Abstract
Hypermethylated in cancer (HIC-1), a new candidate tumor suppressor gene located in 17p13.3, encodes a protein with five C2H2 zinc fingers and an N-terminal broad complex, tramtrack, and bric à brac/poxviruses and zinc-finger (BTB/POZ) domain found in actin binding proteins or transcriptional regulators involved in chromatin modeling. In the human B cell lymphoma (BCL-6) and promyelocityc leukemia (PLZF) oncoproteins, this domain mediates transcriptional repression through its ability to recruit a silencing mediator of retinoid and thyroid hormone receptor (SMRT)/nuclear receptor corepressor (N-CoR)-mSin3A-histone deacetylase (HDAC) complex, a mechanism shared with numerous transcription factors. HIC-1 appears unique because it contains a 13-aa insertion acquired late in evolution, because it is not found in its avian homologue, γF1-binding protein isoform B (γFBP-B), a transcriptional repressor of the γF-crystallin gene. This insertion, located in a conserved region involved in the dimerization and scaffolding of the BTB/POZ domain, mainly affects slightly the ability of the HIC-1 and γFBP-B BTB/POZ domains to homo- and heterodimerize in vivo, as shown by mammalian two-hybrid experiments. Both the HIC-1 and γFBP-B BTB/POZ domains behave as autonomous transcriptional repression domains. However, in striking contrast with BCL-6 and PLZF, both HIC-1 and γFBP-B similarly fail to interact with members of the HDAC complexes (SMRT/N-CoR, mSin3A or HDAC-1) in vivo and in vitro. In addition, a general and specific inhibitor of HDACs, trichostatin A, did not alleviate the HIC-1- and γFBP-B-mediated transcriptional repression, as previously shown for BCL-6. Taken together, our studies show that the recruitment onto target promoters of an HDAC complex is not a general property of transcriptional repressors containing a conserved BTB/POZ domain.
The broad complex, tramtrack, and bric à brac (BTB) or poxviruses and zinc-finger (POZ) domain is an evolutionarily conserved protein–protein interaction domain found in developmentally regulated transcription factors and actin-binding proteins (1–4). In most cases, the BTB/POZ domain is associated with C2H2 zinc-finger motifs in proteins involved in transcriptional regulation through chromatin modeling as, for example, the Drosophila GAGA factor (2). However, many BTB/POZ and zinc-finger proteins are transcriptional repressors such as Drosophila tramtrack and vertebrates zinc finger protein with interacting domain (ZID), zinc finger protein 5 (ZF5), γF1-binding protein isoform B (γFBP-B), BCL-6-associated zinc finger protein (BAZF), BTB and CNC homology protein 2 (BACH2), promyelocytic leukemia (PLZF), and B cell lymphomas 6 (BCL-6) (1, 5–13).
The biological properties of BTB/POZ proteins are contingent on the protein–protein interaction properties of this domain. In numerous examples, the BTB/POZ domain has been clearly shown to mediate homodimerization (14–16) or even multimerization (17). It can also participate in heterophilic interactions, notably with the corepressors silencing mediator of retinoid and thyroid hormone receptor (SMRT) and nuclear receptor corepressor (N-CoR), as shown for BACH2, BCL-6, and PLZF (9, 18–28). SMRT and N-CoR, originally isolated as corepressors of some unliganded nuclear receptors, are components of a larger multiprotein complex including, among others, mSin3A/B and histone deacetylase (HDAC)-1/HDAC-2 (29–37). The recruitment of an HDAC-containing complex has emerged during the past 2 years as a common transcriptional repression mechanism used by a still-growing list of transcription factors belonging to various functional classes, including nuclear hormone receptors (19, 35), Mad and Mxi (19, 31–33), YY1 (30), Rb (38), CBF1 (39), the methyl CpG-binding protein MeCP2 (40–41), the t(8;21) fusion protein AML1-ETO, and the normal ETO protein (42–43), c-ski (44) and the BTB/POZ and zinc-finger transcriptional repressors BACH2 (9) and PLZF, as well as the t(11;17) fusion protein PLZF-RARα (22–28) and BCL-6 (20–21, 27, 45). Whereas some transcription factors seem to preferentially recruit only some components of the repressing complex, as Rb, which interacts directly with HDAC-1 (38), the PLZF and BCL-6 BTB/POZ domains are involved in direct interactions with SMRT/N-CoR, mSin3A, and HDAC-1 (20–28). Strong structural support for these observations has been recently brought by the crystal structure of the BTB/POZ domain from PLZF, which has revealed a tightly interlaced dimer with an extensive hydrophobic region and has precisely defined peptidic motifs involved in the scaffolding and dimerization processes. In addition, a surface-exposed groove lined with conserved residues has been made evident at the dimer interface and has been proposed to represent a putative site of interaction with nuclear corepressors and/or other nuclear proteins (3).
Among all the BTB/POZ proteins known so far, the candidate tumor suppressor gene hypermethylated in cancer (HIC-1), identified because of its association with a “CpG island” located at 17p13.3 that is aberrantly hypermethylated and transcriptionally inactivated in several common types of human cancers (46), appeared unique. First, HIC-1 encodes a typical nuclear BTB/POZ protein with five Krüppel-like C2H2 zinc-finger motifs in its C-terminal part and an N-terminal BTB/POZ domain, but containing a unique 13-aa insertion rich in alanine (8 of 13 residues) (46). On the basis of the sequence alignment of BTB/POZ proteins and the PLZF structure, this insertion is located in a loop between β strand β5 and helix α5, which are involved in scaffolding and dimerization of the domain (3). Second, this specific insertion is only very partially conserved in its avian homologue, γFBP-B, which has been isolated as a sequence-specific transcriptional repressor of the γF-crystallin gene during embryonic development (7). This insertion is also absent in the zebrafish HIC-1 gene but found in the murine HIC-1 gene, suggesting that it has been acquired late in evolution (47–48). Such a sequence divergence is quite unusual in BTB/POZ domains, as exemplified by zinc finger protein 5, where the murine and avian BTB/POZ domains differ only by a conservative mutation (49).
Here, we address the functional properties of HIC-1 and investigate the influence of this HIC-1-specific insertion on the ascribed properties of the BTB/POZ domains. By mammalian two-hybrid experiments, we demonstrate that the HIC-1 and γFBP BTB/POZ domains are able to homo- and heterodimerize, and both appear as autonomous transcriptional repression domains in the context of GAL4 chimeras. The specific insertion has only a slight inhibitory effect on the dimerization properties. Strikingly, we demonstrate that in sharp contrast with the BTB/POZ transcriptional repressors BACH2, BCL-6 and PLZF, HIC-1 and γFBP-B are similarly unable to interact in vivo either with the SMRT/N-CoR corepressors or with mSin3A, strongly suggesting that deacetylase activity is not required for repression by HIC-1 and γFBP-B. In GST pull-down experiments, HIC-1 fails to interact with SMRT and HDAC-1, in sharp contrast with BCL-6. Consistent with these results, we further demonstrate that the HIC-1 and γFBP-B repressing potential on transcription is not compromised by the specific HDAC inhibitor trichostatin A (TSA) or by sodium butyrate, in striking contrast with BCL-6. Our studies show that recruitment on target promoters of an HDAC complex is not required for full HIC-1- and γFBP-B-mediated transcriptional silencing and thus is not a general strategy for transcriptional repressors containing a conserved BTB/POZ domain.
Materials and Methods
Plasmids.
The HIC-1 and γFBP-B BTB/POZ domains corresponding to residues 1–131 in γFBP-B and to residues 1–140 in HIC-1 were amplified by PCR by using oligonucleotides flanked by convenient restriction sites. After cloning, a PCR product verified by nucleotide sequencing was digested by BamHI and SstI and cloned into the plasmid pSG5424, containing the GAL4 DB-domain or digested by XhoI and BglII and cloned into the pSG-FNV (Flag-NLS-VP16 AD domain) (20). HIC-1 and γFBP-B full-size constructs were obtained by inserting an intronless genomic fragment in the BTB/POZ domain chimeras by using convenient restriction sites. Details of these constructions are available on request. All these GAL4 and VP16 chimeras were sequenced to confirm the reading frame. The GAL4-BTB/POZ-BCL-6 and the VP16-SMRT (194–657) constructs have been previously described (20). The GAL4-mSin3A (PAH1–4) and (PAH1–2), the VP16-PLZF (full size), the VP16-SMRT (full size), and the GAL4-SMRTα (1–1031) constructs were kindly provided by R. J. Lin and R. M. Evans (the Salk Institute, San Diego, CA) (23, 35, 36). The VP16-N-CoR (1019–2061) construct was a gift from K. D. Huynh and V. J. Bardwell (University of Minnesota, Minneapolis, MN) (21). The pG5LUC vector from the CheckMate mammalian two-hybrid kit (Promega) was used as a GAL4-responsive reporter plasmid.
Repression and Mammalian Two-Hybrid Assays.
HeLa or rabbit kidney (RK13) cells were maintained in Dulbecco medium supplemented with 10% fetal calf serum. The day before transfection, they were plated at 50–60% confluence in 6-well plates. For transfection, cells were incubated with 1.0 μg of plasmid DNA and 4 μl of polyethyleneimine (Euromedex, Souffelweyersheim, France) for 6 hr in 1 ml of OptiMEM (11, 45) and then in fresh complete medium. For reporter assays, detailed transfection conditions are indicated in the relevant figure legend. The pSG5-β-galactosidase (β-gal) vector (50 ng) was cotransfected in each assay to correct for variations in transfection efficiency. Cells were rinsed in PBS 48 hr after transfection and lysed in universal lysis buffer (Promega). Luciferase and β-gal activities were measured by using, respectively, beetle luciferin (Promega) and the Galacto-light Kit (Tropix, Bedford, MA) with a Berthold (Nashua, NH) chemioluminometer. Results presented are the means of at least three transfections.
For experiments using HDAC inhibitors, 24 hr after transfection triplicate transfected cells were either untreated or treated with 300 nM TSA (Biomol, Plymouth Meeting, PA) or 5 mM sodium butyrate (NaBu) (Sigma) for a further 24 hr before harvesting.
GST Pull-Down Assays.
SMRT residues previously shown to interact with the BCL-6 BTB/POZ domain (20) were cloned in frame with GST in the pGEX2TK. The GST-HDAC1 (27) construct was provided by M. Privalski (University of California, Davis, CA) through the courtesy of S. Schreiber (Harvard University, Cambridge, MA). GST pull-down experiments were performed as previously described (27).
Results
The HIC-1 and γFBP-B BTB/POZ Domains Interact in a Mammalian Two-Hybrid Assay.
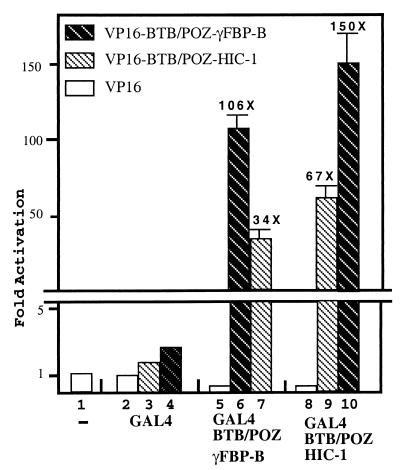
To address the functional role of the HIC-1-specific insertion located between the β5 strand and the α5 helix conserved in the BTB/POZ domain and shown in PLZF to be involved in dimerization, we first reasoned that this insertion may affect the dimerization potential of the HIC-1 BTB/POZ domain. To explore this hypothesis, we prepared mammalian expression constructs for GAL4 DNA-binding- and VP16 activator-tagged BTB/POZ domains of HIC-1 (codons 1–140) and γFBP-B (codons 1–131) and carried out mammalian two-hybrid assays in HeLa cells. Transfection of the HIC-1 or γFBP-B BTB/POZ-containing bait construct in combination with the VP16 activation domain alone resulted in a similar ≈5-fold repression of the GAL4 responsive luciferase reporter gene as compared with the GAL4 DNA-binding and VP16 activation domain combination (Fig. 1, lanes 2, 5, and 8), suggesting that these BTB/POZ domains are autonomous transrepressing domains as already shown for BCL-6 and PLZF (10–13) (see below). When the HIC-1 or γFBP-B BTB/POZ bait constructs were transfected with their homologous VP16-tagged constructs, we observed a significant transcriptional activation resulting from their in vivo homodimerization (Fig. 1, lanes 6 and 9), as already shown for BCL-6 or LRF/FBI (14, 16). However, we noticed that the transcriptional activation level achieved (67-fold vs. 106-fold) was significantly lower for the HIC-1 homodimers than for the γFBP-B homodimers (Fig. 1, lanes 6 and 9). As a control, Western blot analyses of transfected Cos-1 cells demonstrated that these chimeras were equally produced in vivo (data not shown).
Figure 1.
The HIC-1 and γFBP-B BTB/POZ domains are able to homo- and heterodimerize in the mammalian two-hybrid assay. The HIC-1 and γFBP-B BTB/POZ domains were fused either to the GAL4 DNA-binding domain or to the VP16 activator domain. Luciferase and β-gal assays were performed on total extracts from HeLa cells that have been transfected with 750 ng of the GAL4 reporter gene, pG5LUC (CheckMate mammalian two-hybrid kit, Promega), 100 ng of the indicated bait- and activator-tagged expression constructs, and 50 ng of the pSG5 β-gal construct as a control for transfection efficiency. Results represent the average of a triplicate experiment in which the luciferase activity was normalized to β-gal activities.
Previous reports have emphasized the possible heterodimerization between BTB/POZ domains in vitro (1) and between related BTB/POZ proteins in vivo (8, 16). The HIC-1 and γFBP-B BTB/POZ domains were also able to heterodimerize in vivo. However, for the two possible heteromeric combinations, or when they were compared with their respective homomeric combinations, an inhibitory effect mediated by the VP16-HIC-1 construct was clearly observed. The GAL4-HIC-1/VP16-γFBP-B combination appeared five times more efficient in heterodimerization than the reciprocal GAL4-γFBP-B/VP16-HIC-1, as reflected by the level of transcriptional activation obtained (Fig. 1, lanes 7 and 10). In the HIC-1 homodimerization context, the replacement of the VP16-HIC-1 by the VP16-γFBP-B BTB/POZ construct resulted in a significant increase of transcriptional activation (Fig. 1, lanes 9 and 10). Reciprocally, in the γFBP-B homodimerization context, the VP16-HIC-1 BTB/POZ construct induced a ≈3-fold decrease of the observed transcriptional activation (Fig. 1, lanes 6 and 7).
In conclusion, our results show that the HIC-1 and γFBP-B BTB/POZ domains can both homodimerize and heterodimerize in vivo, and that the dimerization property ascribed to the BTB/POZ domains is slightly affected by the presence of the HIC-1-specific insertion.
The Putative Human Tumor Suppressor Gene HIC-1 Encodes a Transcriptional Repressor.
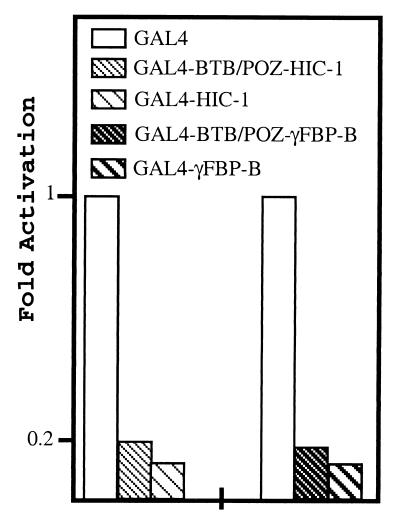
Sustained expression of the murine γF crystallin gene during lens development requires the binding of a strong lens-specific activator to a motif called γF1 in its promoter. Screening of a chicken lens cDNA library with the γF1 element led to the isolation of γFBP-B, which is in fact a BTB/POZ transcriptional repressor of the γF1 element (7). Thus, to examine the role of its human homologue HIC-1 in transcriptional regulation and to address the HIC-1 and γFBP-B repression mechanism, we used GAL4 chimeras and a GAL4-responsive reporter (pG5LUC), because the exact binding sites for γFBP-B and HIC-1 are not fully characterized. Transfection of expression plasmids encoding the full-length γFBP-B or HIC-1 proteins fused to the GAL4 DNA-binding domain in RK13 cells induced a similar 8-fold transcriptional repression of the pG5LUC reporter gene (Fig. 2). This repression strictly depends on the presence of the GAL4 sites, because it is not observed with a CMV-LUC reporter (data not shown) and is thus specific. From this experiment, we can infer that HIC-1 is also a transcriptional repressor indistinguishable, within the limits of this assay, from γFBP-B.
Figure 2.
Repression by the full-length HIC-1 and γFBP-B and by their isolated BTB/POZ domains. The transcriptional repression elicited by the above indicated GAL4 chimeras (200 ng) was measured by transfection assays in RK13 cells with 750 ng of the pG5LUC reporter gene (Promega) and normalized to the β-gal activity of a cotransfected pSG5 β-gal construct.
As already suggested by our mammalian two-hybrid experiment, the HIC-1 and γFBP-B BTB/POZ domains displayed an autonomous and transferable repressive activity. Indeed, when the HIC-1 and γFBP-B BTB/POZ domains fused to the GAL4 DNA-binding domain were transfected in RK13 cells, a ≈5- or 6-fold repression of the reporter, respectively, was observed (Fig. 2). When the two GAL4-BTB/POZ chimeras were directly compared by transient transfection in two different cell types (HeLa and RK13), we noticed that the γFBP-B BTB/POZ domain represses transcription slightly more efficiently that the corresponding HIC-1 construction (data not shown). Thus, the HIC-1 and γFBP-B BTB/POZ domains appear as similar autonomous repression domains.
The HIC-1 and γFBP-B BTB/POZ Domains Interact Neither with the Corepressors N-CoR and SMRT nor with mSin3A in a Mammalian Two-Hybrid Assay.
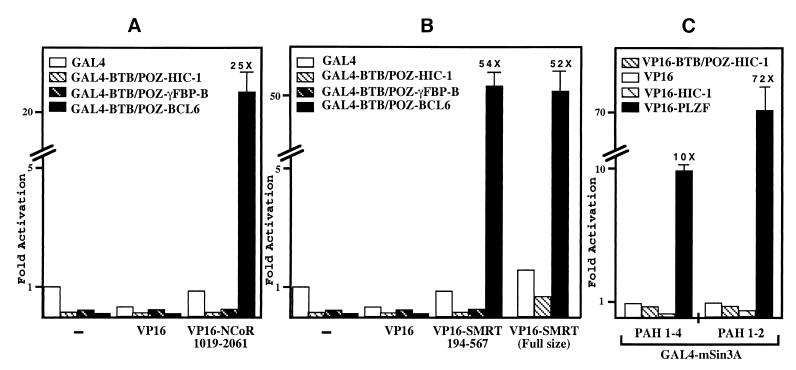
Having established that the full-length HIC-1 and γFBP-B proteins, as well as their isolated BTB/POZ domains, are able to similarly repress transcription in the context of GAL4 chimeras, we next investigated in more detail their mechanism of repression. Previous work from several groups has unambiguously established that the BTB/POZ transcriptional repressors BACH2, BCL-6, and PLZF interact directly with the SMRT and/or its relative N-CoR proteins (9, 20–28). To determine whether the BTB/POZ domains of HIC-1 and γFBP-B could also associate with these corepressors, we performed an in vivo interaction assay with the human N-CoR and SMRT cDNA fragments identified in two independent yeast two-hybrid screens by using the BCL-6 BTB/POZ domain as a bait (20, 21). When the VP16 activation domain fused to human N-CoR sequences corresponding to amino acids 1019–2061 (21) or to amino acids 194–657 of SMRT (20) was transiently transfected in RK13 together with various GAL4 fusion proteins, significant transcriptional activation of a GAL4 responsive reporter gene was observed only with the GAL4-BTB/POZ domain of BCL-6 (Fig. 3 A and B). In contrast, the GAL4 DNA-binding domain gave a low background level and the HIC-1 or γFBP-B BTB/POZ GAL4 chimeras still repressed the reporter gene in the presence of VP16 N-CoR and VP16-SMRT (Fig. 3 A and B).
Figure 3.
N-CoR, SMRT, and mSin3A interact with BCL-6 but not with HIC-1 and γFBP-B. (A–C) Luciferase and β-gal assays were performed on total extracts from RK13 cells that have been transiently transfected with 750 ng of the pG5LUC reporter gene, 100 ng of the indicated GAL4-BTB/POZ bait- and VP16 activator-tagged expression constructs, and 50 ng of the pSG5 β-gal construct as a control for transfection efficiency. Results represent the average of a triplicate experiment in which the luciferase activity was normalized to β-gal activity.
Finally, to exclude a possible interaction outside of the previously described repressing domains, we have also shown that the HIC-1 BTB/POZ domain, as well as the HIC-1 and γFBP-B full-length proteins, failed to interact in the same mammalian two-hybrid assay with a VP16-SMRT full-size construct (35) (Fig. 3B and data not shown). In addition, the HIC-1 BTB/POZ domain or full-length protein fused to the VP16 activation domain did not interact with the recently characterized new N-terminal domain of SMRT (GAL4-SMRTα 1–1031) (36) (data not shown).
The transcriptional repressors BCL-6 and PLZF are engaged in multiple interactions with various components of the HDAC-repressing complex, namely N-CoR/SMRT, mSin3A, or HDAC-1 itself, principally but not exclusively via their BTB/POZ domains (20–28). However, interaction with N-CoR/SMRT is not an absolute prerequisite for the recruitment of an HDAC-repressing complex (31, 33, 38). Thus, during evolution of the BTB/POZ domains, HIC-1 and γFBP-B could have retained only a more restricted subset of these interactions. We have thus tested whether mSin3A, which is used as a common link between SMRT/N-CoR and HDAC, could interact with HIC-1. In our mammalian two-hybrid assay, we used two GAL4-mSin3A constructs, either the full-length mSin3A containing the four paired amphipathic helix (PAH1–4) domains and the HDAC interaction domain located next to PAH3 (33) or the N-terminal portion of mSin3A (PAH1–2), because it strongly interacts with PLZF (23). We failed to detect any significant interaction between these two GAL4-mSin3A constructs and the VP16-HIC-1 BTB/POZ or the VP16-HIC-1 (full-size) constructs (Fig. 3C). As a positive control of interaction, we have shown in the same experiment that VP16-PLZF (full size) interacted strongly with these two GAL4-mSin3A constructs (Fig. 3C), as previously described (23).
Thus, our results demonstrate that HIC-1 and γFBP-B failed to interact in mammalian two-hybrid assays with major components of the HDAC complex, N-CoR, SMRT, and mSin3A, which are recruited by the related BTB/POZ transcriptional repressors BCL-6 and PLZF.
BCL-6, but not HIC-1, Can Associate with SMRT and HDAC-1 in Vitro.
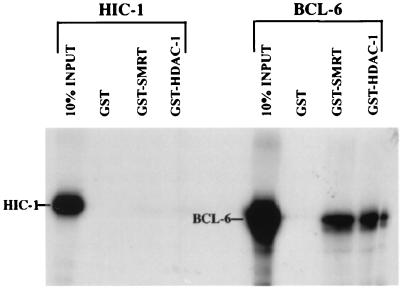
To confirm the results obtained in vivo in our mammalian two-hybrid assays, we next performed glutathione S-transferase (GST) pull-down experiments in vitro. HIC-1 and BCL-6 proteins were translated in vitro in presence of [35S]methionine and incubated with purified GST alone, GST-SMRT (residues 194–657, corresponding to the SMRT region interacting with BCL-6) (20), or GST-HDAC-1 (full size). After extensive washing, the eluted proteins were subjected to electrophoresis, followed by autoradiography. In vitro-translated BCL-6 was specifically brought down by GST-SMRT and GST-HDAC-1 as expected (20, 45) (Fig. 4). However, under similar conditions, GST, GST-SMRT, and GST-HDAC-1 failed to pull down the radiolabeled HIC-1 protein. Thus, the lack of interaction observed in the two-hybrid assay in vivo has been confirmed by an alternative biochemical approach in vitro.
Figure 4.
SMRT and HDAC-1 interact with BCL-6 but not with HIC-1 in vitro. In vitro-translated [35S]methionine-labeled HIC-1 and BCL-6 were subjected to a GST pull-down analysis with GST, GST-SMRT (194–657), and GST-HDAC-1(full-size) fusion proteins. Ten percent of each input protein was loaded on the same gel for electrophoresis. The image was edited from the same x-ray film.
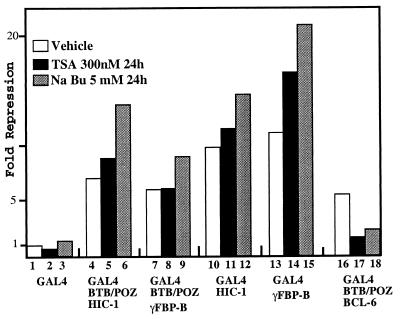
HIC-1 and γFBP-B Transcriptional Repression Is Unaffected by the HDAC Inhibitors, TSA and Sodium Butyrate.
Although they failed to interact with N-CoR/SMRT, mSin3A, or HDAC-1, HIC-1 and γFBP-B could still recruit another member of the HDAC family either directly or through interaction with distinct or even unknown corepressors. However, regardless of the mechanism used, if an HDAC is a mediator of HIC-1 and/or γFBP-B transcriptional silencing, then TSA, which is a highly specific and general inhibitor of HDACs, would be expected to relieve, at least partially, their transcriptional repression (50–53). To test this hypothesis, we measured the effects of these two inhibitors on the repression of a GAL4-responsive gene elicited by the HIC-1 and γFBP-B BTB/POZ domains or by the HIC-1 and γFBP-B full-size proteins fused to the GAL4 DNA-binding domain. Strikingly, the transcriptional repression mediated by all these GAL4 chimeras was not substantially reduced when transfected RK13 cells were treated with 300 nM TSA for 24 hr before harvesting (Fig. 5). We even note that TSA, which is a general and highly specific inhibitor of HDAC, slightly increased their repression potential on the pG5LUC reporter gene. To validate our experimental system, we have shown in the same experimental conditions that TSA alleviated repression by the GAL4-BTB/POZ-BCL-6 chimera by 70%, consistent with HDAC being required for full BCL-6-mediated repression, as previously described (Fig. 5) (45). Treatment of the transfected cells with another HDAC inhibitor (5 mM NaBu) for 24 hr before harvesting clearly increased the repression mediated by the GAL4-HIC-1 and GAL4-γFBP-B chimeras, whereas the repression mediated by the GAL4-BTB/POZ-BCL-6 chimera was again drastically reduced (≈60%), as expected (45) (Fig. 5). Even though in the millimolar range NaBu is known to induce side effects not directly linked to histone hyperacetylation (50–51), the differential response exhibited by HIC-1 and γFBP-B vs. BCL-6 highlights the differences in the repression mechanisms used by these three BTB/POZ proteins.
Figure 5.
Differential effects of HDAC inhibitors on the repressing potential of HIC-1, γFBP-B, and BCL-6. RK13 cells were transiently transfected in triplicate with 200 ng of the indicated GAL4-chimeras and 750 ng of the pG5LUC reporter. Twenty-four hours later, the cells were treated with 300nM TSA (from a ×1,000 concentrated DMSO dissolved stock) (black box) or 5 mM NaBu (grey box) or mock treated with an equal volume of DMSO (vehicle, open box) for a further 24 hr before harvesting. The luciferase activity was normalized to the β-gal activity of a cotransfected pSG5 β-gal construct (50 ng).
In conclusion, these experiments demonstrate that recruitment of a HDAC complex is not required for full HIC-1- and γFBP-B-mediated transcriptional silencing.
Discussion
The human and murine HIC-1 and the avian γFBP-B BTB/POZ domains differ notably by the presence of a specific insertion located in a loop between the conserved β5 strand and α5 helix, known from the PLZF structure to be involved in dimerization and scaffolding of the domain (3). Here we attempted to explore, using the physiologically relevant mammalian two-hybrid system, the functional significance of this specific insertion. As predicted from this structural analysis, we report here that this insertion significantly hinders, but does not abolish, the homo- and heterodimerization potential of the HIC-1 BTB/POZ domain, as compared with the γFBP-B BTB/POZ domain (Fig. 1). By contrast, point mutants of BCL-6 that totally fail to self interact, to repress transcription, and to interact with the SMRT/N-CoR corepressors, have recently been described (21). It is noteworthy that these mutations fall within the conserved N-terminal α1 helix, which participates in the formation of the interlaced dimer and, as a consequence, in the formation of the inner rim of the central groove where the interaction with the corepressors has been proposed to occur (3). The β5 strand-loop-α5 helix is clearly excluded from this central cavity and affects only slightly the transcriptional repression potential of HIC-1 as compared with γFBP-B. Indeed, the HIC-1 and γFBP-B BTB/POZ domains fused to the GAL4 DNA-binding domain appear as very similar autonomous transcriptional repressing domains.
Efficient transcriptional silencing by PLZF and BCL-6 requires the recruitment of a SMRT/N-CoR-mSin3A/B-HDAC complex through multiple and also clearly distinctive physical interactions between members of the repressing complex and various regions of these BTB/POZ proteins (20–28). We thus wanted to define whether HIC-1 and γFBP-B share the same repression mechanism, and how they could interact with this complex in comparison with PLZF and BCL-6. Surprisingly, we failed to detect any significant interaction between HIC-1 and γFBP-B and the commonly used members of the repressing complex, SMRT/N-CoR and mSin3A. Because HIC-1 and γFBP-B behave similarly, this effect is not caused by the presence of the specific insertion but is a property of these two phylogenetically related proteins. Because negative results have to be interpreted with caution, we always included in our assays a GAL4 chimera with a similar PLZF and/or BCL-6 domain as a positive control of interaction. In all instances, they nicely reproduced works previously published, thus fully validating our results. To finally rule out the involvement of deacetylase in the HIC-1 and γFBP-B transcriptional repression, we took advantage of the existence of highly specific, general, and potent inhibitors of this activity, such as TSA (50–53). A 24-hr treatment with 300 nM TSA drastically counteracts the repression elicited by a GAL4-BTB/POZ BCL-6 chimera on a GAL4 responsive gene (45), whereas, under the same conditions, the repressing potential of the HIC-1 and γFBP-B GAL4 chimeras was preserved, thus excluding the participation of HDAC. Similarly, sodium butyrate, which is a more pleiotropic inhibitor of deacetylase and hence of BCL-6-mediated repression, exacerbates the HIC-1- and γFBP-B-repressing potential. At the concentration required (5 mM), NaBu is known to have side effects, including in particular dephosphorylation of histones and other nuclear proteins, such as Rb (54). In HIC-1 and γFBP-B, this putative NaBu-induced dephosphorylation could increase the half-life of these proteins by interfering with degradation pathways conserved among BTB/POZ proteins (55) and/or could stabilize their interaction with partner(s) involved in repression.
Numerous transcription factors in vertebrates and yeast share with the BTB/POZ proteins PLZF and BCL-6 the property to recruit a SMRT/N-CoR-mSin3A/B-HDAC, indicating that the recruitment of an HDAC-repressing complex represents an ancestral and widely used mechanism by which eukaryotes repress transcription. The BTB/POZ domain already present in the ancestor of the crown group eukaryote has undergone independent expansion in plants and various animal lineages (4). During this process, some transcriptional repressors such as HIC-1 and γFBP-B could have developed alternative strategies to the establishment of repressive chromatin structure through targeted histone deacetylation, as exemplified by PLZF and BCL-6. The transcriptional mechanism(s) used by HIC-1 and γFBP-B is still elusive, but several not mutually exclusive hypotheses could be proposed: they could interact with other repressing complexes devoid of HDAC activity; they could negatively interfere with components of the basal transcriptional machinery, as shown for SMRT/N-CoR with TFIIB and TAFII32 (26, 56), or they could participate in the establishment of heterochromatin structure (57). Clearly, the characterization of the HIC-1- and γFBP-B-specific partners by yeast two-hybrid screens will be required to unravel their repression mechanism.
Acknowledgments
We thank Pr. Dominique Stehelin for constant interest and support and J. Coll, B. Vandenbunder, and S. Saule for critical reading of the manuscript. We are indebted to R. J. Lin and R. M. Evans, K. D. Huyinh and V. J. Bardwell, M. Privalski, W. M. Yang and E. Seto, and C. Hassig and S. L. Schreiber for generously providing molecular clones. This work was supported by funds from Centre National de la Recherche Scientifique, the Pasteur Institute, and Association pour la Recherche contre le Cancer. S.D. holds a fellowship from the Ligue Nationale contre le Cancer.
Abbreviations
- TSA
trichostatin A
- SMRT
silencing mediator of retinoid and thyroid hormone receptor
- N-CoR
nuclear receptor corepressor
- HDAC
histone deacetylase
- BTB/POZ
broad complex, tramtrack, and bric à brac/poxviruses and zinc finger
- HIC-1
hypermethylated in cancer
- γFBP-B
γF1-binding protein isoform B
- BCL-6
B cell lymphomas 6
- PLZF
promyelocytic leukemia
- β-galactosidase
β-gal
- PAH1–4
paired amphipathic helix domains
- BACH2
BTB and CNC homology protein 2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bardwell V J, Treisman R. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 2.Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. Cell Growth Diff. 1995;6:1193–1198. [PubMed] [Google Scholar]
- 3.Ahmad K F, Engel C, Privé G G. Proc Natl Acad Sci USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin E V. J Mol Biol. 1999;285:1353–1361. doi: 10.1006/jmbi.1998.2394. [DOI] [PubMed] [Google Scholar]
- 5.Xiong W C, Montell C. Genes Dev. 1993;7:1085–1096. doi: 10.1101/gad.7.6.1085. [DOI] [PubMed] [Google Scholar]
- 6.Numoto M, Niwa O, Kaplan K K, Wong K K, Merell K, Kamiya K, Yanagihara K, Calame K. Nucleic Acids Res. 1993;21:3767–3775. doi: 10.1093/nar/21.16.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q, Shalaby F, Puri M C, Tang S, Breitman M L. Dev Biol. 1994;165:165–177. doi: 10.1006/dbio.1994.1243. [DOI] [PubMed] [Google Scholar]
- 8.Okabe S, Fukuda T, Ishibashi K, Kojima S, Okada S, Hatano M, Ebara M, Saisho H, Tokuhisha T. Mol Cell Biol. 1998;18:4235–4244. doi: 10.1128/mcb.18.7.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muto A, Hoshino H, Madisen L, Yanai N, Obinata M, Karasuyama H, Hayashi N, Nakauchi H, Yamamoto M, Groudine M, Igarashi K. EMBO J. 1998;17:5734–5743. doi: 10.1093/emboj/17.19.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J Y, English M A, Ball H J, Yeyati P L, Waxman S, Licht J D. J Mol Biol. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 11.Deweindt C, Albagli O, Bernardin F, Dhordain P, Quief S, Lantoine D, Kerckaert J P, Leprince D. Cell Growth Differ. 1997;6:1495–1503. [PubMed] [Google Scholar]
- 12.Seyfert V L, Allman D, He Y, Staudt L M. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- 13.Chang C, Ye B H, Chaganti R S K, Dalla-Favera R. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhordain P, Albagli O, Ansieau S, Koken M H K, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J P, Leprince D. Oncogene. 1997;11:2689–2697. [PubMed] [Google Scholar]
- 15.Davies J M, Hawe N, Kabarowski J, Huang Q-H, Zhu J, Brand N J, Leprince D, Dhordain P, Cook M, Morris-Kay G, Zelent A. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- 16.Morrison D J, Pendergast P S, Stravopoulos P, Colmenares S U, Kobayashi R, Hernandez N. Nucleic Acids Res. 1999;27:1251–1262. doi: 10.1093/nar/27.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsani K R, Hadjibagheri M A N, Verrijzer P C. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 19.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 20.Dhordain P, Albagli O, Lin R J, Ansieau S, Quief S, Leutz A, Kerckaert J P, Evans R M, Leprince D. Proc Natl Acad Sci USA. 1997;94:10762–10769. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh D K, Bardwell V J. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- 22.Hong S H, Davie G, Wong C-W, Dejean A, Privalski M L. Proc Natl Acad Sci USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 24.Grignani F, DeMatteis S, Nervi C, Tomassoni L, Gelmetti V, Croce M, Fanelli M, Ruthardt M, Ferrara F F, Zamir I, et al. Nature (London) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 25.He L Z, Guidez F, Triboli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi P P. Nat Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 26.Wong C-W, Privalski M L. Mol Cell Biol. 1998;18:5500–5510. doi: 10.1128/mcb.18.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong C-W, Privalski M L. J Biol Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- 28.David G, Alland L, Hong S-W, Wong C-W, DePinho R A, Dejean A. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- 29.Taunton J, Hassig C, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 30.Yang W-M, Inouye C, Zheng Y, Bears D, Seto E. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 32.Hassig C A, Fleisher T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 33.Laherty C D, Yang E-M, Sun J, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 35.Nagy L, Kao H Y, Chakravarty D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 36.Ordentlich P, Downes M, Xie W, Genin A, Spinner N B, Evans R E. Proc Natl Acad Sci USA. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laherty C D, Billin A N, Lavinsky R, Yochum G S, Bush A C, Sun J M, Mullen T-M, Davie J R, Rose D W, Glass C K, et al. Mol Cell. 1998;2:33–42. doi: 10.1016/s1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 38.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 39.Kao H-Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesh T. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 41.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 42.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Rosenfeld M G, et al. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci P G, Lazar M A. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nomura T, Khan M M, Kaul S C, Dong H D, Wadwha R, Colmenares C, Kohno I, Ishii S. Genes Dev. 1999;13:412–423. doi: 10.1101/gad.13.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhordain P, Lin R J, Quief S, Lantoine D, Kerckaert J P, Evans R M, Albagli O. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makos-Wales M, Biel M, El Deiry W, Nelkin B D, Issa J P, Cavenee W K, Kuerbitz S J, Baylin S B. Nat Med. 1995;1:570–577. doi: 10.1038/nm0695-570. [DOI] [PubMed] [Google Scholar]
- 47.Grimm C, Spörle R, Schmid T E, Adler I-D, Adamski A, Schughart K, Graw J. Hum Mol Genet. 1999;8:697–710. doi: 10.1093/hmg/8.4.697. [DOI] [PubMed] [Google Scholar]
- 48.Guerardel C, Deltour S, Leprince D. FEBS Lett. 1999;451:253–256. doi: 10.1016/s0014-5793(99)00583-9. [DOI] [PubMed] [Google Scholar]
- 49.Bhathal H S, Stumph W E. Biochem Biophys Acta. 1996;1308:1114–1118. doi: 10.1016/0167-4781(96)00094-2. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida M, Horinouchi S, Beppu T. BioEssays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 51.Hassig C A, Schreiber S L. Curr Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 52.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 53.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz B, Avivi-Green C, Polak-Charcon S. Mol Cell Biochem. 1998;188:21–30. [PubMed] [Google Scholar]
- 55.Niu H, Ye B H, Dalla-Favera R. Genes Dev. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muscat G E O, Burke L J, Downes M. Nucleic Acids Res. 1998;26:2899–2907. doi: 10.1093/nar/26.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M. J Biol Chem. 1998;273:2669–2674. doi: 10.1074/jbc.273.41.26698. [DOI] [PubMed] [Google Scholar]