Abstract
Analyzing the pathways by which retinoic acid (RA) induces promyelocytic leukemia/retinoic acid receptor α (PML/RARα) catabolism in acute promyelocytic leukemia (APL), we found that, in addition to caspase-mediated PML/RARα cleavage, RA triggers degradation of both PML/RARα and RARα. Similarly, in non-APL cells, RA directly targeted RARα and RARα fusions to the proteasome degradation pathway. Activation of either RARα or RXRα by specific agonists induced degradation of both proteins. Conversely, a mutation in RARα that abolishes heterodimer formation and DNA binding, blocked both RARα and RXRα degradation. Mutations in the RARα DNA-binding domain or AF-2 transcriptional activation region also impaired RARα catabolism. Hence, our results link transcriptional activation to receptor catabolism and suggest that transcriptional up-regulation of nuclear receptors by their ligands may be a feedback mechanism allowing sustained target-gene activation.
In acute promyelocytic leukemia (APL), the t(15;17) translocation fuses a nuclear receptor, RARα, to a nuclear matrix protein, PML (1). PML/RARα transgenic mice show impaired neutrophilic differentiation and develop leukemia, demonstrating that expression of the fusion protein suffices to initiate this malignancy (2). PML/RARα impairs both nuclear receptor-induced differentiation and PML-triggered apoptosis, likely accounting for the differentiation block and the unrestrained growth of the leukemic cells (3, 4). Inhibition of the retinoic acid (RA) response appears to involve the stabilization of corepressor proteins–histone deacetylase complexes on RA response elements (5–8). The PML protein, which is localized on nuclear subdomains (PML nuclear bodies), has growth-suppressive and proapoptotic properties (9–16). PML/RARα expression delocalizes nuclear body proteins, which was proposed to account for apoptosis resistance (17).
RA promotes differentiation of APL cells and induces clinical remissions in patients (18). Arsenic trioxide (AS) also induces remissions, through combined induction of apoptosis and differentiation (19). Both RA and AS treatments trigger PML/RARα degradation and nuclear body restoration (20–23), initially suggesting that the therapeutic action of these two drugs could be due to the down-regulation of the oncogenic fusion protein. However, data to support this hypothesis are conflicting (24, 25). Nevertheless, PML/RARα degradation is most likely responsible for the dramatic cross-facilitation of RA and AS effects, either in vitro or in vivo (4, 26).
Two classes of retinoic acid receptors, the RARs and the RXRs, have been identified whose natural ligands are trans-retinoic acid (trans-RA) and 9-cis retinoic acid (9-cis-RA), respectively. Studies using retinoic acid response elements (RARE) have shown that most elements bind a RAR/RXR heterodimer. All nuclear receptors are characterized by a highly conserved structure including a zinc-finger DNA-binding domain and a hormone-binding domain. The major ligand-dependent activation domain (AF-2) is present in a highly conserved α-helix (AF-2 AD core) which mediates ligand-induced interactions with transcriptional coactivator proteins. Whereas some of these proteins harbor domains suggestive for an involvement in chromatin remodeling (such as transcription intermediary factor 1α or TIF1α), another one is the 26S proteasome regulatory subunit 1 (SUG-1) (27). The exact contribution of these proteins to the function of nuclear receptors is still unclear.
Dissecting the pathways involved in PML/RARα degradation, we confirm the implication of caspases, which become activated during RA- or AS-treatment of APL cells, and cleave a specific site in the PML moiety of PML/RARα. We demonstrate that RARα itself, like RARα fusion proteins, is catabolized by the proteasome after exposure to RA. RARα degradation requires a functional receptor, because mutations in the DNA-binding domain, AF-2 function, and RXR dimerization interface severely impair RA-induced receptor degradation. The whole RAR/RXR heterodimer is degraded when activated by specific agonists for either receptor. These findings provide a direct link between ligand-dependent transcriptional activation and nuclear receptor catabolism.
Materials and Methods
Protein Analysis.
Cells (NB4, CHO, COS-6) were grown as previously described (26). Immunofluorescence and Western blot analysis were performed as before by using specific rabbit polyclonal antibodies: anti-RARα (RPαF′) or anti-RARα (C-20) (Santa Cruz Biotechnology) or the EGFP monoclonal antibody (CLONTECH). In vitro transcription/translation with T7 polymerase and rabbit reticulocyte lysates was performed according to the manufacturer's instructions (Promega). Immunoprecipitations were performed by using standard procedures(28) with an anti-RARα monoclonal antibody Ab-9αF (29) or anti-PML monoclonals (M. C. Guillemin, unpublished results).
Vectors and Transfections.
RARα-SRαMSVtkNeo was constructed by cloning the RARα EcoRI fragment of pSG5-RARα into the EcoRI site of the SRαMSVtkNeo plasmid (30) and stable CHO-RARα clones were Neo-selected from transfected CHO cells. pEGFP-RARα was constructed by cloning the MscI–ApaI RARα fragment of pSG5-RARα in the Ecl1 136II-ApaI site of pEGFP-c1 vector (CLONTECH). RARα (380, 383 RR), and RARα194T195P were created by site-directed mutagenesis (31) and sequenced. All other RARα mutants were described elsewhere. Gel-shift analysis was perfomed by using the RARβ RARE, as previously described (32). COS-6 cells were transiently transfected by using the fugene procedure (Boehringer Mannheim).
Chemicals.
Retinoids: AM580 was synthesized by CIRD-Galderma (Sophia Antipolis, Valbonne, France). Trans-RA and 9-cis-RA were purchased from Sigma. BMS 453 and SR 11237 were kindly provided by H. Gronemeyer (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France). Inhibitors: LLnL, N-ethylmaleimide (NEM), E64 ester, PAO, and cycloheximide (CHX) were from Sigma. Leupeptin, PefablocR, TPCK, and TLCK were from Boehringer Mannheim. Lactacystin and MG132 were from Biomol (Plymouth Meeting, PA). z-VAD-fmk was from Bachem.
Results
In APL Cells, RA Down-Regulates Expression of Both PML/RARα and RARα Proteins.
PML/RARα was shown to be rapidly catabolized in response to RA in NB4 cells, either in a proteasome- or caspase-dependent manner (21, 23, 24). A kinetic analysis showed that RA caused a biphasic PML/RARα degradation (Fig. 1A). A rapid decrease in PML/RARα expression was observed within 1 h, whereas a second degradative step occurred after 12 h and was characterized by the appearance of a 90-kDa PML/RARα cleavage product (ΔPML/RARα). RA also induced a progressive disappearance of RARα expressed from the nonrearranged allele, clearly visible after 12 h (Fig. 1A). Northern blot analysis demonstrated that RA did not down-regulate the PML/RARα transcript, and rather up-regulated RARα (not shown), as described (33), suggesting involvement of posttranscriptional mechanisms in PML/RARα and RARα down-regulation.
Figure 1.
(A) RA triggers PML/RARα and RARα down-regulation in NB4 cells by Western blot analysis. ΔPML/RARα is a cleavage product of the fusion, and * denotes a nonspecific protein sometimes detected with the RARα antibody. (B) Comparison of RARα agonists (trans-RA, AM580, 9-cis-RA, and TTNPB) or antagonists (Roche Molecular Biochemicals; RO-415253, RO-61–8431, and RO-40–8757) for their ability to modulate PML/RARα and RARα expression. NB4 cells were treated for 2 days with the various compounds; all were at a 10−6 M concentration. (C) Kinetics of PML/RARα down- regulation induced by AS (10−6 M). (D) Effect of protease inhibitors on a decrease in PML/RARα.. NB4 cells were treated as indicated for 8 h (allowing for the decrease in PML/RARα, but not RARα, expression). Note the presence of higher molecular weight species with AS and inhibitors, which could represent a PML/RARa-Pic1 modification adduct (◂). (E) PML/RARα is degraded by caspases through a specific site in PML. 35S-labeled in vitro translated PML/RARα (Upper) or PML (Lower) were incubated with recombinant caspase 1, 3, 4, 6, 7, or 8 for 4 h and analyzed by PAGE. (F) PML is a functional caspase target in vivo. CHO cells overexpressing PML were treated with z-VAD, etoposide (a DNA-damaging agent that activates caspases), or both for 24 h. PML is cleaved (◂) in a z-VAD-reversible manner.
Synthetic retinoids with RARα agonist activities (AM580, 9-cis-RA, tetrahydro-tetramethyl-naphthalenyl-propenyl benzoic acid; TTNPB) also induced PML/RARα and RARα down-regulation. In contrast, RARα antagonists all stabilized both RARα and PML/RARα (Fig. 1B), suggesting that endogenous retinoids present in the serum suffice to induce a baseline level of catabolism. PML/RARα degradation was always associated with the restoration of PML nuclear bodies as assessed by immunofluorescence detection of the PML or Sp100 proteins (not shown). Association of a RXRα agonist to a RARα antagonist (BMS453, SR11237) was previously shown to induce NB4 differentiation, coupled to PML nuclear bodies reformation (34). This treatment was also associated to PML/RARα down-regulation, coupled to the appearance of ΔPML/RARα (not shown). Finally, although AM580 or TTNPB (10−6 M) induced the same kinetics of NB4 differentiation that 10−6 M RA, these two compounds induced the complete degradation of PML/RARα only 24–48 h later than RA does (not shown), suggesting that receptor activation does not strictly parallel its down-regulation by synthetic retinoids (see below).
Caspases Cleave PML and PML/RARα.
Therapeutic doses of arsenic trioxide (10−6 M) also induce rapid PML/RARα catabolism (20). In this setting, RARα expression was unaffected and ΔPML/RARα was rarely detectable even after a 2-day exposure (Fig. 1C), suggesting that the degradative pathways triggered by the two drugs are not identical. Yet, protease inhibitors (lactacystin, MG132, and LLnL), thiol reagents (TPCK, TLCK, N-ethylmaleimide, and PAO), or caspase inhibitors (z-VAD and Ac-Asp-Glu-Val-Asp) all significantly (but not completely) blocked both RA- or AS-induced PML/RARα degradation at 8 h (Fig. 1D and not shown). Taking the inhibitory effect of z-VAD on PML/RARα degradation, recombinant caspases were then assayed for their ability to cleave in vitro-translated PML/RARα.. Caspase 1, 6, and 7 degraded PML/RARα in a z-VAD-reversible manner (not shown), whereas caspase 3, 4, and 8 did not (Fig. 1E). The same caspases also degraded PML (Fig. 1E), but not RARα (not shown). When assaying a set of C-terminal PML mutants, we identified a caspase site at D523. This finding is consistent with a previously mapped site in PML/RARα (24). PML-overexpressing cells were then treated with z-VAD, etoposide (which induces caspase activation), or both. Etoposide led to the disappearance of full-size PML (Fig. 1F) and z-VAD restored PML expression, demonstrating that PML is a caspase target in vivo.
RA-Induced Degradation of RARα in Non-APL Cells.
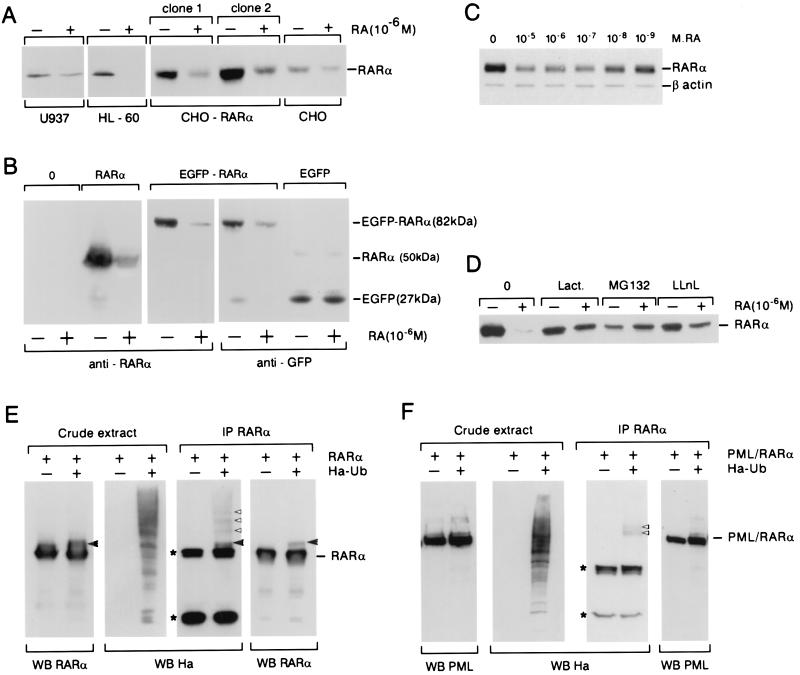
To ascertain whether a RA-induced decrease in RARα expression was also observed in other cellular systems, CHO clones overexpressing RARα1, U937, HL-60, or COS cells were or were not treated with 10−6 M RA overnight. Again, RA sharply decreased RARα protein expression detected by Western blotting (Fig. 2A). In transiently transfected COS-6 cells, RARα1 expression was sharply decreased on RA exposure (Fig. 2B). RA induced a decrease in the intensity of RARα fluorescence but did not change the number of transfected cells. Therefore, RA does not induce the death of RARα-overexpressing cells and we conclude that RA induces RARα degradation. Because transfected PML/RARα or PML zinc finger/RARα were also degraded after RA treatment (ref. 35 and unpublished work), we hypothesized that the presence of a moiety RARα suffices to cause RA-triggered degradation of RARα fusion proteins. Hence, cDNAs encoding RARα and EGFP were fused in frame. Western blot analysis of transfected COS cells demonstrated that the chimeric EGFP-RARα protein was catabolized in response to RA, whereas the EGFP protein was not (Fig. 2B). This experiment predicts that all oncogenic RARα fusion proteins will be degraded after RA exposure.
Figure 2.
Endogenous or transfected RARα is catabolized after RA exposure. (A) RARα is down-regulated after an overnight exposure to 10−6 M RA in U937 and HL-60 cells as well as in parental CHO cells or clones overexpressing RARα1 from a retroviral vector. (B) COS cells were transfected with expression vectors encoding RARα, EGFP-RARα, or EGFP and were or were not treated with RA overnight. Western blot analysis was performed with anti-RARα antibodies (Left) or anti-GFP antibodies (Right). (C) COS cells were transfected with RARα, translation was blocked with CHX, and various doses of RA applied for 5 h as indicated. An actin control is provided. (D) Proteasome inhibitors (10−6 M) block RA-induced RARα catabolism. (E and F) RARα or PML/RARα degradation is associated to receptor ubiquitination. COS cells were cotransfected with expression vectors encoding RARα or PML/RARα and (or and not) Ha-tagged ubiquitin. Cells exposed to RA for 3 h either were or were not immunoprecipitated (IP). Western blot analysis was performed by using anti-RARα, anti-PML, or anti-Ha as indicated. *, Heavy and light chains of the mouse antibodies used for immunoprecipitation; ◂, a RARα–ubiquitin adduct observed with both anti-Ha and anti-RAR antibodies, whereas those marked by ▹ are visible with anti-Ha alone.
To assess whether RARα catabolism requires RA target gene activation, RARα degradation was analyzed in the presence of CHX. After 5 h, RA induced a 10-fold decrease in the level of the receptor, demonstrating that degradation of RARα occurs independently of de novo protein synthesis (not shown). Because the apparent rate of RARα degradation is accelerated in such a CHX chase setting, these conditions were used thereafter. A dose-response analysis demonstrated a progressive RARα degradation from 10− 9 M to 10− 5 M (Fig. 2C), corresponding to the doses required for receptor activation. Protein catabolism often involves proteasome degradation of ubiquitin-tagged molecules. Indeed, all three proteasome inhibitors tested (lactacystin, MG132, and ALLN) blocked RA-induced RARα degradation (Fig. 2D). To assess whether RA exposure is associated to ubiquitination of RARα, COS cells were transfected with expression vectors for RARα with or without HA-tagged ubiquitin and were treated or not with RA for 3 h. Probing a RARα immunoprecipitate with an anti-HA antibody demonstrated the ubiquitin tagging of RARα (Fig. 2E). Identical results were obtained for PML/RARα (Fig. 2F). However, no obvious enhancement was observed after RA exposure, possibly reflecting either the presence of endogenous retinoids in the serum or the extreme lability of polyubiquitin adducts. Hence, RA-triggered proteasomal degradation of RARα is associated to its ubiquitination, as described for many other proteins (36).
Mutational Analysis of RA-Induced RARα Degradation.
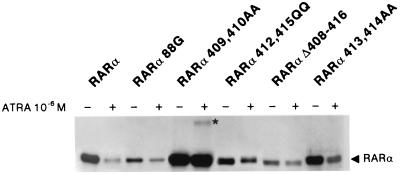
A set of RARα mutants were transfected in COS cells and treated with or without RA for 12 h. RARα degradation was assessed by Western blotting (Fig. 3 and data not shown). Deletion of the N-terminal A region (Δ 1–60) or mutation in the N-COR binding site (194–195TP) did not induce significant differences with wild-type receptor with respect to RA-induced degradation. In contrast, a mutation in the DNA-binding domain (C88G) that completely abolishes DNA-binding in gel-shift assays, severely impaired RA-induced RARα degradation. Similarly, deletion of the core sequence required for AF-2 activation (amino acids 408–416), also blocked RARα catabolism (Fig. 3). Point mutations were then introduced in this region. Amino acids 409 and 410 are critical for degradation, whereas residues 412 and 415 impair, but do not block catabolism. Finally, residues 413 and 414 do not affect RARα catabolism. For the AF-2 mutants that do not degrade, the RARα doublet often shifted to its upper form, possibly corresponding to a ligand-induced change in phosphorylation (not shown). RARα amino acids 409–410 were consistently expressed at much higher levels than the wild-type, and high molecular weight species with a ladder-like appearance were observed after trans-RA exposure (star in Fig. 3). Hence, some of the highly conserved acidic and hydrophobic residues in the AF-2 activation domain are required for ligand-induced degradation.
Figure 3.
Analysis of RA-induced degradation of RARα mutants. COS cells were transfected with expression vectors for the indicated RARα mutants, separated in equal parts, exposed to CHX, and treated or not treated with 10−6 M trans-RA for 6 h. Extracts were analyzed by Western blot. *, A modified RARα protein, possibly a polyubiquitin adduct.
Role of RXR Dimerization.
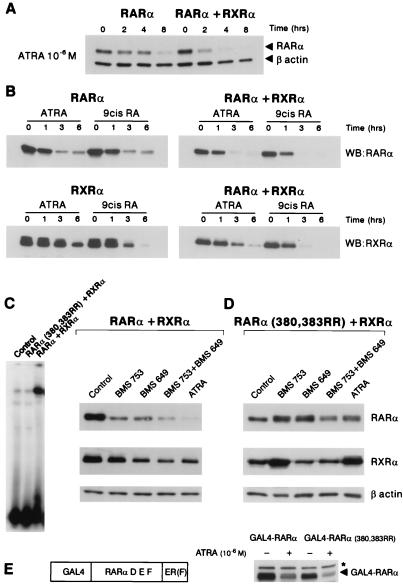
Kinetic analyses comparing the degradation of RARα alone or in association with RXRα in transfected HeLa cells demonstrated that RXRα accelerated trans-RA-induced RARα degradation (Fig. 4A). However, the magnitude of this acceleration depended very much on the cell line used (see Fig. 4B below), possibly reflecting variations in the endogenous RXR content. COS cells were then transfected with RARα, RXRα, or both and treated with trans-RA or 9-cis-RA (Fig. 4B). When the receptors were separately overexpressed, both retinoids degraded RARα, whereas 9-cis-RA degraded RXRα much more efficiently than trans-RA. When receptors were coexpressed, either ligand degraded both RARα and RXRα (Fig. 4B). These observations strongly suggest that the DNA-binding RAR/RXR heterodimer complex is the substrate for degradation.
Figure 4.
RARα and RXRα catabolism requires their heterodimerization. (A) RXR accelerates RA-induced RARα degradation in transfected HeLa cells. Degradation was assessed in a CHX chase as in Fig. 3. A β-actin control is provided. (B) Kinetics of RARα, RXRα, or RARα/RXRα degradation in the presence of trans- or 9-cis-RA (10−6 M). COS cells were transfected with RXRα, RARα, or both, translation was blocked with CHX, and cells were exposed to retinoids for 0, 1, 3, or 6 h as indicated. No significant variations in the levels of RARα or RXRα were observed during the chase in the absence of retinoids (not shown). (C) Gel-shift analysis of RARα/RXRα or RXRα/RARα(380,383RR)-transfected COS cell extracts on a RARβ RARE. (D) COS cells were transfected with RXRα and either RARα (Left) or RARα(380,383RR) (Right). Specific agonists for RARα (BMS 753) or RXRα (BMS 649) or RA were added for 5 h. (E) Schematic representation of the GAL4-RARα fusion used. The D, E, and F domains of RARα (amino acids 154–462) were fused to a GAL4 DNA-binding domain (N-terminal) and to an estrogen receptor tag [domain F, ER(F)]. Mutations that abolish RXR binding were introduced in this expression vector, yielding GAL4-RARα(380,383 RR). COS cells were transfected with either of these expression vectors, treated or not treated with trans-RA for 6 h after CHX exposure, and analyzed by Western blot. ◂, Fusion proteins; *, nonspecific protein.
Because 9-cis-RA binds both RARα and RXRα, we then examined the effect of receptor selective synthetic retinoids (BMS 753 and BMS 649 for RARα or RXRα, respectively) on the catabolism of these receptors (Fig. 4D). By using the same experimental setting, we demonstrate that in the presence of a specific RARα agonist, RXRα was also catabolized and vice versa. Exposure to both agonists resulted in additive effects on degradation (Fig. 4D Left). Note, however, that similar concentrations of trans-RA induced a more rapid catabolism of the receptors.
By using the crystal structure of the RARα/RXRα ligand binding domain heterodimer, contact positions between the two proteins were identified at residues 380 and 383 (D. Moras, personal communication). These mutations did not impair RA binding or AF-2 activation function as demonstrated by RA-induced transactivation of a chimeric GAL4-RARα construct (see below). When coexpressed with RXRα, this RARα point mutant (380, 383 RR) was completely defective in DNA-binding on a DR5 RARE by gel-shift analysis (Fig. 4C), suggesting that these mutations inhibit RXRα binding and hence target the DNA sequence binding. RARα(380,383RR) was cotransfected with RXRα in COS cells and treated with RARα and RXRα-specific agonists as above. Inhibition of heterodimerization blocked the degradation of both RARα and RXRα, at least during this 5-h assay (Fig. 4D). RARα agonist (BMS 753 and trans-RA) even appeared to stabilize RXRα; however, even when coexpressed with RARα(380,383RR), RXRα degradation could still be triggered by 9-cis-RA (not shown). Because these two changes abolish both RXR and DNA binding, the 380,383 RR mutations were introduced in a GAL4-RARα construct in which RARα D, E, and F domains (encompassing the RXR binding site and AF-2) are tethered to DNA by the GAL4 DNA-binding domain (Fig. 4E). COS cells were then transfected with a CAT reporter gene containing a GAL4 binding site and either of these two GAL4-RARα expression vectors. Analysis of CAT activity demonstrated that GAL4-RARα-(380, 383RR) could, like GAL4-RARα, activate transcription in a RA-dependent manner (not shown). Western blot analysis demonstrated that both GAL4-RARα and GAL4-RARα(380,383 RR) were degraded in response to RA exposure, independently of RXRα coexpression (Fig. 4E and data not shown). Hence, the inability of the RARα mutant defective in RXR binding to be degraded likely results from its inability to bind DNA.
Discussion
We demonstrate that RA directs proteasome-mediated degradation of RARα, thereby accounting for the degradation of APL-associated fusion proteins. The domains required for transcriptional activation are also required for receptor catabolism, providing a striking illustration of how transcription factors are turned off following activation.
Our findings demonstrate that RA degrades PML/RARα by two distinct pathways: caspase activation and direct proteasome targeting, accounting for the biphasic degradation in Fig. 1A and also for the fact that both caspase and proteasome inhibitors were previously shown to antagonize PML/RARα catabolism (21, 24, 25). Both RA and AS induce the activation of caspases (not shown), accounting for the fact that z-VAD partially inhibited both RA- and AS-induced PML/RARα degradation. Inhibitor studies suggest that arsenic also targets PML/RARα through an as yet unidentified proteasome-dependent pathway (Fig. 1D). This unidentified pathway may be the one responsible for AS-induced PML degradation (20), because PML/RARα, like PML, is targeted onto nuclear bodies (NBs) after AS treatment (20). Alternatively, arsenic appears to alter RARα phosphorylation and induces its progressive depletion (20, 26). Such AS-induced RARα alterations may also be implicated in PML/RARα catabolism.
What is the relevance of PML/RARα degradation to the therapeutic response? RA resistance of some APL cell lines is associated with failure to degrade the fusion protein (37, 38); yet, AS-induced PML/RARα degradation (or RA-induced PLZF/RARα catabolism) is not sufficient to induce terminal differentiation or apoptosis (26, 34, 39). However, in all of these cases, a moderate maturation was noted which may result from PML/RARα degradation and the relief of the differentiation block. Use of caspase inhibitors suggested that RA-induced terminal differentiation can proceed without full PML/RARα degradation (not shown; ref. 24). Finally, up-regulation of PML/RARα, but not RARα, by proteasome inhibitors partially restored RA response in an NB4 subline (25). Interpretation of these findings is complex because partial degradation of the fusion often occurs, and in some studies, differentiation is assessed only by gene activation rather than by morphologic or functional assays. In that sense, PML/RARα and RARα appear to control distinct sets of target genes (37), not all of which may be involved in differentiation. The balance between the active receptor and its dominant-negative counterpart is a highly dynamic one, because RARα2, but not PML/RARα will be transcriptionally induced after RA exposure. Yet, our studies strongly suggest that degradation of PML/RARα, like that of RARα, is a postactivation phenomenon. Nevertheless, PML/RARα degradation by RA or AS likely plays a key role in the synergy between the two agents, that cross-facilitate each others effects (4, 26).
Signaling molecules need to be turned off after activation. Cytoplasmic transmembrane receptors are often internalized following ligand binding and later recycled to the cell surface. For the nuclear factor κ B (NFκB) response pathway, the active p50/p65 complex will transcriptionally activate its IκB inhibitor. Finally, for nuclear transcription factors (such as AP1), activation often induces the ubiquitin-dependent degradation of the activated complex. In many cases, a specific phosphorylation will recruit ubiquitin-conjugating enzymes (36). Following the covalent binding of poly-ubiquitin residues, the protein will be degraded by the proteasome. A similar scheme is likely to occur here: one of the first modifications induced by RA exposure is a shift toward higher molecular weight species that might represent receptor phosphorylation and the presence of ubiquitin adducts. Interestingly, AF-2 was previously shown to bind the SUG-1 proteasome component (27) in a hormone-enhanced manner. The estrogen and vitamin D3 receptors were also shown to be catabolized by the proteasome after ligand exposure (40, 41). Finally, the progesterone receptor PR is down-regulated on progesterone exposure (42). The very high conservation of the AF-2 AD core domain would be consistent with a common degradative pathway involving AF-2/SUG-1 interactions. In that respect, note that the AF-2 mutants that are degraded in response to RA are those previously shown to retain SUG-1 binding (27); yet, formal proof for SUG-1 involvement would require inhibition of RARα degradation by dominant-negative SUG-1 constructs.
Our results demonstrate that inhibition of RXR binding abrogates RARα degradation through the blockage of DNA binding. The requirement for DNA binding and AF-2 for degradation suggests that some allosteric signal is transduced from the DNA-binding domain to AF-2. Interestingly, in the case of the glucocorticoid receptor, similar allosteric signals were proposed to modulate the function of AF-2 (43). Alternatively, involvement into a transcriptionally active complex could induce specific modifications, in particular transcription factor II H-induced AF-1 phosphorylations (44) in the B domain that could be a signal for subsequent degradation. Altogether, the requirement for AF-2 function and DNA-binding strongly suggests that degradation is a postactivation step only affecting transcriptionally active receptors, providing a feedback mechanism on receptor activity.
Apart from the major activation function AF-2, RARs may control target gene expression through their ligand-independent AF-1 activation and corepressor-induced histone deacetylase-mediated transcriptional repression. RA-induced receptor catabolism could potentially relieve transcriptional repression or abrogate AF-1 activation mediated by RARα isoforms that are efficiently degraded, yet not transcriptionally activated by RA (such as RARα1 or RARγ). This could prove to be a novel mechanism by which RA could activate or repress target genes. Such ligand-induced catabolism sheds a new light on the common observation that many nuclear receptors (in particular the RARs) are transcriptionally up-regulated by their ligand (32). Such transcriptional up-regulation may provide a way to circumvent the degradation feedback mechanism and hence allow sustained target gene activation.
Acknowledgments
We thank P. Chambon for his kind gift of reagents, support, and helpful discussions. We are most grateful to D. Moras (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France) for communication of unpublished data on the crystal structure of RAR/RXR heterodimers. We thank Gilles Despouy for his help with ATRA-binding assays. We thank D. Bohnann for the gift of the HA-ubiquitin expression vector. Bristol-Myers Squibb is gratefully acknowledged for providing receptor-specific ligands. We thank all members of the lab for helpful discussions and critical reading of the manuscript. M.G. and J.Z. were supported by the Ligue Nationale Française Contre le Cancer. J.Z. was also supported by Programme de Recherche Avancée. E.K. was supported by Fondation Chateaubriand and Institut National de la Santé et de la Recherche Médicale. We thank members of LPH for their help. This work was supported by grants from Association pour la Recherche sur le Cancer.
Abbreviations
- RA
retinoic acid
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- PML
promyelocytic leukemia
- APL
acute promyelocytic leukemia
- AF-2
ligand-dependent activation domain
- AS
arsenic trioxide
- RARE
retinoic acid response element
- CHX
cycloheximide
- EGFP
enhanced green fluorescent protein
References
- 1.de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 2.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quignon F, Chen Z, de Thé H. Biochim Biophys Acta. 1997;1333:M53–M61. doi: 10.1016/s0304-419x(97)00025-5. [DOI] [PubMed] [Google Scholar]
- 4.Lallemand-Breitenbach V, Guillemin M-C, Janin A, Daniel M-T, Degos L, Kogan S C, Bishop J M, de Thé H. J Exp Med. 1999;189:1043–1052. doi: 10.1084/jem.189.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin R J, Nagy L, Inoue S, Shao W L, Miller W H, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 6.Grignani F, de Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, et al. Nature (London) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 7.He L-Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 8.Guidez F, Ivins S, Zhu J, Soderstrom M, Waxman S, Zelent A. Blood. 1998;91:2634–2642. [PubMed] [Google Scholar]
- 9.Koken M H M, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyck J A, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 11.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 12.Mu Z M, Chin K V, Liu J H, Lozano G, Chang K S. Mol Cell Biol. 1994;14:6858–6867. doi: 10.1128/mcb.14.10.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quignon F, de Bels F, Koken M, Feunteun J, Ameisen J-C, de Thé H. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z-G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 15.Daniel M-T, Koken M, Romagné O, Barbey S, Bazarbachi A, Stadler M, Guillemin M-C, Degos L, Chomienne C, de Thé H. Blood. 1993;82:1858–1867. [PubMed] [Google Scholar]
- 16.Koken M H M, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix M K, Sobczak-Thépot J, Juhlin L, Degos L, Calvo F, de Thé H. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- 17.Grignani F, Ferrucci P, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C, Nicoletti I, et al. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 18.Huang M, Ye Y, Chen R, Chai J, Lu J, Zhoa L, Gu L, Wang Z. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- 19.Chen G-Q, Zhu J, Shi X-G, Ni J-H, Zhong H-J, Si G-Y, Jin X-L, Tang W, Li X S, Xong S M, et al. Blood. 1996;88:1052–1061. [PubMed] [Google Scholar]
- 20.Zhu J, Koken M H M, Quignon F, Chelbi-Alix M K, Degos L, Wang Z Y, Chen Z, de Thé H. Proc Natl Acad Sci USA. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida H, Kitamura K, Tanaka K, Omura S, Miyazaki T, Hachiya T, Ohno R, Naoe T. Cancer Res. 1996;56:2945–2948. [PubMed] [Google Scholar]
- 22.Muller S, Matunis M J, Dejean A. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raelson J V, Nervi C, Rosenauer A, Benedetti L, Monczak Y, Pearson M, Pelicci P G, Miller W H. Blood. 1996;88:2826–2832. [PubMed] [Google Scholar]
- 24.Nervi C, Ferrara F F, Fanelli M, Rippo M R, Tomassini B, Ferrucci P F, Ruthardt M, Gelmetli Y, Gambacorti-Passerini C, Diverio D, et al. Blood. 1998;92:2244–2251. [PubMed] [Google Scholar]
- 25.Fanelli M, Minucci S, Gelmetti V, Nervi C, Gambacorti-Passerini C, Pelicci P G. Blood. 1999;93:1477–1481. [PubMed] [Google Scholar]
- 26.Gianni M, Koken M H M, Chelbi-Alix M K, Benoit G, Lanotte M, Chen Z, de Thé H. Blood. 1998;91:4300–4310. [PubMed] [Google Scholar]
- 27.vom Baur E, Zechel C, Heery D, Heine M J S, Garnier J M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 29.Rochette-Egly C, Oulad-Abdelghani M, Staub A, Pfister V, Scheuer I, Chambon P, Gaub M-P. Mol Endocrinol. 1995;9:860–871. doi: 10.1210/mend.9.7.7476969. [DOI] [PubMed] [Google Scholar]
- 30.Muller A, Young J, Pendergast A M, Ponderl M, Landau N, Littman D, Witte O. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1994. [Google Scholar]
- 32.de Thé H, Vivanco-Ruiz M D M, Tiollais P, Stunnenberg H, Dejean A. Nature (London) 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- 33.Chomienne C, Balitrand N, Ballerini P, Castaigne S, de Thé H, Degos L. J Clin Invest. 1991;88:2150–2154. doi: 10.1172/JCI115547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J-Y, Clifford J, Zusi C, Starrett J, Tortolani D, Ostrowski J, Reczek P R, Chambon P, Gronemeyer H. Nature (London) 1996;382:819–822. doi: 10.1038/382819a0. [DOI] [PubMed] [Google Scholar]
- 35.Koken M H M, Reid A, Quignon F, Chelbi-Alix M K, Davies J M, Kabarowski J H S, Zhu J, Dong S, Chen S-J, Chen Z, et al. Proc Natl Acad Sci USA. 1997;94:10255–10260. doi: 10.1073/pnas.94.19.10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laney J D, Hochstrasser M. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 37.Shao W, Benedetti L, Lamph W W, Nervi C, Miller W H J. Blood. 1997;89:4282–4289. [PubMed] [Google Scholar]
- 38.Duprez E, Lillehaug J R, Naoe T, Lanotte M. Oncogene. 1996;12:2451–2459. [PubMed] [Google Scholar]
- 39.Koken M H M, Daniel M-T, Gianni M, Zelent A, Licht J, Buzyn A, Minard P, Degos L, Varet B, de Thé H. Oncogene. 1999;18:1113–1118. doi: 10.1038/sj.onc.1202414. [DOI] [PubMed] [Google Scholar]
- 40.Nawaz Z, Lonard D M, Dennis A P, Smith C L, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masuyama H, MacDonald P N. J Cell Biochem. 1998;71:429–440. [PubMed] [Google Scholar]
- 42.Milgrom E, Luu Thi M T, Atger M, Bavlieu E E. J Biol Chem. 1973;248:6366–6374. [PubMed] [Google Scholar]
- 43.Lefstin J A, Thomas J R, Yamamoto K. Genes Dev. 1994;23:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- 44.Rochette-Egly C, Adam S, Rossignol M, Egly J-M, Chambon P. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]