Abstract
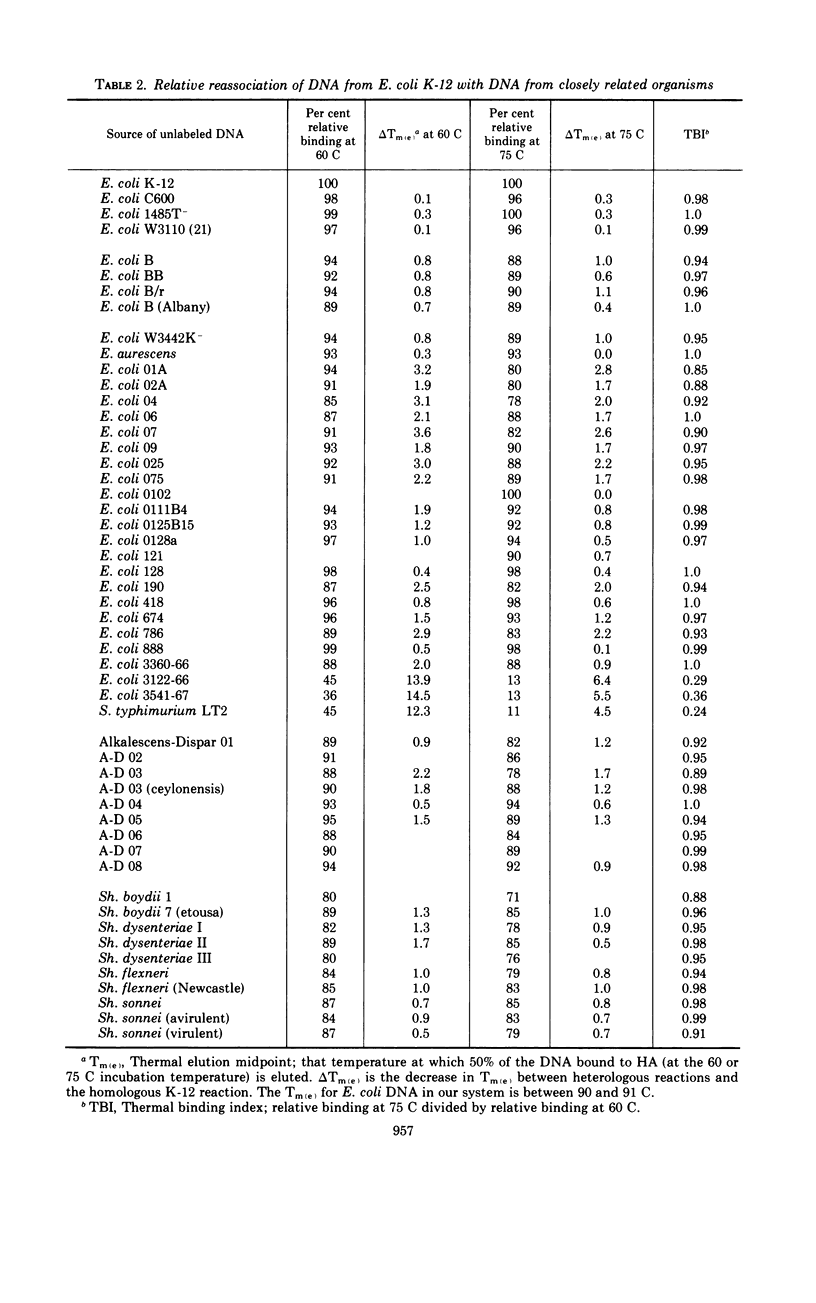
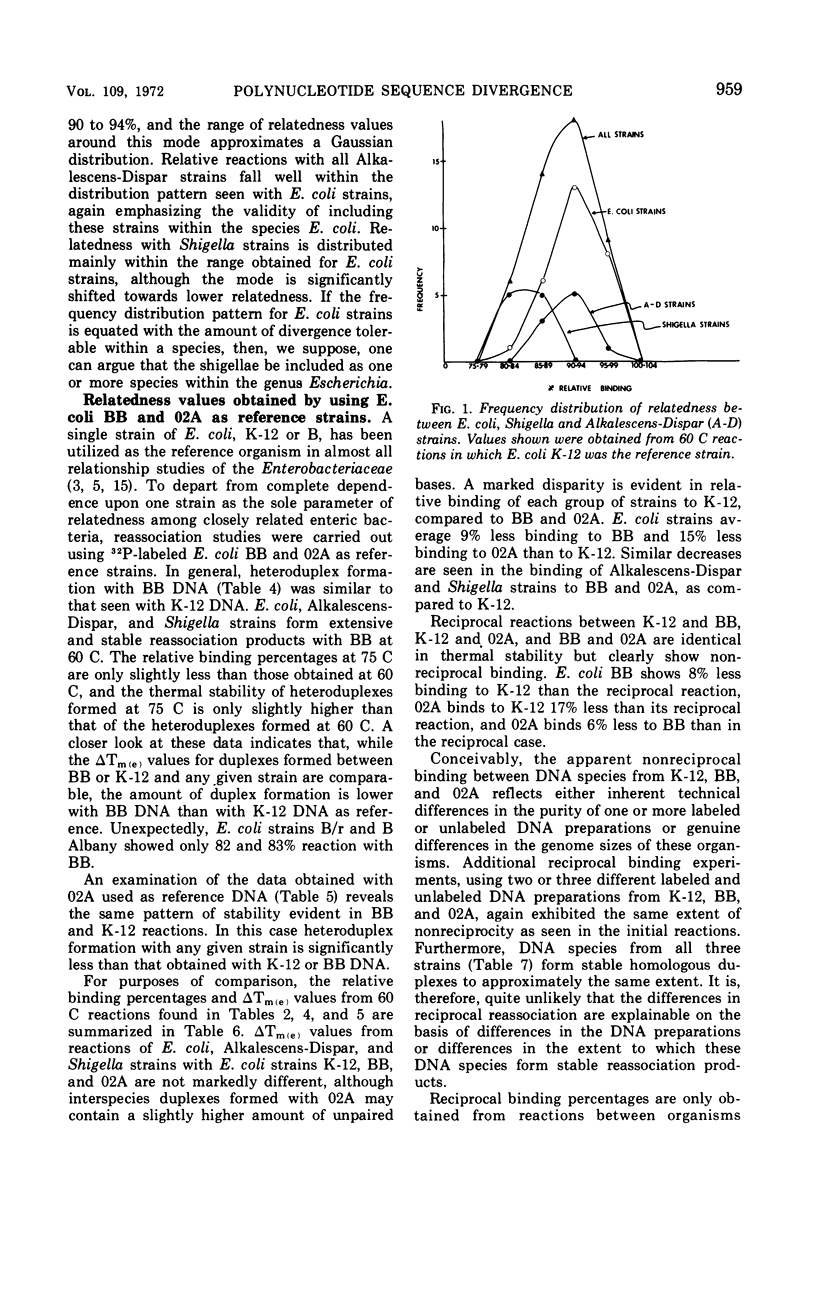
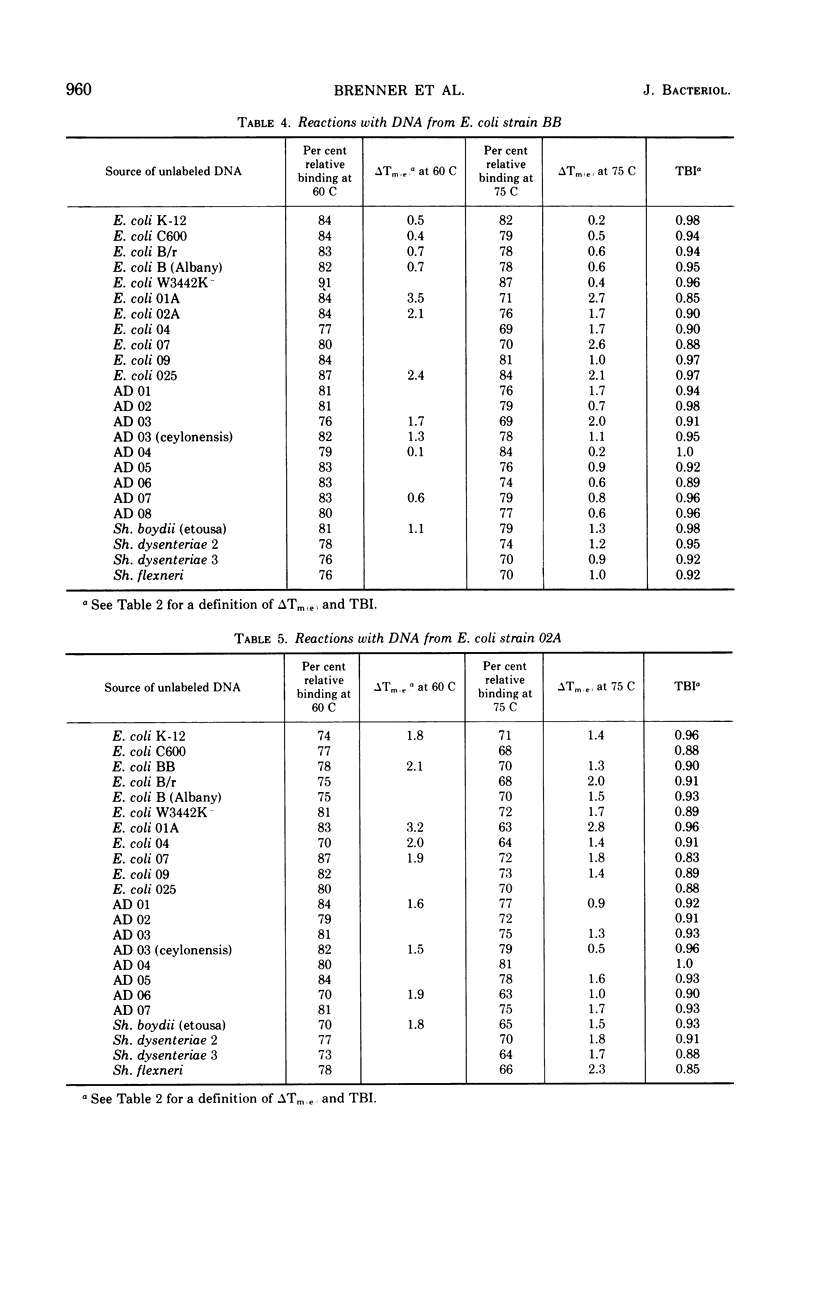
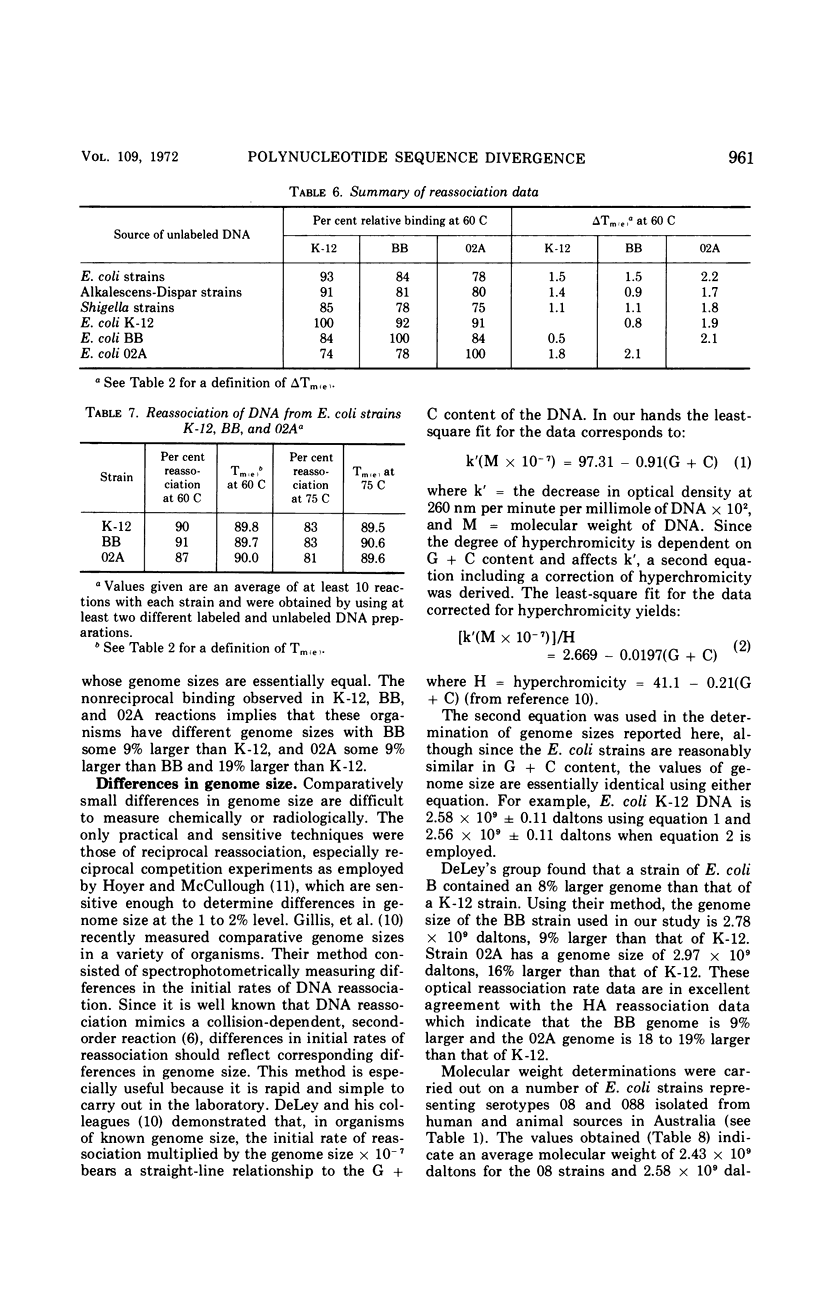
Polynucleotide sequence similarity tests were carried out to determine the extent of divergence present in a number of Escherichia coli strains, obtained from diverse human, animal, and laboratory sources, and closely related strains of Shigella, Salmonella, and the Alkalescens-Dispar group. At 60 C, relative reassociation of deoxyribonucleic acid (DNA) from the various strains with E. coli K-12 DNA ranged from 100 to 36%, with the highest level of reassociation found for three strains derived from K-12, and the lowest levels for two “atypical” E. coli strains and S. typhimurium. The change in thermal elution midpoint, which indicates the stability of DNA duplexes, ranged from 0.1 to 14.5 C, with thermal stability closely following the reassociation data. Reassociation experiments performed at 75 C, at which temperature only the more closely related DNA species form stable duplexes, gave similar indications of relatedness. At both temperatures, Alkalescens-Dispar strains showed close relatedness to E. coli, supporting the idea that they should be included in the genus Escherichia. Reciprocal binding experiments with E. coli BB, 02A, and K-12 yielded different reassociation values, suggesting that the genomes of these strains are of different size. The BB genome was calculated to be 9% larger than that of K-12, and that of 02A 9% larger than that of BB. Calculation of genome size for a series of E. coli strains yielded values ranging from 2.29 × 109 to 2.97 × 109 daltons. E. coli strains and closely related organisms were compared by Adansonian analysis for their relatedness to a hypothetical median strain. E. coli 0128a was the most closely related to this median organism. In general, these data compared well with the data from reassociation experiments among E. coli strains. However, anomalous results were obtained in the cases of Shigella flexneri, S. typhimurium, and “atypical” E. coli strains.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BAUTZ E. K., BAUTZ F. A. THE INFLUENCE OF NONCOMPLEMENTARY BASES ON THE STABILITY OF ORDERED POLYNUCLEOTIDES. Proc Natl Acad Sci U S A. 1964 Dec;52:1476–1481. doi: 10.1073/pnas.52.6.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNS K. I., THOMAS C. A., Jr ISOLATION OF HIGH MOLECULAR WEIGHT DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:476–490. doi: 10.1016/s0022-2836(65)80004-3. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Johnson K. E., Citarella R. V., Falkow S. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol. 1969 May;98(2):637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Rake A. V., Johnson K. E. Batch procedure for thermal elution of DNA from hydroxyapatite. Anal Biochem. 1969 Apr 4;28(1):447–459. doi: 10.1016/0003-2697(69)90199-7. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Martin M. A., Hoyer B. H. Deoxyribonucleic acid homologies among some bacteria. J Bacteriol. 1967 Aug;94(2):486–487. doi: 10.1128/jb.94.2.486-487.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T. Measurement of 32P activity in a liquid scintillation counter without the use of scintillator. Anal Biochem. 1968 Jan;22(1):70–73. doi: 10.1016/0003-2697(68)90260-1. [DOI] [PubMed] [Google Scholar]
- Colwell R. R. Polyphasic taxonomy of the genus vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J Bacteriol. 1970 Oct;104(1):410–433. doi: 10.1128/jb.104.1.410-433.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis M., De Ley J., De Cleene M. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970 Jan;12(1):143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Hoyer B. H., McCullough N. B. Homologies of deoxyribonucleic acids from Brucella ovis, canine abortion organisms, and other Brucella species. J Bacteriol. 1968 Nov;96(5):1783–1790. doi: 10.1128/jb.96.5.1783-1790.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LISTON J., WIEBE W., COLWELL R. R. QUANTITATIVE APPROACH TO THE STUDY OF BACTERIAL SPECIES. J Bacteriol. 1963 May;85:1061–1070. doi: 10.1128/jb.85.5.1061-1070.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird C. D., McConaughy B. L., McCarthy B. J. Rate of fixation of nucleotide substitutions in evolution. Nature. 1969 Oct 11;224(5215):149–154. doi: 10.1038/224149a0. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- MCCARTHY B. J., BOLTON E. T. An approach to the measurement of genetic relatedness among organisms. Proc Natl Acad Sci U S A. 1963 Jul;50:156–164. doi: 10.1073/pnas.50.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler R. J., Mandel M. Quantitative aspects of deoxyribonucleic acid renaturation: base composition, state of chromosome replication, and polynucleotide homologies. J Bacteriol. 1971 May;106(2):608–614. doi: 10.1128/jb.106.2.608-614.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]


