Abstract
Protein acylation is an important way in which a number of proteins with a variety of functions are modified. The physiological role of the acylation of cellular proteins is still poorly understood. Covalent binding of fatty acids to nonintegral membrane proteins is thought to produce transient or permanent enhancement of the association of the polypeptide chains with biological membranes. In this paper, we investigate the functional role for the palmitoylation of an atypical membrane-bound protein, yeast protoporphyrinogen oxidase, which is the molecular target of diphenyl ether-type herbicides. Palmitoylation stabilizes an active heat- and protease-resistant conformation of the protein. Palmitoylation of protoporphyrinogen oxidase has been demonstrated to occur in vivo both in yeast cells and in a heterologous bacterial expression system, where it may be inhibited by cerulenin leading to the accumulation of degradation products of the protein. The thiol ester linking palmitoleic acid to the polypeptide chain was shown to be sensitive to hydrolysis by hydroxylamine and also by the widely used serine-protease inhibitor phenylmethylsulfonyl fluoride.
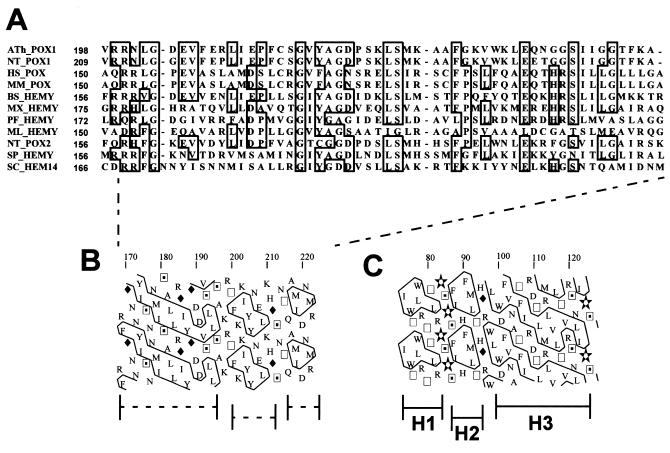
Protoporphyrinogen oxidase is a mitochondrial membrane-bound flavoprotein that catalyzes the penultimate reaction in the heme biosynthesis pathway, the oxygen-dependent aromatization of protoporphyrinogen IX to protoporphyrin IX (1). This enzyme has been shown to be the molecular target of diphenyl ether-type herbicides in plants (2, 3). The biochemical characteristics of the protein are those of an intrinsic protein, but sequence analysis of cloned yeast, mammalian, or plant protoporphyrinogen oxidases failed to reveal any typical membrane-spanning α-helical structure (4–9). Studies of the biogenesis of the yeast enzyme have shown that the protein is synthesized as a precursor that is rapidly converted to the active form of protoporphyrinogen oxidase, but that this maturation does not involve the removal of a N-terminal mitochondrial targeting peptide (6, 10). Although the sequence of protoporphyrinogen oxidase contains several hydrophobic regions, none of those is longer than 15 uncharged residues, and they are therefore unlikely to form membrane spanning segments; the largest hydrophobic domain in the protein is very homologous to the βαβ-dinucleotide (Rossman)-binding fold found in many flavoproteins. Anchoring to the membrane may involve amphipathic helical domains that could be responsible for insertion of the protoporphyrinogen oxidase into the inner mitochondrial membrane, as described for prostaglandin H2 synthase-1, a monotopic membrane-bound protein of the endoplasmic reticulum (11). The conserved domain A in protoporphyrinogen oxidases (12) (Fig. 1A) may well represent such an anchoring structure, and hydrophobic cluster analysis (13) of this domain (Fig. 1B) shows striking similarities to the membrane-anchoring domain of prostaglandin H2 synthase (Fig. 1C). Another possibility, compatible with a posttranslational modification of the protein leading to the shift in electrophoretic mobility initially attributed to the proteolytic cleavage of a putative presequence previously described (10), is that protoporphyrinogen oxidase is anchored to the inner mitochondrial membrane by a different mechanism, such as acylation. This prompted us to look for a potential posttranslational modification of the polypeptide chain that could contribute to both the highly hydrophobic nature of the protein and the difference in electrophoretic mobility of the precursor and the mature forms of the protein. Studies on the biogenesis of the Ras oncogene products showed that acylation of these proteins may alter their electrophoretic mobility and hydrophobicity (14). We therefore investigated the possibility that protoporphyrinogen oxidase is an acylated protein.
Figure 1.
(A) Sequence alignment of the putative membrane-anchoring domain in protoporphyrinogen oxidases (clustal w). (B) Two-dimensional graph representation [Hydrophobic Cluster Analysis (13)] of the peptide sequence of yeast protoporphyrinogen oxidase putative membrane-anchoring domain. (C) Two-dimensional graph representation (Hydrophobic Cluster Analysis) of the peptide sequence of sheep prostaglandin H2-synthase-1 membrane-anchoring domain determined from the crystal structure of the protein (11). H1, H2, and H3 are the amphipathic helices in interaction with the lipid bilayer.
Materials and Methods
Materials.
Protoporphyrin IX, disodium salt, was purchased from Serva. l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (EC 3.4.21.4) from bovine pancreas used either as a soluble enzyme (Type XIII, 10,000–13,000 units⋅mg−1) or attached to agarose beads (75–100 units⋅ml−1 packed gel), cerulenin (2, 3-epoxy-4-oxo-7, 10 dodecadieneamide) and PMSF were obtained from Sigma; acifluorfen, sodium salt from ChemService, West Chester, PA. Diphenyleneiodonium chloride was obtained from the Alexis, San Diego, CA. All inhibitors were dissolved in DMSO to give 10-mM stock solutions. [3H]Myristic acid and [3H]palmitic acid were from Amersham France, and [3H]palmitoleic acid was from IsotopChim, Ganagobie-Peyruis, France.
Analytical Methods on Recombinant Yeast Protoporphyrinogen Oxidase.
Protoporphyrinogen oxidase was overproduced and purified from yeast cells as in ref. 6. The purified protein (1 mg⋅ml−1) was delipidated either by (i) a modified Folch procedure or (ii) treatment with sodium hydroxylamine. In the first method, 1 vol of protein solution was vortexed with 4 vol methanol and centrifuged for 10 sec at 9,000 × g. One vol of chloroform was added and the samples were centrifuged for 10 sec at 9,000 × g. Proteins in the chloroform phase were precipitated by 3 vol of methanol and collected by centrifugation for 2 min at 9,000 × g. The pelleted proteins were dissolved in Laemmli buffer and analyzed by SDS/PAGE. In the second method, 1 vol of protein solution was mixed with 1 vol sodium hydroxylamine (2 M, pH 8.0) and incubated for 1 hr at 4°C.
Chemical analysis of lipids associated to the protein was performed on a Hewlett–Packard gas-phase chromatography system after saponification and extraction.
Trypsin was assayed at 25°C in 0.05 M potassium phosphate buffer, pH 7.3, containing 0.05 M KCl. Proteolysis of native or deacylated protoporphyrinogen oxidase was started by adding either 25 N-α-benzoyl-l-arginine ethyl ester (BAEE) units of soluble trypsin (2% final concentration wt/wt) or 2.5 BAEE units of immobilized trypsin to the incubation medium. One BAEE unit of trypsin catalyzes the production of an ΔOD253 of 0.001 per min in 3.2 ml at pH 7.6 and 25°C. The reaction was started by adding trypsin, and protoporphyrinogen oxidase activity was determined during trypsin proteolysis. The degree of trypsin cleavage was assessed by SDS/PAGE. When immobilized trypsin was used, the incubation mixture was centrifuged for 1 min at 13,000 × g, and protoporphyrinogen oxidase activity measured on the supernatant. Control experiments with BAEE as the substrate showed that trypsin was slightly inhibited by hydroxylamine at the concentrations used in the deacylation experiments. Control experiments with BSA as the substrate showed that the trypsin was fully active under these assay conditions.
CD measurements were performed with a Jasco (Easton, MD) J-710 spectropolarimeter by using a 0.1-cm pathlength quartz cuvette. The temperature of the cell was controlled by a programmable Peltier thermoelectric system that allows thermal-ramping (melting-curve) experiments as well as automated scans at preset temperatures with temperature monitored within the cuvette. All spectra were recorded at 20°C and analyzed on accumulation of 10 spectra. The secondary structure of the protein was estimated from the CD spectra by using commercial software (Jasco) with the data of Yang et al. (15) as references. The software searches for a theoretical spectrum that best fits to experimental data either considering the estimated protein concentration (finite mode) or by fitting the protein concentration (infinite mode). The two fitting modes provided very similar results in our study, indicating that the estimation of protein concentration was correct. Thermal denaturation experiments were done with temperature increases of 1°C/min. The incubation mixture was 0.05 M potassium phosphate buffer, pH 7.3, containing 0.5% n-octyl glucoside. The assays were started at 20°C 10 μM purified native enzyme or hydroxylamine-treated enzyme incubated with or without acifluorfen (10 to 100 μM), 4-nitro-2,2′-diphenyleneiodenium (20 to 200 μM).
In Vitro Labeling with Tritiated Fatty Acids.
Yeast cells were grown at 30°C on yeast extract/peptone/dextrose medium to an OD600 of 6, concentrated 10 times by centrifuging, and resuspended in fresh yeast extract/peptone medium. The cell suspensions were incubated for 3–12 h at 30°C with tritiated fatty acids in ethanol (0.1 mCi/ml cells). Cells were collected by centrifuging, broken with glass beads in a 0.1 M potassium phosphate buffer, pH 7.3, and a fraction enriched in mitochondrial membranes prepared by differential centrifugation. The membrane-bound proteins were solubilized in a 0.1 M potassium phosphate buffer, pH 7.3, containing 0.1 M KCl, 1 mM EDTA and 2% (wt/vol) N-octyl glucoside, and analyzed by SDS/urea/PAGE followed by fluorography of the dried gels. Because of the labile nature of the acyl-protein bond, samples were not reduced before electrophoresis, and the gels were not fixed in acetic acid/ethanol before fluorography.
The Escherichia coli strain BL21(DE3) transformed with the plasmid pSBETa-HEM14 (16) overproduced yeast protoporphyrinogen oxidase when the cells were grown in a M63 synthetic medium supplemented with 0.4% glucose to an OD600 of 1.5 and the gene expression induced with 1 mM isopropylthiogalactoside for 18 h. In labeling experiments, the tritiated fatty acids (0.1 mCi/ml cells) were added to the cell suspension 30 min after beginning the induction of protoporphyrinogen oxidase production. Cells were collected by centrifuging, sonicated in a 0.1 M potassium phosphate buffer, pH 7.3, and the membrane fraction prepared by differential centrifugation. The membrane-bound proteins were solubilized and processed as described above. The in vivo effects of cerulenin on protoporphyrinogen oxidase stability were analyzed by immunodetection of the protein. Cerulenin (0 to 50 μg/ml cell suspension; stock 22.4 mM in DMSO) was added to the cell suspension 30 min after beginning the induction of protoporphyrinogen oxidase production. Control experiments were run with the soluble yeast enzyme, coproporphyrinogen oxidase, cloned in pSBETa and expressed in the same conditions as protoporphyrinogen oxidase.
Miscellaneous.
Published procedures were used for SDS/PAGE (17). Size exclusion chromatography analysis of protoporphyrinogen oxidase was performed by using a TSK-3000 column (0.75 × 30 cm) equilibrated in 0.05 M potassium phosphate buffer, pH 7.3, containing 0.5% N-octyl glucoside. The flow rate was 0.6 ml⋅min−1. Protoporphyrinogen oxidase was assayed by measuring the rate of appearance of protoporphyrin fluorescence (18) at 30°C. The incubation mixture was 0.1 M potassium phosphate buffer, pH 7.3, saturated with air, containing 2 μM protoporphyrinogen IX, 3 mM palmitic acid (in DMSO 0.5% vol/vol final concentration), 5 mM DTT, 1 mM EDTA, and 0.3 mg/ml (final concentration) Tween 80, to ensure maximum fluorescence of the protoporphyrin IX. Protoporphyrinogen was prepared by reducing protoporphyrin IX hydrochloride dissolved in KOH/EtOH (0.04 N/20%) with freshly prepared 3% sodium amalgam (19). Coproporphyrinogen oxidase was measured as described (20). Protein concentrations of solutions diluted with 0.1 N NaOH were measured by the microscale Bradford technique on solution. One unit of activity is the amount of enzyme that oxidizes 1 nmol protoporphyrinogen IX to protoporphyrin IX per hour at 30°C.
Results and Discussion
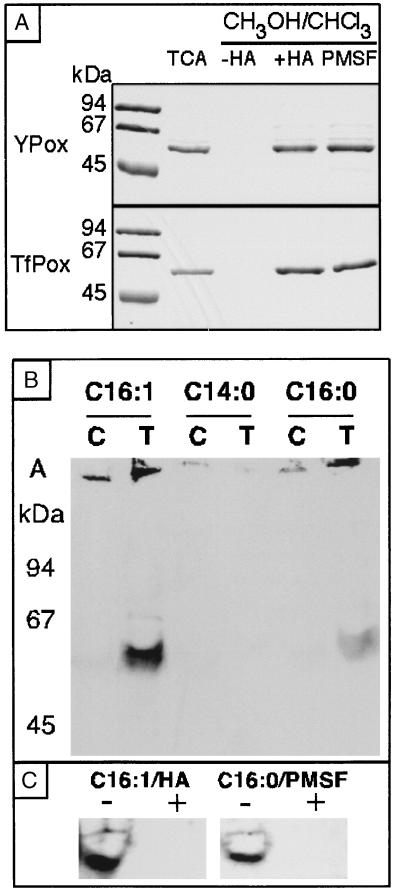
Protoporphyrinogen oxidase was overproduced in yeast cells and purified to homogeneity. Attempts to remove excess detergents and lipids from the purified protein by chloroform extraction of the lipids and precipitation of the protein with methanol showed that the protein was extracted in the lipid phase (Fig. 2A). This suggested that the protein may be acylated, and we investigated this possibility further. The N-terminal peptide sequence of the protein is identical to that deduced from the gene sequence and starts with a methionine, which rules out the possibility of the myristoylation of the protein as this requires a glycine residue in the N-terminal position. The protein lacks any consensus sequence for isoprenylation in its C terminus (C-A-A-X, where A is an aliphatic residue and X, the C-terminal amino acid residue of the protein). We therefore evaluated the potential of the protein for palmitoylation. Palmitoylation generally involves the thioesterification of cysteine residues (21). Thioesters are labile and may be hydrolyzed under mild conditions (1 M hydroxylamine, pH 8.0, 4°C). Treatment of the purified protoporphyrinogen oxidase in this way made it possible to recover the protein as a soluble polypeptide that was no longer extracted in the lipid phase (Fig. 2A). Chemical analysis of the lipids associated with the purified protein showed that the enzyme preparations contained many phospholipids, principally phosphatidylethanolamine and phosphatidylserine, at a molar ratio of over 10:1 (phospholipids/protein). Palmitoleic acid was found bound to the protein in almost stoichiometric amounts, together with smaller quantities of palmitic acid (10% by mass of lipid). The acylation of protoporphyrinogen oxidase was confirmed in in vivo labeling experiments where a wild-type yeast strain, a HEM14-null mutant strain, and this mutant transformed by a multicopy plasmid overproducing protoporphyrinogen oxidase, were grown in the presence of [3H]myristic acid, [3H]palmitic acid or [3H]palmitoleic acid. Protoporphyrinogen oxidase was not labeled in the presence of [3H]myristic acid but did incorporate palmitic acid and to an even greater extent palmitoleic acid, confirming that this fatty acid is the major component involved in modifying the protein. Some mitochondrial proteins are myristoylated (22, 23) or may be sequestered to the inner membrane through a phosphatidylinositol (24). Protoporphyrinogen oxidase is a new member of the recently described class of yeast proteins that are subjected to palmitoylation (25) and is the first mitochondrial enzyme of this class.
Figure 2.
(A) Coomassie blue staining of protoporphyrinogen oxidase purified from yeast cells (Ypox) or from transformed E. coli cells (TfPox) on an SDS/PAGE gel in the pellet of proteins precipitated with trichloracetic acid and in the pellet of proteins precipitated with methanol/chloroform without hydroxylamine treatment (−HA), with prior hydroxylamine treatment (+HA), or with prior PMSF treatment (+PMSF). (B) SDS/PAGE and fluorographic analysis of recombinant yeast protoporphyrinogen oxidase after in vivo labeling of the transformed (T) E. coli BL21(DE3) with tritiated fatty acids: C16:1 [3H]palmitoleic acid; C14:0 [3H]myristic acid; C16:0 [3H]palmitic acid. In control experiments (C), the labeled cells were transformed by the pSBETa expression vector without insert. A, aggregated proteins. (C) SDS/PAGE and fluorographic analysis of recombinant yeast protoporphyrinogen oxidase after in vivo labeling with [3H]palmitoleic acid (C16:1) or [3H]palmitic acid (C16:0) (−) followed by hydroxylamine treatment (+) or PMSF treatment (+).
When the yeast protoporphyrinogen oxidase gene was expressed in E. coli, the protein was overproduced as a fully active and flavinylated membrane-bound protein (16). The purified recombinant protoporphyrinogen oxidase exhibited hydrophobicity properties similar to those of the original enzyme and was recovered in the lipid phase of the chloroform/methanol extraction procedure; treatment of the protein with hydroxylamine restored the solubility properties of the hydrophilic core of the protein (Fig. 2A). This suggested that protoporphyrinogen oxidase is subject to the same type of posttranslational modification in E. coli as in yeast mitochondria. In vivo labeling of the bacterial cells overproducing recombinant protoporphyrinogen oxidase with radiolabeled fatty acids showed that the protein was efficiently acylated by [3H]palmitoleic acid and to a lesser extent by [3H]palmitic acid, but not by [3H]myristic acid (Fig. 2B). The labeling was completely removed by hydroxylamine treatment of the protein (Fig. 2C). Palmitoylation specific to protoporphyrinogen oxidase occurs both in yeast cells and in E. coli, and this raises some questions about the molecular basis of this type of posttranslational modification. Several components of the protein acylation machinery in yeast have been characterized in the course of studies on the biogenesis of the Ras proteins, which have to undergo myristoylation, farnesylation, and palmitoylation to be fully activated. Both myristoylation and farnesylation are known to be catalyzed by specific enzymes or enzyme complexes in yeast (26, 27), but the proper myristoylation of heterologous proteins in E. coli requires the coexpression of several components of the eukaryotic modification enzymes (28). In yeast, the palmitoylation of the Ras2 protein is affected by mutations in the SHR5 gene, but the overall palmitoylation activity of cell-free extracts from an SHR5-null mutant is similar to that of a wild-type strain (29). No homologue of SHR5 or of any other eukaryotic protein–palmitoyl transferase is found in E. coli, but this organism contains some palmitoyl transferase activity, which is involved at least in lipid-A biosynthesis (30). The similar pattern of acylation of overproduced protoporphyrinogen oxidase in both yeast and E. coli argues against a target-specific modification enzyme, because no homologue of HEM14 is found in E. coli. However, it is more likely that the protoporphyrinogen oxidase polypeptide chain readily exhibits uncatalyzed palmitoyl—CoA-dependent acylation similar to that reported for model cysteine-containing peptides (31).
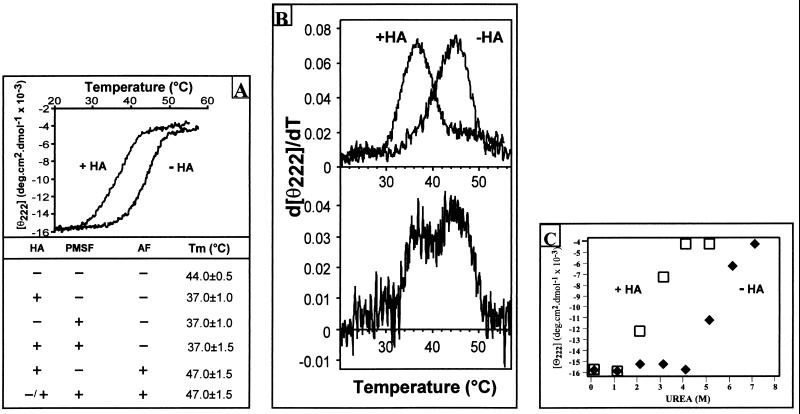
The physicochemical and enzymatic properties of the purified native recombinant protoporphyrinogen oxidase and of the deacylated protein were compared. No significant change in the enzyme activity was observed as a result of deacylation. The Vmax (80 μmol⋅h−1⋅mg−1 protein), the Km for protoporphyrinogen (0.1 μM), and the reactivity of the enzyme toward various classes of inhibitors (diphenyl ethers and diphenyleneiodonium derivatives) were unchanged. The effects of deacylation on the structure of the protein were analyzed by CD measurement of the changes in protein ellipticity after hydroxylamine treatment. No significant change in the overall content of secondary structures was observed. However, the deacylated protein appeared to be significantly more heat labile than the native protein, with Tm reduced from 44°C to 37°C (Fig. 3A), as reported for deacylated rhodopsin (32). The deacylated protein still constitutes a single folding unit, as shown by the single peak in the first derivative plot of the melting curves (Fig. 3B). Melting analysis of protoporphyrinogen oxidase appears to be an extremely powerful tool for evaluating the degree of acylation of the protein, and mixtures of acylated and deacylated protein can be clearly identified from the two peaks in the first derivative plot of the composite melting. To further investigate the possible effects of acylation on protoporphyrinogen oxidase stability, we observed solvent denaturation of the native and hydroxylamine treated enzymes in the presence of urea. Although the Cm were very different (5.5 M urea for the native protein vs. 2 M urea for the deacylated protein), the m-slope of the denaturation curves (Fig. 3C) was not significantly different, suggesting little or no influence of deacylation on the unfolded or denaturated state of the protein (33–35). This was confirmed by analyzing the oligomeric status of the acyl and deacyl protein. We have previously shown that purified native yeast protoporphyrinogen oxidase is a monomer as determined by size exclusion chromatography in size-exclusion chromatography (SEC)-HPLC experiments (6). The hydroxylamine-treated enzyme exhibited the same chromatographic behavior as the native enzyme (elution time 8.2 ± 0.1 min), suggesting that the protein remains monomeric after deacylation. This implies that the changes in stability of the deacyl protein are not related to oligomerization. A high degree of oligomerization was, however, observed when heat-denaturated native or deacylated protein was subjected to SEC-HPLC.
Figure 3.
(A) Thermal denaturation of yeast protoporphyrinogen oxidase measured by CD. The purified native enzyme (10 μM) used either was the untreated native enzyme (−HA) or had been treated with hydroxylamine (+HA). Temperature increases of 60°C⋅h−1 were used, and the CD was measured at 222 nm. (Lower) The table summarizes the Tm values of the enzyme on sample protein after (+) or without (−) prior treatment with hydroxylamine or PMSF and measured in the absence (−) or presence (+) of 100 μM acifluorfen. (B) First derivative of the melting curves shown in A, measured on a freshly prepared enzyme (Upper) or on an enzyme preparation stored for 3 mo at −20°C (Lower). (C) Solvent denaturation of yeast protoporphyrinogen oxidase measured by CD. The purified native enzyme (10 μM) used either was the untreated native enzyme (−HA) or had been treated with hydroxylamine (+HA). The CD was measured at 222 nm as a function of urea concentration.
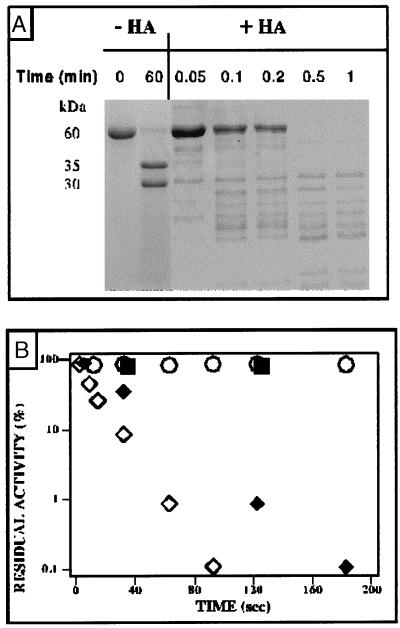
The functional role for acylation may be more than stabilizing transient or permanent enhancement of the association of the polypeptide chains with biological membranes, and the concept has emerged recently that the fatty acylation of proteins may play an essential part in modulating the reactivity of key components of major signal transduction cascades (21, 36–39). The most striking feature of deacylated protoporphyrinogen oxidase is its extreme sensitivity to proteases. We have recently demonstrated that yeast protoporphyrinogen oxidase is organized into two functional domains, which are structured to form a single folding unit that is extremely resistant to proteases (trypsin, endo-Glu proteinase, or carboxypeptidases A, B, and Y) (12). This basic protein exhibits a single trypsin cleavage site in a loop connecting what appeared to be two compact folding domains. However, once the protein had been deacylated, trypsin rapidly degraded the polypeptide chain to form a large number of small peptides (Fig. 4A). The half-life of the enzyme activity dropped from >2 h to <20 sec as a result of the action of trypsin (Fig. 4B). Control experiments showed that the activity of trypsin itself was not enhanced but in fact slightly inhibited by hydroxylamine treatment. The increase in sensitivity to proteases observed with deacylated protoporphyrinogen oxidase is several orders of magnitude greater than that reported for gastric mucus glycoprotein, a marker protein in cystic fibrosis, which is secreted to protect the mucosal epithelium. Gastric mucus glycoprotein is a heavily glycosylated and palmitoylated protein, with more than 20 fatty acids per polypeptide chain (40, 41), which appears to be slightly more susceptible to pepsin when depalmitoylated than when fully acylated.
Figure 4.
Trypsin proteolysis of recombinant yeast protoporphyrinogen oxidase with: (A) Coomassie blue staining of proteins on an SDS/PAGE gel. Purified yeast protoporphyrinogen oxidase was incubated with 25 units of trypsin, and aliquot fractions of the digestion mixture without (−HA) or after (+HA) prior treatment with hydroxylamine were analyzed by SDS/PAGE as a function of incubation time. (B) Protoporphyrinogen oxidase activity measured as a function of incubation time with 25 units trypsin without hydroxylamine treatment (○) or with hydroxylamine treatment (◊) and in the presence of 2.5 units immobilized trypsin without (■) or with (⧫) hydroxylamine treatment.
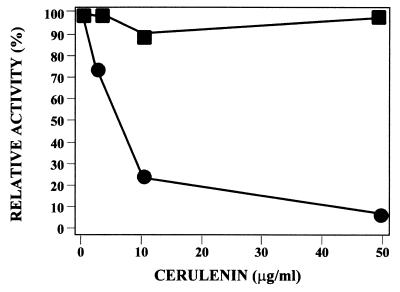
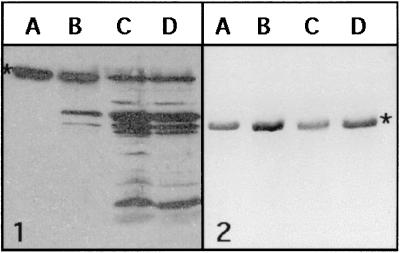
To evaluate whether acylation may play a role in the regulation of protoporphyrinogen oxidase stability in vivo, we overproduced the protein in bacterial cells in the presence of cerulenin, a potent inhibitor of β-ketoacyl-acyl carrier protein synthase (42). This antibiotic has also been shown to inhibit the acylation of viral glycoproteins (43, 44), hormone-releasing factor (45), or HLA (subtype D)-associated invariant chain (46). Protoporphyrinogen production is severely affected by cerulenin in a dose-dependent manner (Fig. 5). Immunodecoration of total proteins from control and treated cells shows accumulation of large amounts of degradation products in the latter case (Fig. 6-1). Attempts to label in vivo protoporphyrinogen oxidase with [3H]palmitic acid in the presence of a high concentration of cerulenin failed (data not shown). The T7-based expression system used to overproduce yeast protoporphyrinogen oxidase was not affected by the presence of cerulenin, as shown by the similar levels of activity and protein found when yeast coproporphyrinogen oxidase, a soluble enzyme, was overproduced in E. coli under the same experimental conditions (Fig. 6-2). Therefore, we conclude that acylation of protoporphyrinogen oxidase plays an important role in stabilizing the protein both in vitro and in vivo.
Figure 5.
Effect of cerulenin on recombinant protoporphyrinogen oxidase activity (●) and coproporphyrinogen oxidase activity (■) in E. coli cells overproducing the yeast proteins. Cerulenin was added to the cell suspension 30 min after isopropyl-d-thiogalactoside.
Figure 6.
Immunodecoration with rabbit polyclonal antibodies raised to yeast protoporphyrinogen oxidase (1) or to yeast coproporphyrinogen oxidase (2) of total proteins from E. coli cells overproducing the proteins in the absence (lane A) or in the presence of cerulenin, 2.5 (lane B), 10 (lane C), or 50 (lane D) μg/ml cell suspension. Proteins were separated by SDS/PAGE and transferred to nitrocellulose sheets. The titer of antibodies was 1/10,000 for antiyeast protoporphyrinogen oxidase and 1/3,000 for antiyeast coproporphyrinogen oxidase. Peroxidase-conjugated anti-rabbit IgG secondary antibodies were used for chemiluminescent detection.
Experiments were carried out to map the acylation sites on [3H]palmitate-labeled protoporphyrinogen oxidase. The labeled native protein was subjected to trypsin treatment, and the two large peptides generated resolved by SDS/PAGE. Labeling was detected by fluorography. Surprisingly, no radioactivity associated with either peptide was recovered. At first, we considered whether trypsin could have esterase activity and could have deacylated the protein, but experiments led to a more trivial explanation. In the proteolysis experiments, the action of trypsin was stopped by adding PMSF, a standard inhibitor of serine proteases, to the reaction medium. However, the addition of PMSF to the labeled protoporphyrinogen oxidase caused the deacylation of the enzyme very efficiently (Fig. 2C). The sulfhydryl reagents 5,5′-dithiobis(2-nitrobenzoate) and 4-chloromercuribenzoate were found to be as effective as PMSF in deacetylating protoporphyrinogen oxidase and induced marked trypsin sensitivity in the treated enzyme. The effect of PMSF on protoporphyrinogen oxidase was similar to that produced by hydroxylamine treatment, in terms of changes in the hydrodynamic properties of the protein and the reduction of the Tm of the protein (Fig. 3A). Using trypsin immobilized to agarose beads allows removal of the protease of samples without adding PMSF, giving essentially the same results as those described before. The half-life of protoporphyrinogen oxidase was slightly longer (45 sec), but the experiments were carried out with less trypsin, and agarose beads may lower the catalytic efficiency of the protease or substrate accessibility to its active site. This rules out a trivial artifact in the experiments because of the presence of the trypsin inhibitor. PMSF action leads to a paradoxical situation in which an additive intended to protect a protein against proteases may actually make the target more labile and promote its degradation. Sodium and ammonium fluoride (100 μM) also induced deacylation of protoporphyrinogen oxidase. However, neither PMSF nor fluoride anion allows the hydrolysis of thiol esters in buffered neutral solution in model compounds such as α-toluenethiol acetate (R. Mornet and M. Dias, personal communication). Because PMSF is readily hydrolyzed in water, giving the fluoride anion, it can be expected that the thiol ester in protoporphyrinogen oxidase is decomposed through a mechanism resulting from the activation of the carbonyl group by hydrogen bonding to a residue of the polypeptide chain. It would then allow the attack of the fluoride ion, giving the free thiol and the acyl fluoride that is then readily hydrolyzed. The finding that the use of PMSF causes the deacylation of protoporphyrinogen oxidase may be relevant to many studies of acylated proteins and should be taken into account in designing studies to investigate the function of this important posttranslational modification of proteins. The molecular basis of the switch from a protease-resistant to a protease-sensitive form of the polypeptide chain of the protoporphyrinogen oxidase after deacylation remains unclear. Data from CD estimations of secondary structure (40.0 ± 1.5% α-helix, 23.5 ± 2.5% β-sheet, 18.0 ± 2.0% β-turn, and 18.5 ± 2.5% random coil for both native and deacylated protein) clearly indicate that deacylation does not induce any massive structural transformation, such as that observed with the scrapie prion protein, PrP, where a conformational change from α-helices to β-sheets is associated with the pathogenicity of PrPSC and its resistance to proteolysis (47). The presence of bound fatty acid associated with yeast protoporphyrinogen oxidase probably locks the protein into a conformation, exposing the maximum surface area to interact with the lipid bilayer. Removing the fatty acid may relax the tensed protein and destructure a micellar form of the protein, reducing the protection of the many positively charged residues of the protein by phospholipid polar heads, thus making it more accessible to the action of trypsin. This biological situation has several similarities to that described during the activation of the complement proteins C3 and C4. A key step in the elimination of pathogens from the body involves the covalent binding of complement proteins C3 and C4 to the surface of pathogens. Proteolytic activation then brings about a conformational change in these proteins, as a result of which an internal thioester is exposed and may react with amino or hydroxyl groups on the target surface to form amide or ester bonds or may be hydrolyzed. Dodds et al. (48) have reported that the binding of the human C4A isotype involves a direct reaction between amino nucleophiles and the thioester, whereas that of the C4B isotype involves a two-step mechanism. In contrast, the binding of the C4B isotype, and probably that of C3, to hydroxyl nucleophiles involves a histidine residue, which attacks the thioester to form an intramolecular acyl-imidazole bond. The thiolate anion released then acts as a base to catalyze the binding of hydroxyl nucleophiles, including water, to the acyl function (49). This mechanism enables complement proteins to bind to the hydroxyl groups of the carbohydrates which are found on all biological surfaces, including the components of bacterial cell walls. In addition, the rapidity with which the thioester is hydrolyzed helps to contain this very damaging reaction within the immediate proximity of the activation site.
Protoporphyrinogen oxidase is degraded extremely rapidly by means of a simple reversible modification, and this makes it an ideal target for a finely tuned regulation of tetrapyrrole synthesis, which may be relevant to a better understanding of the pathophysiology of variegate porphyria, in which the enzyme is partially or completely defective, or of plants treated with the inhibitors of protoporphyrinogen oxidase. This model of the regulation of biological activity by means of the rapid degradation of an acylated/deacylated target will be investigated in other acylated proteins or enzymes.
Acknowledgments
We thank Prof. René Mornet, Dr. M. Dias, and Dr. O. Ploux for many helpful discussions. The English text was edited by Dr. O. Parkes. This work was supported by grants from the Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT) (Actions Coordonnées Concertées-Sciences de la Vie no. 5), the Centre National de la Recherche Scientifique (Programme Physique et Chimie du Vivant), and the Association de Recherche sur le Cancer (ARC no. 4025).
References
- 1.Camadro J M, Arnould S, Le Guen L, Santos R, Matringe M, Mornet R. In: Peroxidizing Herbicides. Böger P, Wakabayashi K, editors. Berlin, Heidelberg: Springer; 1998. pp. 245–277. [Google Scholar]
- 2.Matringe M, Camadro J M, Labbe P, Scalla R. Biochem J. 1989;260:231–235. doi: 10.1042/bj2600231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witkowski D A, Halling B P. Plant Physiol. 1989;90:1239–1242. doi: 10.1104/pp.90.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dailey H A, Dailey T A. J Biol Chem. 1996;271:8714–8718. doi: 10.1074/jbc.271.15.8714. [DOI] [PubMed] [Google Scholar]
- 5.Lermontova I, Kruse E, Mock H P, Grimm B. Proc Natl Acad Sci USA. 1997;94:8895–8900. doi: 10.1073/pnas.94.16.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camadro J M, Labbe P. J Biol Chem. 1996;271:9120–9128. doi: 10.1074/jbc.271.15.9120. [DOI] [PubMed] [Google Scholar]
- 7.Nishimura K, Taketani S, Inokuchi H. J Biol Chem. 1995;270:8076–8080. doi: 10.1074/jbc.270.14.8076. [DOI] [PubMed] [Google Scholar]
- 8.Taketani S, Yoshinaga T, Furukawa T, Kohno H, Tokunaga R, Nishimura K, Inokuchi H. Eur J Biochem. 1995;230:760–765. doi: 10.1111/j.1432-1033.1995.0760h.x. [DOI] [PubMed] [Google Scholar]
- 9.Birchfield N B, Latli B, Casida J E. Biochemistry. 1998;37:6905–6910. doi: 10.1021/bi973026k. [DOI] [PubMed] [Google Scholar]
- 10.Camadro J M, Thome F, Brouillet N, Labbe P. J Biol Chem. 1994;269:32085–32091. [PubMed] [Google Scholar]
- 11.Picot D, Loll P J, Garavito R M. Nature (London) 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 12.Arnould S, Camadro J M. Proc Natl Acad Sci USA. 1998;95:10553–10558. doi: 10.1073/pnas.95.18.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callebaut I, Labesse G, Durand P, Poupon A, Canard L, Chomilier J, Henrissat B, Mornon J P. Cell Mol Life Sci. 1997;53:621–645. doi: 10.1007/s000180050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock J, Magee A, Childs J, Marshall C. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 15.Yang J T, Wu C S, Martinez H M. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 16.Arnould S, Takahashi M, Camadro J M. Biochemistry. 1998;37:12818–12828. doi: 10.1021/bi980713i. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Camadro J M, Matringe M, Scalla R, Labbe P. In: Target Assays for Modern Herbicides and Related Phytotoxic Compounds. Böger P, Sandmann G, editors. CRC/Lewis; 1993. pp. 29–34. [Google Scholar]
- 19.Poulson R, Polglase W J. J Biol Chem. 1975;250:1269–1274. [PubMed] [Google Scholar]
- 20.Labbe P, Camadro J M, Chambon H. Anal Biochem. 1985;149:248–260. doi: 10.1016/0003-2697(85)90502-0. [DOI] [PubMed] [Google Scholar]
- 21.Milligan G, Parenti M, Magee A I. Trends Biochem Sci. 1995;20:181–187. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- 22.Stucki J W, Lehmann L H, Siegel E. J Biol Chem. 1989;264:6376–6380. [PubMed] [Google Scholar]
- 23.Vassilev A O, Plesofsky-Vig N, Brambl R. Proc Natl Acad Sci USA. 1995;92:8680–8684. doi: 10.1073/pnas.92.19.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller G, Bandlow W. Biochemistry. 1989;28:9968–9973. doi: 10.1021/bi00452a014. [DOI] [PubMed] [Google Scholar]
- 25.Casey W M, Gibson K J, Parks L W. J Biol Chem. 1994;269:2082–2085. [PubMed] [Google Scholar]
- 26.Goodman L E, Judd S R, Farnsworth C C, Powers S, Gelb M H, Glomset J A, Tamanoi F. Proc Natl Acad Sci USA. 1990;87:9665–9669. doi: 10.1073/pnas.87.24.9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He B, Chen P, Chen S Y, Vancura K L, Michaelis S, Powers S. Proc Natl Acad Sci USA. 1991;88:11373–11377. doi: 10.1073/pnas.88.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knoll L J, Gordon J I. J Biol Chem. 1993;268:4281–4290. [PubMed] [Google Scholar]
- 29.Jung V, Chen L, Hofmann S L, Wigler M, Powers S. Mol Cell Biol. 1995;15:1333–1342. doi: 10.1128/mcb.15.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brozek K A, Bulawa C E, Raetz C R. J Biol Chem. 1987;262:5170–5179. [PubMed] [Google Scholar]
- 31.Quesnel S, Silvius J R. Biochemistry. 1994;33:13340–13348. doi: 10.1021/bi00249a021. [DOI] [PubMed] [Google Scholar]
- 32.Traxler K W, Dewey T G. Biochemistry. 1994;33:1718–1723. doi: 10.1021/bi00173a014. [DOI] [PubMed] [Google Scholar]
- 33.Shortle D, Meeker A K. Proteins. 1986;1:81–89. doi: 10.1002/prot.340010113. [DOI] [PubMed] [Google Scholar]
- 34.Shortle D, Meeker A K. Biochemistry. 1989;28:936–944. doi: 10.1021/bi00429a003. [DOI] [PubMed] [Google Scholar]
- 35.Shortle D. FASEB J. 1996;10:27–34. doi: 10.1096/fasebj.10.1.8566543. [DOI] [PubMed] [Google Scholar]
- 36.Cox A D, Der C J. Curr Opin Cell Biol. 1992;4:1008–1016. doi: 10.1016/0955-0674(92)90133-w. [DOI] [PubMed] [Google Scholar]
- 37.Mumby S M. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin S, Aderem A. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 39.Marshall C J. Science. 1993;259:1865–1866. doi: 10.1126/science.8456312. [DOI] [PubMed] [Google Scholar]
- 40.Slomiany A, Slomiany B L, Witas H, Aono M, Newman L J. Biochem Biophys Res Commun. 1983;113:286–293. doi: 10.1016/0006-291x(83)90464-3. [DOI] [PubMed] [Google Scholar]
- 41.Slomiany A, Witas H, Aono M, Slomiany B L. J Biol Chem. 1983;258:8535–8538. [PubMed] [Google Scholar]
- 42.Buttke T M, Ingram L O. Biochemistry. 1978;17:5282–5286. doi: 10.1021/bi00617a031. [DOI] [PubMed] [Google Scholar]
- 43.Schlesinger M J, Malfer C. J Biol Chem. 1982;257:9887–9890. [PubMed] [Google Scholar]
- 44.Kotwal G J, Ghosh H P. J Biol Chem. 1984;259:4699–4701. [PubMed] [Google Scholar]
- 45.Saermark T, Jacobsen C, Magee A, Vilhardt H. J Mol Endocrinol. 1990;4:51–59. doi: 10.1677/jme.0.0040051. [DOI] [PubMed] [Google Scholar]
- 46.Koch N, Hammerling G J. J Biol Chem. 1986;261:3434–3440. [PubMed] [Google Scholar]
- 47.Pan K M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, et al. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodds A W, Ren X D, Willis A C, Law S K. Nature (London) 1996;379:177–179. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- 49.Law S K, Dodds A W. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]