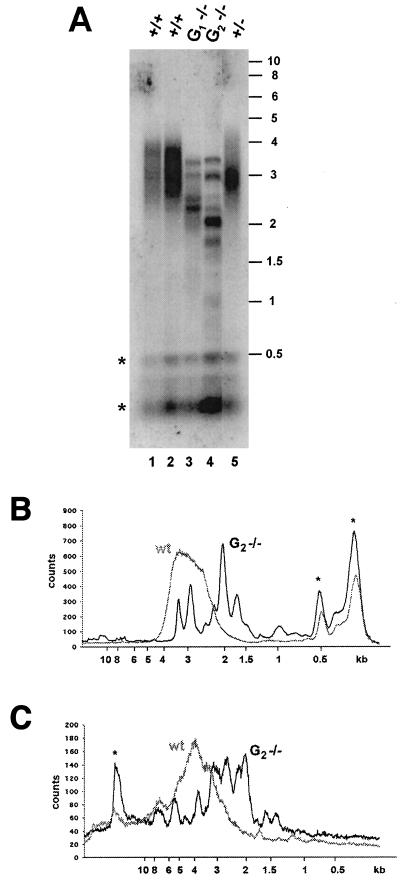
Figure 4.
Progressive telomere shortening in telomerase-deficient plants. (A) TRF analysis. Shown are results with Tru9I digestion of DNA from rosette leaves (lane 1) or floral buds and siliques (lane 2) of wild-type plants and rosette leaves of heterozygous plants (lane 5). Results with rosette leaf DNA from first-generation (G1) and second-generation (G2) plants homozygous AtTERT disruption are shown in lanes 3 and 4. (B) Quantitative analysis of TRF data from wild-type (wt) and G2 homozygous mutant plants digested with Tru9I or HaeIII (C) are shown. * indicate interstitial DNA with homology to the telomeric repeat. The rate of telomere shortening was calculated by comparing the size of the smallest telomere fragments in each generation of the mutants with telomeres from wild-type plants.