varying amounts of unlabeled competing probes (self-competition, hsp 27, and DR-4) was conducted to compare the binding affinity of the AgEcRE-60 with respect to known EcREs. EMSAs were performed with AaEcR/AaUSP and AgEcRE-60 as a probe in the absence (−) or presence of increasing amounts of AgEcRE-60, hsp27 EcRE, or DR-4. The position of the complex formed by the heterodimer AaEcR/AaUSP is indicated by an arrow.
After establishing the ability of the AgEcRE-60 to bind AaEcR/AaUSP, we became interested in seeing whether this binding site was able to serve as an in vivo EcRE in A. gambiae. We therefore tested the ability of AgEcRE-60 to bind proteins present in nuclear extracts of adult A. gambiae. A specific complex was detected under these conditions (Fig. 5, lane 2). This complex seemingly represents the natural EcR/USP of A. gambiae (AgEcR/AgUSP) because its specificity was confirmed by competition analysis (Fig. 5, lanes 3–5) and its identity was confirmed by USP antibodies, which specifically supershifted the complex (Fig. 5, lane 6). However, EcR antibody was unable to supershift the complex (Fig. 5, lane 7) (also see Discussion). We also confirmed that, in the absence of 20E, the binding affinity of AgEcR/AgUSP to AgEcRE-60 clearly was diminished in the binding reaction (Fig. 5, lane 8). Moreover, the mobility of the A. gambiae EcR/USP complex exhibited the same mobility as that of the in vitro-translated AaECR/AaUSP (lane 9).
Figure 5.
Nuclear protein extracts from A. gambiae contain the ecdysone receptor complex EcR/USP. Nuclear extracts from whole adults of A. gambiae were examined by EMSA with AgEcRE-60 as a probe. The retarded complex resulting from the specific interaction between AgEcR/AgUSP and AgEcRE-60 is indicated by an arrow (lane 2). One hundred-fold molar excess of unlabeled probe (lane 3) or hsp27 EcRE (lane 4) was included as a specific competitor (SP). The same molar excess of unlabeled double-stranded oligonucleotide of unrelated sequence (NS) was included in lane 5. The supershift assay was performed with anti-USP (lane 6) and anti-EcR (lane 7). The effect of the absence of 20E in the binding reaction is presented in lane 8. As a control, a binding reaction using in vitro-translated AaEcR/AaUSP was included (lane 9). The positions of the supershifted complex (USP) is indicated by a solid circle. It should be noted that the antibodies are derived from species different than A. gambiae (26); anti-USP is derived from D. melanogaster and the anti-EcR is derived from A. aegypti, which could explain the partial supershift of the A. gambiae double-stranded oligonucleotide.
Discussion
Our study describes the genomic structure of an arthropod prophenoloxidase gene. The genomic structure revealed that AgPPO1 gene is composed of five exons and four introns of variable lengths. The putative prophenoloxidase-activating enzyme cleavage site NRFG is situated in exon 1. Interestingly, the two highly homologous copper-binding sites, CuA and CuB, are encoded by two separate exons (exons 2 and 3), which could have resulted from a possible gene duplication of the copper-binding site. This result may have some evolutionary significance in that it has been reported that the copper-binding domains of arthropod PPOs have evolved from those of an ancestral arthropod hemocyanin (34–36). All arthropod hemocyanins are structurally similar and can be divided into three domains (37). In arthropod hemocyanins, the CuA- and CuB-binding sites are always located in domain 2 (37). Amino acid sequence alignment between AgPPO1 and the arthropod hemocyanins shows that AgPPO1 has corresponding domains. Like arthropod hemocyanins, the copper-binding sites (CuA and CuB) of AgPPO1 are also located in domain 2. The localization of the copper-binding site in the same domain further establishes a phylogenetic link between these copper proteins. Furthermore, AgPPO1 also has four introns and five exons and 25% of sequence identity to the hemocyanin-related, insect-storage-protein genes (38).
Primary structure of the promoter region of the AgPPO1 gene allowed us to identify putative immune regulatory motifs: two κB-like motifs and a GATA motif commonly found in the promoters of inducible immune genes of insects (39), as well as putative complete IFN-stimulated responsive elements (ISRE) not yet detected in insect immune genes, although ISRE half-sites have been detected in the promoters of the Drosophila diptericin gene and the peptidoglycan recognition protein gene in Bombyx mori (40, 41). However, in AgPPO1, these immune regulatory motifs appear nonfunctional, at least under the experimental conditions tested, because microbial or parasitic challenge could not induce any AgPPO1 transcriptional activity. Our results corroborate those obtained by Müller et al. (8), who showed that bacteria-challenged PPO-producing A. gambiae cells did not up regulate any of the six PPO genes, whereas other immune-responsive genes encoding Gram-negative-binding protein (GNBP) and defensin were up-regulated (8). The lack of PPO-gene up-regulation after bacteria challenge already has been documented in the fall webworm, Hyphantria cunae (42). In contrast to bacteria, microfilarial-worm (Onchocerca spp.) injection into field-collected blackflies Simulium damnosum s.l. was shown to induce PPO gene up-regulation (43). On the contrary, microfilarial-worm (Dirofilaria immitis) inoculation into the mosquito Armigeres subalbatus did not provoke any PPO gene up-regulation (44). Like A. subalbatus inoculated with D. immitis microfilaria, A. gambiae 4a-3B cells were unable to up-regulate AgPPO1 when inoculated with W. bancrofti microfilaria. The failure of microbial cell wall components and parasites to up-regulate AgPPO1 does not necessarily diminish its putative immune protein function. Unlike most other immune effector molecules, prophenoloxidases are constitutively expressed during different developmental stages and are maintained as inactive zymogens in the hemolymph or cuticle until activated by microbial cell wall components and parasites via the prophenoloxidase cascade (3). Constitutive synthesis of an immune protein zymogen may be even more effective in the insect's resistance to microbial/parasitic invasion, because mRNA transcription and de novo synthesis of prophenoloxidase (80 kDa) would require a much longer period of time than an antibacterial peptide such as cecropin (4 kDa).
Apart from its role in insect immunity gene regulation, the GATA motif also has been implicated in the hormonal regulation of insect developmental genes (45). Recently, Dittmer and Raikhel (46) identified functional GATA motifs along with potential hormone-response elements in the upstream sequences of a cathepsin D-like aspartic protease cDNA from the mosquito A. aegypti. However, the potential hormone-response elements were unable to bind A. aegypti EcR/USP (46).
In addition to the GATA motif, we also found two putative EcREs in the promoter of AgPPO1. At present, the consensus sequence of EcREs has been identified only in D. melanogaster and Manduca sexta ecdysteroid-responsive genes (26, 30). In the present study, we show that physiological concentrations of 20E (200 nM) could up-regulate AgPPO1 gene expression in PPO-producing 4a-3B cells, further suggesting that these putative EcREs could play a functional role in AgPPO1 gene regulation. It is well known that 20E flux regulates essential processes in mosquito development, molting, metamorphosis, reproduction, and, in particular, vitellogenesis 24 hr after a blood meal (25, 30). Very recently, Müller et al. (8) showed in A. gambiae 24 hr after blood feeding (i.e., during egg formation) that several PPO genes, including PPO1, were up-regulated and that PPO5 gene was down-regulated, whereas PPO6 expression was not affected by blood feeding.
To better understand the mechanism of 20E responsiveness in the AgPPO1 gene, we conducted a functional study of the putative EcREs. The analysis of the AgEcRE-60 shows that it can act as an IR-1 (it matches 10 of 13 positions regarding the consensus) or as a DR-1 (10 of 13 positions). In this sense, several groups demonstrate that the heterodimer EcR/USP can bind to a different repertoire of EcREs including inverted and directed repeats (20, 21, 25). Although most of the natural EcREs described in D. melanogaster show an IR structure [hsp27 (18); Fbp1 (47); Sgs-4 (48); Lsp-2 (49); Eip 28/29 (50); hsp23 (51)], D'Avino et al. (21) demonstrated that in ng genes the natural EcRE is a DR-12. In addition to these findings regarding Drosophila, Lan et al. (26) recently has identified one putative IR-1 EcRE in Manduca sexta, which was able to bind to the EcR/USP complex. In mosquitoes, two EcREs were discovered in the promoter regions of vitellogenin and vitellogenic carboxypeptidase and two vitellogenic genes of the yellow fever mosquito A. aegypti, which present DR-1 and DR-2 structures, respectively (A.A.M. and A.R., unpublished results). Several lines of evidence showed that these A. aegypti EcREs are acting as truly functional elements. It has been demonstrated that either IR or DR can mediate ecdysteroid responsiveness in insects, as well as in mammalian cells (20, 25), and that the level of ecdysone-dependent transactivation from the presence of different DR and IR EcREs is correlated with their binding affinities (25). It is tempting to speculate that, in our case, AgPPO1 gene expression can be achieved as a result of synergistic hormone induction (AaEcR/AaUSP), transcription factors like NF-κB, and tissue-specific factors such as GATA and CACCC. Indeed, a putative CACCC element is tightly clustered with AgEcRE-60 and a putative GATA-binding site is situated 93 bp upstream.
Significantly, the EcRE described here was able to bind not only to in vitro-produced EcR and USP proteins but also to what appears to be the heterodimeric receptor present in the nuclear extracts of A. gambiae. The existence of this receptor already has been described in Drosophila (49, 51), A. aegypti (25), and M. sexta (26), among others. Although the heterologous antibodies raised against EcR of A. aegypti were unable to supershift the A. gambiae heterodimer, the identity of the complex was partially confirmed through the EcR/USP complex by using heterologous antibodies raised against D. melanogaster USP and by the clear enhancement of the intensity of the complex because of the presence of 20E in the binding reaction. Previously, it has been demonstrated that 20E increases the binding of the heterodimeric EcR/USP to EcRE sequences (23). Because USP alone is not able to bind 20E (23), it appears that the binding activity detected in nuclear extracts of A. gambiae is due to the heterodimer EcR/USP. The above results strongly suggest the functionality of this EcRE in A. gambiae and the role of 20E in AgPPO1 gene regulation. We assume that AgPPO1 participates in melanin synthesis as other phenoloxidases, but given that the most intense transcriptional activity is observed during embryo formation and egg maturation, we question whether it could participate in other physiological processes. Further investigation is warranted to determine other putative functions of prophenoloxidases.
Acknowledgments
We extend special thanks to M. Coluzzi and R. Dallai for their discussions and support and to W. B. Neale for his critical reading and editing. We also appreciate the technical assistance of S. Perrot. P. gallinaceum ookinetes were kindly supplied by A. Raibaud (Laboratoire de Biochimie et Biologie Moléculaire des Insectes, Institut Pasteur) and W. bancrofti was supplied by C. Plichart and P. Esterre (Unité d'Immunologie, Institut L. Malardé, Papaeete, French Polynesia). This study received financial support from Institut Pasteur, the European Galileo program (P.T.B. and R. Dallai, University di Siena), Yonsei University Grant 1996 (W.-J.L.), National Institutes of Health Grant AI-36959 (A.R.), and The European Union Training and Mobility of Researchers Network on Insect–Parasite Interactions (F.C.K). H.-M.M. was supported by the Deutsche Forschungsgemeinschaft. S.-J.H. is the recipient of a French Government scholarship. K.D.M. was supported by a European Human Capital and Mobility fellowship and by grants from United Nations Development Program/WorldBank/WHO Special Program for Research and Training in Tropical Diseases. D.M. is the recipient of a postdoctoral research grant from the Spanish Ministry of Education and Culture, and A.G.O.M. is the recipient of a postdoctoral Pierre and Marie Curie research grant from European Union.
Abbreviations
- EcREs
ecdysteroid response elements
- 20E
20-hydroxyecdysone, PPO, prophenoloxidase
- EcR/USP
ecdysteroid receptor–Ultraspiracle complex
- IR
inverted
- DR
directed
- EMSA
electrophoretic mobility-shift assay
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF031626).
References
- 1.Bodine J H, Allen T H. J Cell Comp Physiol. 1941;18:151–160. [Google Scholar]
- 2.Mason H S. Adv Enzymol. 1955;16:105–184. doi: 10.1002/9780470122617.ch3. [DOI] [PubMed] [Google Scholar]
- 3.Ashida M, Brey P T. In: Molecular Mechanisms of Immune Responses in Insects. Brey P T, Hultmark D, editors. London: Chapman & Hall; 1998. pp. 135–172. [Google Scholar]
- 4.Collins F H, Sakai R K, Vernick K D, Paskewitz S, Seeley D C, Miller L H, Collins W E, Campbell C C, Gwadz R W. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 5.Zheng L, Cornel A J, Wang R, Erfle H, Voss H, Ansorge W, Kafatos F C, Collins F H. Science. 1997;276:425–428. doi: 10.1126/science.276.5311.425. [DOI] [PubMed] [Google Scholar]
- 6.Jiang H, Wang Y, Korochkina S E, Benes H, Kanost M R. Insect Biochem Mol Biol. 1997;27:393–699. doi: 10.1016/s0965-1748(97)00045-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee W J, Ahmed A, della Torre A, Kobayashi A, Ashida M, Brey P T. Insect Mol Biol. 1998;7:41–50. doi: 10.1046/j.1365-2583.1998.71047.x. [DOI] [PubMed] [Google Scholar]
- 8.Müller H M, Dimopoulos G, Blass C, Kafatos F C. J Biol Chem. 1999;274:11727–11735. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 9.Ashida M. Arch Biochem Biophys. 1971;144:749–762. doi: 10.1016/0003-9861(71)90383-3. [DOI] [PubMed] [Google Scholar]
- 10.Ashida M, Yoshida H. Insect Biochem. 1988;18:11–19. [Google Scholar]
- 11.Hiruma K, Riddiford L M. Dev Biol. 1988;130:87–97. doi: 10.1016/0012-1606(88)90416-2. [DOI] [PubMed] [Google Scholar]
- 12.Riddiford L M. In: in Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Kerkut G A, Gilbert L I, editors. Vol. 8. New York: Plenum; 1985. pp. 37–84. [Google Scholar]
- 13.Bownes M. Annu Rev Entomol. 1986;31:507–531. [Google Scholar]
- 14.Hagedorn H H. In: Ecdysone from Chemistry to Mode of Action. Koolman J, editor. New York: Thieme; 1989. pp. 279–289. [Google Scholar]
- 15.Segraves W A. Semin Cell Biol. 1994;5:105–113. doi: 10.1006/scel.1994.1014. [DOI] [PubMed] [Google Scholar]
- 16.Henrich V C, Brown N E. Insect Biochem Mol Biol. 1995;25:881–897. doi: 10.1016/0965-1748(95)00030-y. [DOI] [PubMed] [Google Scholar]
- 17.Thummel C S. Cell. 1995;83:871–877. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- 18.Riddihough G, Pelham H R B. EMBO J. 1987;6:3729–3734. doi: 10.1002/j.1460-2075.1987.tb02707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherbas L, Lee K, Cherbas P. Gene Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- 20.Antoniewski C, Mugat B, Delbac F, Lepesant J A. Mol Cell Biol. 1996;16:2977–2986. doi: 10.1128/mcb.16.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Avino P P, Crispi S, Cherbas L, Furia M. Mol Cell Endocrinol. 1995;113:1–9. doi: 10.1016/0303-7207(95)03584-t. [DOI] [PubMed] [Google Scholar]
- 22.Yao T P, Forman B M, Jiang Z, Cherbas L, Chen J D, McKewon M, Cherbas P, Evans R M. Nature (London) 1993;336:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 23.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 24.Thomas H E, Stunnenberg H G, Steward A F. Nature (London) 1993;362:471–475. doi: 10.1038/362471a0. [DOI] [PubMed] [Google Scholar]
- 25.Wang S F, Miura K, Miksicek R J, Segraves W A, Raikhel A S. J Biol Chem. 1998;273:27531–27540. doi: 10.1074/jbc.273.42.27531. [DOI] [PubMed] [Google Scholar]
- 26.Lan Q, Hiruma K, Hu X, Jindra M, Riddiford L M. Mol Cell Biol. 1999;19:4897–4906. doi: 10.1128/mcb.19.7.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathiopoulos K D, della Torre A, Predazzi V, Petrarca V, Coluzzi M. Proc Natl Acad Sci USA. 1998;95:12444–12449. doi: 10.1073/pnas.95.21.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritisch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Salazar C E, Mills-Hamm D, Kumar V, Collins F H. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapitskaya M, Wang S F, Cress E D, Dhadialla T S, Raikhel A S. Mol Cell Endocrinol. 1996;121:119–132. doi: 10.1016/0303-7207(96)03847-6. [DOI] [PubMed] [Google Scholar]
- 31.Miura K, Wang S F, Raikhel A S. Mol Cell Endocrinol. 1999;156:111–120. doi: 10.1016/s0303-7207(99)00136-7. [DOI] [PubMed] [Google Scholar]
- 32.Schule R, Muller M, Otsuka-Murakami H, Renkawitz R. Nature (London) 1988;33:87–90. doi: 10.1038/332087a0. [DOI] [PubMed] [Google Scholar]
- 33.Strähle U, Schmid W, Schütz G. EMBO J. 1988;7:3389–3395. doi: 10.1002/j.1460-2075.1988.tb03212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall M, Scott T, Sugumaran M, Söderhall K, Law J H. Proc Natl Acad Sci USA. 1995;92:7764–7768. doi: 10.1073/pnas.92.17.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujimoto K, Okino N, Kawabata S-I, Iwanaga S, Ohnishi E. Proc Natl Acad Sci USA. 1995;92:7769–7773. doi: 10.1073/pnas.92.17.7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawabata T, Yasahara Y, Ochiai M, Mastsuura S, Ashida M. Proc Natl Acad Sci USA. 1995;92:7774–7778. doi: 10.1073/pnas.92.17.7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linzen B, Soeter M, Riggs A F, Schneider H-J, Schartau W, Moore M D, Yokota E, Behrens P Q, Nakashima H, Takagi T, et al. Science. 1985;229:519–529. doi: 10.1126/science.4023698. [DOI] [PubMed] [Google Scholar]
- 38.Fujii T, Sakurai H, Izumi S, Tomino S. J Biol Chem. 1989;264:11020–11025. [PubMed] [Google Scholar]
- 39.Engström Y. In: Molecular Mechanisms of Immune Responses in Insects. Brey P T, Hultmark D, editors. London: Chapman & Hall; 1997. pp. 211–244. [Google Scholar]
- 40.Georgel P, Meister M, Kappler C, Lemaitre B, Reichhart J M, Hoffmann J A. Biochem Biophys Res Commun. 1993;197:508–517. doi: 10.1006/bbrc.1993.2508. [DOI] [PubMed] [Google Scholar]
- 41.Ochiai M, Ashida M. J Biol Chem. 1999;274:11854–11858. doi: 10.1074/jbc.274.17.11854. [DOI] [PubMed] [Google Scholar]
- 42.Park D S, Shin S W, Kim M G, Park S S, Lee W J, Brey P T, Park H Y. Insect Biochem Mol Biol. 1998;27:983–992. doi: 10.1016/s0965-1748(97)00081-7. [DOI] [PubMed] [Google Scholar]
- 43.Hagen H E, Kläger S L, McKerrox J H, Ham P J. Exp Parasitol. 1997;86:213–218. doi: 10.1006/expr.1997.4165. [DOI] [PubMed] [Google Scholar]
- 44.Cho W L, Liu H S, Lee C H, Kuo C C, Chang T Y, Liu C T, Chen C C. Insect Mol Biol. 1998;7:31–40. doi: 10.1046/j.1365-2583.1998.71049.x. [DOI] [PubMed] [Google Scholar]
- 45.Bodmer R, Venkatesh T V. Dev Gent. 1998;22:181–186. doi: 10.1002/(SICI)1520-6408(1998)22:3<181::AID-DVG1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Dittmer N T, Raikhel A S. Insect Biochem Mol Biol. 1997;27:323–335. doi: 10.1016/s0965-1748(97)00007-6. [DOI] [PubMed] [Google Scholar]
- 47.Antoniewski C, Laval M, Lepesant J M. Insect Biochem Mol Biol. 1993;23:105–114. doi: 10.1016/0965-1748(93)90088-a. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann M, Korge G. EMBO J. 1995;14:716–726. doi: 10.1002/j.1460-2075.1995.tb07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antoniewski C, O'Grady M S, Edmondson R G, Lassieur S M, Benes H. Mol Gen Genet. 1995;249:545–556. doi: 10.1007/BF00290580. [DOI] [PubMed] [Google Scholar]
- 50.Cherbas L, Lee K, Cherbas P. Genes Dev. 1991;5:120–131. doi: 10.1101/gad.5.1.120. [DOI] [PubMed] [Google Scholar]
- 51.Luo Y, Amin J, Voellmy R. Mol Cell Biol. 1991;11:3660–3675. doi: 10.1128/mcb.11.7.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]