Abstract
Pediatric gastroenteritis is a major cause of childhood morbidity and mortality worldwide, especially in developing countries. It has been increasingly recognised that human caliciviruses (HuCV), comprising noroviruses (NoV), and sapoviruses (SaV), are important in both outbreak and non-outbreak settings. This study aimed to characterise caliciviruses detected in the faeces of hospitalized children and children in the community in India. This study examined 350 faecal samples from children presenting to the hospital with acute gastroenteritis and 673 samples collected from children in the community, 500 from children with diarrhea, and 173 samples from children without diarrhea. Strain characterisation was performed by reverse transcription-polymerase chain reaction (RT-PCR) and partial sequencing of the gene encoding the RNA-dependent RNA polymerase (RdRp) and/or a region spanning the open reading frames (ORFs) 1 and 2 (ORF1/ORF2) junction. A total of 68 of 350 specimens (19.4%) from hospitalized children were positive, and SaV and NoV accounted for 5.1 and 15.1% of the infections, respectively. Mixed infections of HuCVs with other enteric pathogens were seen in 9.4% of the total children tested. Sixty-eight out of 673 (10.1%) samples collected from children in the community were positive for caliciviruses, and SaV and NoV accounted for 3.4 and 6.6% of the infections. In the community cohort 55/500 (11%) and 13/173 (7.5%) were from symptomatic and asymptomatic children, respectively, and SaVs accounted for 17/500 (3.4%) and NoVs for 38/500 (7.6%) of the symptomatic infections. This is the first report of genotyping of circulating caliciviruses in both hospital and community in India and has increased the evidence for the role of these viruses in pediatric gastroenteritis in India.
Keywords: gastroenteritis, genotyping, norovirus, sapovirus
INTRODUCTION
With the application of new and sensitive diagnostic techniques, human caliciviruses (HuCVs) have been recognised to be the leading cause of outbreaks of gastroenteritis in adults and an important cause of gastroenteritis in children in developed countries [Fankhauser et al., 1998; Lopman et al., 2003; Moreno-Espinosa et al., 2004]. However, little is known about the role of calicivirus in endemic pediatric gastroenteritis in developing countries. Children under 5 years of age in developing countries experience the highest rates of illness and death due to diarrhea, but despite the importance of gastrointestinal disease as a cause of poor health in young children, many cases are of unknown etiology [Guerrant, 1994].
HuCVs comprise noroviruses (NoVs), previously referred to as “Norwalk-like viruses” or “small round structured viruses” and Sapoviruses (SaVs), which are small (27-32 nm in diameter), single-stranded positive sense RNA viruses recently designated as separate genera in the Caliciviridae family based on phylogenetic analyses and genomic organization [Green et al., 2000]. NoVs that infect humans have been classified into three genetically and antigenically distinct genogroups [Zheng et al., 2006] and SaVs that infect humans into five genogroups [Farkas et al., 2004]. The genome has three open reading frames (ORFs) that encode a polyprotein precursor to nonstructural proteins including the RNA-dependent RNA polymerase (RdRp), and other proteins (ORF 1), the major and minor capsid proteins VP1 and VP2 (ORF 2 and 3) [Jiang et al., 1992, 1993; Lambden et al., 1993; Bertolotti-Ciarlet et al., 2003]. Knowledge about the role of HuCVs in pediatric gastroenteritis in developing countries has been limited by the lack of sensitive and specific diagnostic techniques. HuCVs have not been propagated in cell culture, and direct visualization of viruses by electron microscopy was previously the only technique available before the cloning and characterisation of the Norwalk virus genome allowed the development of molecular detection assays that have been applied in molecular epidemiological studies [Atmar and Estes, 2001].
Despite the advances in diagnostics, there is limited data available from India. Only two studies have been published using molecular methods for detection of HuCVs, from Vellore in 2000 [Kang et al., 2000], and from Delhi in 2002 [Girish et al., 2002]. This study characterises HuCVs in samples collected from hospitalized children with acute gastroenteritis and samples collected from symptomatic and asymptomatic children in the community. Phylogenetic analysis of the regions of the genes encoding the RdRp, capsid, and/or the region spanning the ORF1/ORF2 of the genome of selected SaV and NoV strains has been performed to classify the strains co-circulating in the study populations during a defined surveillance period.
MATERIALS AND METHODS
Study Population
The study was carried out in Vellore in Southern India. Hospital based studies were carried out at the Christian Medical College (CMC) hospital, a 2100 bed referral hospital. A total of 350 faecal samples collected during a period of 3 years (December 2001-2004) from children <5 years age, presenting to the hospital with acute gastroenteritis were examined for caliciviruses. The samples were collected within 24 hr of admission to hospital. For the community based study, 500 samples collected from children with diarrhea and 173 samples from asymptomatic children were randomly selected from a cohort of 452 children who had been recruited for follow-up from birth to the age of 3 years in an urban slum population The community samples were collected every 2 weeks from the cohort of children during the same period as the hospital samples.
Assessment of Severity
Diarrhea was defined as the passage of three watery stools in a 24-hr period. An episode was defined as at least 1 day of diarrhea, preceded and followed by two or more days without diarrhea. The severity of diarrhea was assessed using the 20-point scale of the Vesikari scoring system [Ruuska and Vesikari, 1990] which was originally developed for scoring severity of rotavirus diarrhea but incorporates duration and frequency of diarrhea and vomiting, associated fever and dehydration. To permit calculation of this score, the episode was considered mild for a score ≤5, moderately severe for a score of 6-10 and severe for scores >10.
RNA Extraction and cDNA Synthesis
RNA was extracted from 200 μl of a 10% faecal suspension (100 μl of stool and 900 μl of Minimal Essential Medium) using the method of Boom et al. [1990] and eluted in 50 μl of RNase-free sterile water containing 40 U of RNase inhibitor (RNAsin, Ambion, Huntingdon, Cambridgeshire, UK). Forty microliters of extracted RNA, which was denatured at 95°C for 5 min and immediately chilled in ice for 2 min, was used for generation of complementary DNA (cDNA) in 70 μl reactions by reverse transcription (RT) at 37°C for 1 hr. RT was carried out in the presence of random primers (hexamers; Pd(N)6, Pharmacia Biotech, Little Chalfont, Buckinghamshire, UK), and 400 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen, Life Technologies, Paisley, UK). Following generation of cDNA, the tubes were incubated at 95°C for 5 min and chilled on ice for 2 min. The cDNA was then stored at -20°C until subsequent use in PCRs.
Norovirus RT-PCR
The cDNA was used directly for detecting both genogroups I and II NoVs [Ando et al., 1994]. Single round PCRs were performed using published oligonucleotide primers Ni and E3 which amplify a 113-bp region of the RNA polymerase gene to detect GII NoVs [Green et al., 1995], and SG1 and D1 primers, which amplify 150-bp product of the RNA polymerase gene to detect GI strains [Green et al., 1995]. To amplify the 597-bp NoV GI ORF1-ORF2 junction region, a PCR was carried out using a mixture of three forward primers, GIFF-1, -2 and -3 [Kageyama et al., 2003] and a reverse primer, GISKR [Kojima et al., 2002]. Similarly, to amplify the NoV GII ORF1-ORF2 junction region (468-bp), a PCR was also performed with a mixture of three forward primers, GIIFB -1, -2 and -3 [Kageyama et al., 2003] and the reverse primer G2SKR [Kojima et al., 2002]. PCR was carried out in 50 μl reactions containing 20 pmole primers and 1 U Taq DNA polymerase (Invitrogen, Life Technologies). Products were amplified using the following conditions: 95°C for 2 min, then 40 cycles of 95°C for 30 sec, 48°C for 30 sec, and 72°C for 2 min, followed by 1 cycle of 72°C for 5 min and then held at 15°C. Cycling reaction conditions were similar for amplification of both polymerase and the ORF1-ORF2 junctions regions. Amplification products were examined by gel electrophoresis of the PCR amplicon in 2% agarose gels stained with (0.5 μg/ml) ethidium bromide and photographed under UV light using a BioRad GelDoc system (BioRad, Hemel Hempstead, Hertfordshire, UK).
Sapovirus RdRp-Specific PCR
The cDNA was amplified by hemi-nested PCR using previously described methods with first round primers SR80 and JV33 which amplify a 320-bp region of the RdRp gene (Noel et al., 1999; Vinje et al., 2000] and second round primers SR80 GI 1-3 (5′CTR KCV GAT ATT GGA RAG ATT T 3′) at position 4405-4425 on Manchester virus (GenBank X86560), SR80 GI 2 (5′ AGT CTY TCC ATC TTA GAG AGA 3′) at position 25-45 on Parkville virus (GenBank U73124), SR80 GII 1-2 (5′ GCT GCR TCY TTG KCA ATC CT 3′) at position 4400-4419 on Bristol virus (GenBank AJ249939) and JV33, amplifying a 280-, 293-, and 289-bp region of the RdRp, respectively. Products were amplified using the following conditions: 95°C for 2 min, then 31 cycles of 95°C for 15 sec, 40°C for 45 sec, and 72°C for 1 min, followed by 1 cycle of 72°C for 5 min and then held at 15°C. Amplicons were electrophoresed in agarose gels and stained with ethidium bromide as described above.
ORF1/ORF2 Junction—Specific PCR for SaV
For the SaV genotyping PCR, PCR amplification from cDNA used previously described first round primers SLV5317 and SLV5749 [Yan et al., 2003] which amplified a 432-bp region and second round primers Svpol3′-A (5′ AAG GMR CSY MCA AAA ATA GTG 3′) at position 5144-5164 on Manchester virus (GenBank X86560), SVpol3′-B (5′ GAA GRK RCW MCC AAA TTA GTG 3′) at position 5147-5167 on Bristol virus (GenBank AJ249939), amplifying a 371- and 375-bp region of the RdRp/capsid junction, respectively. Cycling conditions were 94°C for 5 min, then 35 cycles of 94°C for 30 sec, 50°C for 1 min and 72°C for 1 min, followed by 1 cycle of 72°C for 5 min and then hold at 15°C. Electrophoresis was performed as described above.
DNA Sequencing and Analysis
DNA sequencing was performed using a dye terminator cycle sequencing kit and a CEQ 2000 XL DNA analysis system (Beckman Coulter, High Wycombe, Buckinghamshire, UK) directly from the purified amplicons, which were sequenced in both directions using the same oligonucleotide primers used in the PCRs described above. Sequences and phylogenetic analyses were carried out using the Bionumerics 2.5 (Applied Maths, Kortrijk, Belgium) and/or the BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) software packages. Following the classification criteria for NoVs and SaVs, strains were assigned to a genogroup if the polymerase and/or capsid region shared >70% homology at the nt level to those of strains within a recognised genogroup. Genotypes were assigned based on >90% homology at the nt level with other strains within a given genotype.
RESULTS
Detection of HuCVs
The incidence of HuCV infections among hospitalized children and in the community, and their association with other pathogens is described in Table I. HuCVs were detected in 68 (19.4%) samples of the 350 children hospitalized with diarrhea. Eighteen (5.1%) of 350 samples from children hospitalized with acute gastroenteritis were positive for SaV, and 53 (15.1%) for NoV. Multiple pathogens were detected in samples collected from 33 children, this was 9.4% of the total numbers of children tested in the hospital (Table I).
TABLE I.
Incidence of HuCVs Singly and in Association With Other Pathogens in Children Hospitalised for diarrhea, and Children With and Without diarrhea in the Community
| Children hospitalised for diarrhea (n - 350) |
Children in the community with diarrhea (n - 500) |
Children in the community without diarrhea (n - 173) |
||||
|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | |
| HuCV | 68 | 19.4 | 55 | 11 | 13 | 7.5 |
| NoV alone | 27 | 7.7 | 33 | 6.4 | 5 | 2.9 |
| SaV alone | 7 | 2.0 | 13 | 2.6 | 6 | 3.5 |
| SaV + Cryptosporidium | 1 | 0.3 | ||||
| SaV + Rotavirus | 6 | 1.7 | 1 | 0.02 | ||
| SaV + Vibrio | 1 | 0.3 | ||||
| SaV + Giardia | 0 | 0.0 | 3 | 0.06 | ||
| NoV + Cryptosporidium | 1 | 0.3 | ||||
| NoV + Aeromonas | 1 | 0.3 | ||||
| NoV + rotavirus | 21 | 6.0 | 5 | 0.01 | 2 | 0.04 |
| NoV + SaV + rotavirus | 3 | 0.9 | ||||
HuCvs were detected in 55 of 500 (11%) diarrheal samples of children with diarrhea in the community and in 13 out of 173 (7.5%) asymptomatic children. SaVs were detected in 23 (3.4%) samples from children in the community, of which 17 had diarrhea and 6 were asymptomatic. NoV was found in 45 samples (6.6%), of which 38 had diarrhea and 7 were asymptomatic. Nine (16.4%) of 55 children with diarrhea had a second pathogen and there were two asymptomatic multiple infections (Table I).
The age range of children presenting with HuCV-associated diarrhea to the hospital was 8-52 months with a median of 13 months. The median age of detection of HuCVs in children from the community was 12 months with an age range of 0-30 months (P = 0.04). There was no difference in the median age at infection for symptomatic and asymptomatic infection. A larger proportion of HuCV-positive children admitted to hospital with diarrhea were male (60.2%), but this is a reflection of male children constituting approximately 60% of all children brought to the hospital for gastroenteritis in this and previous studies.
In the community, 58.2% of the episodes of HuCV diarrhea were mild, 38.2% were moderate, and 3.6% were severe. In contrast, only 7.5% of HuCV diarrheal cases admitted in the hospital were mild, 39.6% moderate, and 52.8% were severe (P < 0.001). The median Vesikari score was also higher in the hospital as compared to the community (11.0 vs. 5) (P < 0.001). However, there was no significant difference severity of diarrhea in those infected with NoV or SaV alone and those multiply infected with a HuCV and another enteric pathogen in either the hospital or the community (P = 0.2, P = 0.85 and P = 1, P = 0.95, Wilcoxon rank sum test) for NoVs and SaVs in the hospital and the community, respectively (Table II).
TABLE II.
Comparison of Severity of diarrhea Between NoVs and SaVs, and Single Infections With HuCVs and Multiple Infections With HuCVs and Other Enteric Pathogens
| Community median severity score |
Hospital median severity score |
|||
|---|---|---|---|---|
| n | (Inter-quartile range) | n | (Inter-quartile range) | |
| No. of diarrheal episodes | 55 | 5 (5,7)a | 68 | 11.5 (9,13)b |
| No. of NoV infection | 36 | 5 (5,5) | 14 | 10.5 (9,13) |
| No of SaV infection | 17 | 5 (5,7) | 47 | 12 (9,13) |
| Mixed infection with NoV and SaV | — | — | 3 | 12 (11,14) |
| Comparison of severity between single caliciviral or mixed infection (P-value) c | 0.3 | 0.33 | ||
| Infection with calicivirus (either SaV or NoV) alone | 44 | 5 (5,7) | 33 | 11 (9,13) |
| Calicivirus infection with another infection | 9 | 5 (5,7) | 31 | 12 (10,14) |
| Comparison of severity between single and mixed infection (P-value) c | 0.94 | 0.23 | ||
| NoV alone | 31 | 5 (5,7) | 25 | 11 (9,13) |
| NoV with another infection | 5 | 5 (5,7) | 22 | 12 (10,14) |
| Comparison of severity between single and mixed infection (P-value) c | 1 | 0.2 | ||
| SaV alone | 13 | 5 (5,5) | 7 | 11 (8,13) |
| SaV with another infection | 4 | 5 (4,7.5) | 7 | 10 (9,12) |
| Comparison of severity between single and mixed infection(p-value) c | 0.95 | 0.85 | ||
Severity scores available for 53 episodes of diarrhea.
Severity scores available for 64 episodes of diarrhea.
Wilcoxon Rank sum test.
Diversity of SaV Strains
A total of 31 SaV strains from 29 samples were genotyped, 6 from the hospital, and 23 from the community (Table III). GII-1 was the most frequently identified genotype in both settings although the numbers of strains of each genotype were too small for meaningful statistical analysis. Figure 1 shows the diversity of genogroup II SaVs from the hospital and the community. Infections with multiple SaV strains were found in two children (6.4%).
TABLE III.
Distribution of SaV Genotypes in Children in Different Settings in South India
| Community -diarrhea |
Communitya-no diarrhea |
Hospital |
Total |
|||||
|---|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | No | % | |
| GI-1 | 2 | 11.8 | 2 | 25 | 1 | 16.7 | 5 | 16.1 |
| GI-2 | 1 | 5.9 | 1 | 12.5 | 0 | 0.0 | 2 | 6.5 |
| GI-3 | 4 | 23.5 | 1 | 12.5 | 0 | 0.0 | 5 | 16.1 |
| GII-1 | 9 | 52.9 | 2 | 25 | 3 | 50 | 14 | 45.2 |
| GII-2 | 1 | 5.9 | 2 | 25 | 1 | 16.7 | 4 | 12.9 |
| GII-3 | 0 | 0.0 | 0 | 0.0 | 1 | 16.7 | 1 | 3.2 |
| TOTAL | 17 | 100 | 8 | 100 | 6 | 100 | 31 | 100 |
Two children had dual SaV infections, one had GI-2 +GII-1 and the other GII-1 + GII-2.
Fig. 1.
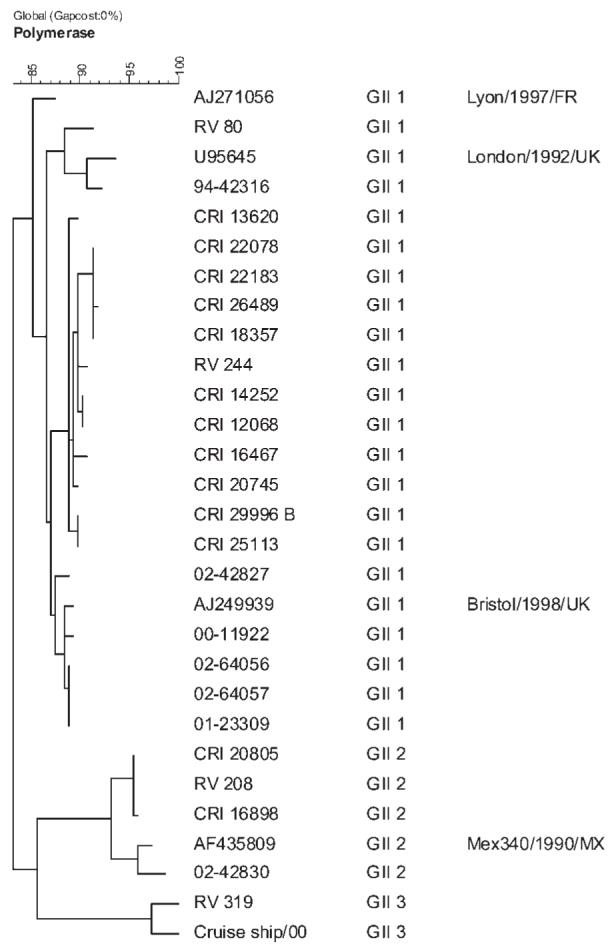
Dendrogram constructed using the maximum parsimony method and 280 nt of the interprimer region, amplified using SR80/JV33, of the polymerase encoding gene of SaVs of Genogroup II. Strain designations or accession numbers for reference strains are indicated in the first column. The genotype is listed in the second column. Sequences from this study are available from the corresponding author.
Diversity of NoV Strains
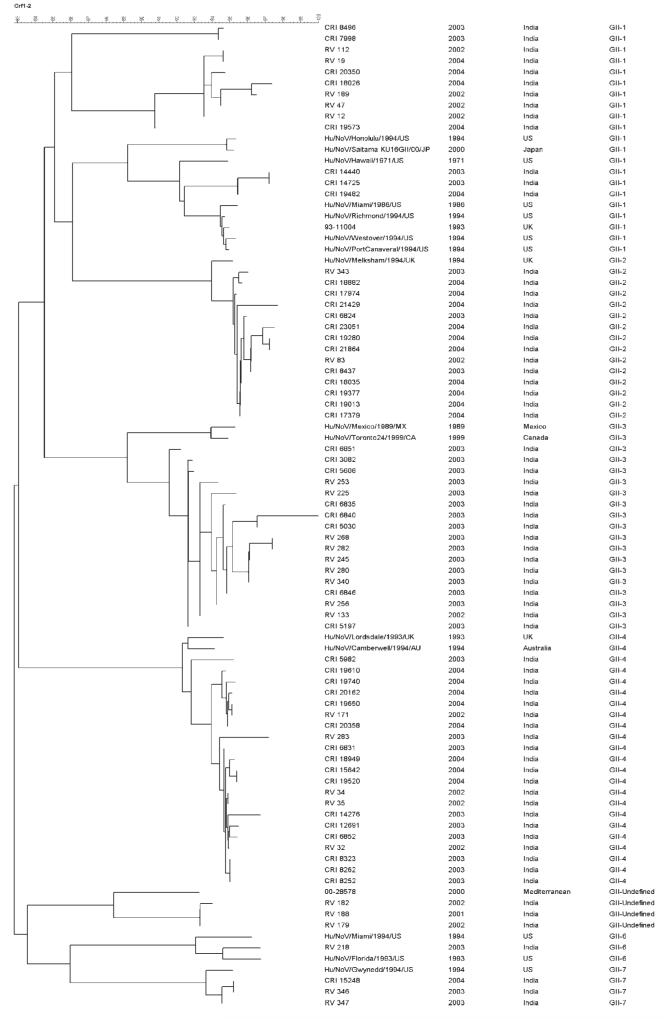
Seventy-three NoV strains from 72 samples were genotyped, 27 from the hospital, and 45 from the community (Table IV). A total of seven genotypes were found (Fig. 2), all but one within genogroup II. The only genogroup I strain found was a GI-3 in a coinfection with a GII-2 strain. In the community, GII-4 was the most frequently identified genotype, followed by GII-2, while in the hospital, GII3 was the most frequently identified genotype followed by GII-1 and GII-4 in equal proportions. However, no statistically significant differences were found in the distribution of genotypes between the hospital and community (P-0.1). Three children in the hospital had NoV strains of unidentified genotype. These strains were most closely related (>92% homology at the nt level) to a strain previously found in the Mediterranean region in the year 2000 and which do not share sufficient homology with any of the recognised genotypes to be classified within them. Figure 2 is the dendrogram constructed using the maximum parsimony method and 300 nt of 5′terminus of the ORF2 region.
TABLE IV.
Distribution of NoV Genotypes in Children in Different Settings in South India
| Community with diarrhea |
Communitya without diarrhea |
Hospital |
Total |
|||||
|---|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | No | % | |
| GII-1 | 8 | 21.1 | 0 | 19.6 | 5 | 18.5 | 13 | 19.2 |
| GII-2 | 10 | 26.3 | 2 | 23.9 | 2 | 7.4 | 14 | 17.8 |
| GII-3 | 4 | 10.5 | 4 | 17.4 | 9 | 33.3 | 17 | 23.3 |
| GII-4 | 15 | 39.5 | 1 | 34.8 | 5 | 18.5 | 21 | 28.8 |
| GII-6 | 0 | 0.0 | 0 | 0.0 | 1 | 3.7 | 1 | 1.4 |
| GII-7 | 1 | 2.6 | 0 | 2.2 | 2 | 7.4 | 3 | 4.1 |
| GI-3 | 0 | 0.0 | 1 | 2.2 | 0 | 0 | 1 | 1.4 |
| GII-Undefined | 0 | 0.0 | 0 | 0.0 | 3 | 11.1 | 3 | 4.1 |
| TOTAL: | 38 | 100 | 8 | 100 | 27 | 100 | 73 | 100 |
One child had a dual NoV infection with GII-2 and GI-3 strains.
Fig. 2.
Dendrogram constructed using the maximum parsimony method and 300 nt of 5′terminus of the ORF2 of NoVs. Strain designations or accession numbers for reference strains are indicated in the first column. The second and third columns refer to the year and place of identification of the strain. The genotype is listed in the fourth column. Sequences from this study are available from the corresponding author.
Repeat infections with HuCVs were identified in nine children in the community. The time between infections ranged from 1 to 12 months. Seven children were symptomatic at the time of the second infection, but the second infecting genotype was different from that associated with the first infection. The only child that had two infections with a genogroup II 2 NoV 6 months apart, was asymptomatic on both occasions. Another child had two asymptomatic infections 11 months apart with SaVs of different genogroups.
DISCUSSION
In this study, HuCVs were detected in 19.4% of the samples analyzed from children with gastroenteritis presenting to the hospital and in 11% of symptomatic and 7.5% of asymptomatic children in the community, respectively. The results from the hospital population are consistent with those from similar studies conducted in other developing and developed countries. In a report from Thailand, 12% of stool specimens were positive for NoVs [Hansman et al., 2004a]. Reports from Malawi, China, Vietnam, Mongolia, and India [Kang et al., 2000; Hansman et al., 2004b, 2005; Dove et al., 2005; Liu et al., 2006], have reported NoVs associated with 6-25% of acute gastroenteritis cases. This contrasts with incidences of 25-30% of rotavirus gastroenteritis in hospitalized children in this region of India [Banerjee et al., 2006], and indicates that NoV disease is less prevalent or less severe than that associated with rotavirus infection. A significantly larger number of children (P < 0.001) in the hospital cohort had HuCV when compared to the symptomatic children in the community.
A significant finding of this study was the incidence of calicivirus in asymptomatic children, with 3.4 and 4% of surveillance samples in the community positive for SaV and NoV, respectively, which has not been reported previously from India.
The incidence of SaV and NoV gastroenteritis in the community was 3.4% (17/500) and 7.6% (38/500), respectively. This is lower than reported previously in community-based studies in children in developed countries which have found NoVs in 18-21% cases of gastroenteritis in children <5 years of age [Pang et al., 1999]. Comparable data on rates of asymptomatic carriage are also not available for this age group but de Wit et al. [2001], in a nested case-control study, reported that NoVs were associated with symptomatic infection in 16.1% of cases of gastroenteritis and asymptomatic infection in 5.2% of controls.
Analysis of clinical disease through severity scores and of sequence diversity showed that disease in the community associated with HuCVs was mild or moderate and that there was no statistically significant difference in the distribution of genotypes between children with diarrhea in the community and those presenting to hospital (P=0.1).
It is also interesting to note that in eight of nine repeat infections identified in the community, the genotypes seen in the second infection were different from those first encountered. This highlights the high incidence and diversity of HuCVs in this setting. Immune responses to homotypic and heterotypic viruses have been insufficiently studied in calicivirus infection, in part due to the diversity of the agents and the lack of appropriate assays. A prospective cohort study aimed at genotyping infecting strains in conjunction with measurement of immune responses would allow dissection of the immune response and the presence or lack of protection from infection and/or disease, but no such studies have been conducted.
In this study, HuCVs from India have been genotyped for the first time. As reported from other parts of the world, genogroup II NoVs were more prevalent in children in the hospital and the community, and belonged to multiple genotypes [de Wit et al., 2001; Lopman et al., 2003]. GII-4 strains which are widely distributed in the world and have recently caused multiple outbreaks in Europe, Asia, and Australia [Bull et al., 2006] were also seen in both the community and the hospital in this study, but interestingly this genotype was more common in the community and was not the most prevalent genotype in the hospital. The majority of the data on health-care associated NoV infections reported worldwide shows infections caused by GII-4 strains, however, most of these studies refer to hospital an/or residential care institution outbreaks, with most involving adults and the elderly. The hospital data presented here is from children and does not represent nosocomial outbreaks, which may explain the greater diversity observed in the hospital. Among the SaVs, genogroup II strains were more common than genogroup I and this finding also differs from reports from the United Kingdom and Japan [Phan et al., 2005; Gallimore et al., 2006].
In summary, these data have described the prevalence of co-circulating HuCVs in children with or without gastroenteritis in Vellore, Southern India and shown that these viruses contribute significantly to the burden of diarrheal disease. Detection of HuCVs in asymptomatic children signifies that these viruses can circulate in the general population in the absence of disease in young children. Asymptomatic infection may be widespread in young children as, although immunity is of short duration, the frequency of infection in this age group may result in fewer symptomatic infections. These asymptomatic infections, however, constitute a significant reservoir for infection in the community, and may act as a source of both endemic and epidemic disease.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust Grant number 063144 and the Indian Council for Medical Research.
The work was performed at the Christian Medical College, Vellore, India.
REFERENCES
- Ando T, Mulders MN, Lewis DC, Estes MK, Monroe SS, Glass RI. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solidphase immune electron microscopy. Arch Virol. 1994;135:217–226. doi: 10.1007/BF01309781. [DOI] [PubMed] [Google Scholar]
- Atmar RL, Estes MK. Diagnosis of noncultivatable gastroenteritis viruses, the human caliciviruses. Clin Microbiol Rev. 2001;14:15–37. doi: 10.1128/CMR.14.1.15-37.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I, Ramani S, Primrose B, Moses P, Iturriza-Gomara M, Gray JJ, Jaffar S, Monica B, Muliyil JP, Brown DW, Estes MK, Kang G. Comparative study of rotavirus epidemiology in children from a community based birth cohort and a hospital in South India. J Clin Microbiol. 2006;44:2468–2474. doi: 10.1128/JCM.01882-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti-Ciarlet A, Crawford SE, Hutson AM, Estes MK. The 3′ end of Norwalk virus mRNA contains determinants that regulate the expression and stability of the viral capsid protein V P1: A novel function for the VP2 protein. J Virol. 2003;77:11603–11615. doi: 10.1128/JVI.77.21.11603-11615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boom R, Sol CJA, Salismans MMM, Jansen CL, Wertheim-van Dillen PME, van den Noordaa J. Rapid and simple method for the purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Tu ET, McIver CJ, Rawlinson WD, White PA. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J Clin Microbiol. 2006;44:327–333. doi: 10.1128/JCM.44.2.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit MA, Koopmans MP, Kortbeek LM, Wannet WJ, Vinje J, vanLeusden F, Bartelds AI, van Duynhoven YT. Sensor, a population-based cohort study on gastroenteritis in the Netherlands: Incidence and etiology. Am J Epidemiol. 2001;154:666–674. doi: 10.1093/aje/154.7.666. [DOI] [PubMed] [Google Scholar]
- Dove W, Cunliffe NA, Gondwe JS, Broadhead RL, Molyneux ME, Nakagomi O, Hart CA. Detection and characterisation of human caliciviruses in hospitalized children with acute gastroenteritis in Blantyre, Malawi. J Med Virol. 2005;77:522–527. doi: 10.1002/jmv.20488. [DOI] [PubMed] [Google Scholar]
- Fankhauser RL, Noel JS, Monroe SS, Ando TA, Glass RI. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J Infect Dis. 1998;178:1571–1578. doi: 10.1086/314525. [DOI] [PubMed] [Google Scholar]
- Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz-Palacios GM, Pickering LK, Jiang X. Genetic diversity among sapoviruses. Arch Virol. 2004;149:1309–1323. doi: 10.1007/s00705-004-0296-9. [DOI] [PubMed] [Google Scholar]
- Gallimore CI, Iturriza-Gomara M, Lewis D, Cubitt D, Cotterill H, Gray JJ. Characterisation of sapoviruses collected in the United Kingdom from 1989 to 2004. J Med Virol. 2006;78:673–682. doi: 10.1002/jmv.20592. [DOI] [PubMed] [Google Scholar]
- Girish R, Broor S, Dar L, Ghosh D. Foodborne outbreak caused by a Norwalk-like virus in India. J Med Virol. 2002;67:603–607. doi: 10.1002/jmv.10145. [DOI] [PubMed] [Google Scholar]
- Green J, Gallimore CI, Norcott JP, Lewis D, Brown DWG. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J Med Virol. 1995;47:392–398. doi: 10.1002/jmv.1890470416. [DOI] [PubMed] [Google Scholar]
- Green KY, Ando T, Balayan MS, Estes MK, Maston DO, Nakata S, Neil JD, Studdert MJ, Thiel HJ. Taxonomy of the caliciviruses. J Infect Dis. 2000;181(Suppl 2):S322–S330. doi: 10.1086/315591. [DOI] [PubMed] [Google Scholar]
- Guerrant RL. Twelve messages from enteric infections for science and society. Am J Trop Med Hyg. 1994;51:26–35. doi: 10.4269/ajtmh.1994.51.26. [DOI] [PubMed] [Google Scholar]
- Hansman GS, Katayama K, Maneekarn N, Peerakome S, Khamrin P, Tonusin S, Okitsu S, Nishio O, Takeda N, Ushijima H. Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J Clin Microbiol. 2004a;42:1305–1307. doi: 10.1128/JCM.42.3.1305-1307.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman GS, Doan LT, Kguyen TA, Okitsu S, Katayama K, Ogawa S, Natori K, Takeda N, Kato Y, Nishio O, Noda M, Ushijima H. Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch Virol. 2004b;149:1673–1688. doi: 10.1007/s00705-004-0345-4. [DOI] [PubMed] [Google Scholar]
- Hansman GS, Kuramitsu M, Yoshida H, Katayama K, Takeda N, Ushijima H, Surenkhand G, Gantolga D, Kuroiwa C. Viral gastroenteritis in Mongolian infants. Emerg Infect Dis. 2005;11:180–182. doi: 10.3201/eid1101.040337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang M, Graham DY, Estes MK. Expression, self-assembly and antigenicity of the Norwalk virus capsid protein. J Virol. 1992;66:6527–6532. doi: 10.1128/jvi.66.11.6527-6532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang M, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195:51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription PCR. J Clin Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang G, Hale AD, Richards AF, Jesudason MV, Estes MK, Brown DW. Detection of ‘Norwalk-like viruses’ in Vellore, southern India. Trans Roy Soc Trop Med and Hyg. 2000;94:681–683. doi: 10.1016/s0035-9203(00)90231-1. [DOI] [PubMed] [Google Scholar]
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- Lambden PR, Caul EO, Ashley CR, Clarke IN. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259:516–518. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- Liu C, Grillner L, Jonsson K, Linde A, Shen K, Lindell AT, Wirgart BZ, Johansen K. Identification of viral agents associated with diarrhoea in young children during a winter season in Beijing, China. J Clin Virol. 2006;35:69–72. doi: 10.1016/j.jcv.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman BA, Reacher MH, Van Duijnhoven Y, Hanon FX, Brown D, Koopmans M. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg Infect Dis. 2003;9:90–96. doi: 10.3201/eid0901.020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Espinosa S, Farkas T, Jiang X. Human caliciviruses and paediatric gastroenteritis. Semin Pediatr Infect Dis. 2004;15:237–245. doi: 10.1053/j.spid.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Noel JS, Fankhauser RL, Ando T, Monroe SS, Glass RI. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J Infect Dis. 1999;179:1334–1344. doi: 10.1086/314783. [DOI] [PubMed] [Google Scholar]
- Pang X, Joensuu J, Vesikari T. Human calicivirus—Associated sporadic gastroenteritis in Finnish children less than two years of age followed prospectively during a rotavirus vaccine trial. Pediatr Infect Dis J. 1999;18:420–426. doi: 10.1097/00006454-199905000-00005. [DOI] [PubMed] [Google Scholar]
- Phan TG, Nguyen TA, Nishimura S, Nishimura T, Yamamoto A, Okitsu S, Ushijima H. Etiologic agents of acute gastroenteritis among Japanese infants and children: Virus diversity and genetic analysis of Sapovirus. Arch Virol. 2005;150:1415–1424. doi: 10.1007/s00705-005-0514-0. [DOI] [PubMed] [Google Scholar]
- Ruuska T, Vesikari T. Rotavirus disease in Finnish children: Use of numerical scores for clinical severity of diarrhoeal episodes. Scand J Infect Dis. 1990;22:259–267. doi: 10.3109/00365549009027046. [DOI] [PubMed] [Google Scholar]
- Vinje J, Green J, Lewis DC, Gallimore CI, Brown DW, Koopmans MP. Genetic polymorphism across regions of the three open reading frames of “Norwalk-like viruses”. Arch Virol. 2000;145:223–241. doi: 10.1007/s007050050020. [DOI] [PubMed] [Google Scholar]
- Yan H, Yagyu F, Okitsu S, Nishio O, Ushijima H. Detection of norovirus (GI, GII), Sapovirus and astrovirus in faecal samples using reverse transcription single-round multiplex PCR. J Virol Methods. 2003;114:37–44. doi: 10.1016/j.jviromet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]