Abstract
Targeting of gene regulatory factors to specific intranuclear sites may be critical for the accurate control of gene expression. The acute myelogenous leukemia 8;21 (AML1/ETO) fusion protein is encoded by a rearranged gene created by the ETO chromosomal translocation. This protein lacks the nuclear matrix-targeting signal that directs the AML1 protein to appropriate gene regulatory sites within the nucleus. Here we report that substitution of the chromosome 8-derived ETO protein for the multifunctional C terminus of AML1 precludes targeting of the factor to AML1 subnuclear domains. Instead, the AML1/ETO fusion protein is redirected by the ETO component to alternate nuclear matrix-associated foci. Our results link the ETO chromosomal translocation in AML with modifications in the intranuclear trafficking of the key hematopoietic regulatory factor, AML1. We conclude that misrouting of gene regulatory factors as a consequence of chromosomal translocations is an important characteristic of acute leukemias.
Modifications in nuclear morphology are a hallmark of tumor cells and are used for diagnosis. However, there is limited knowledge of the subnuclear changes that accompany or contribute to tumor-related alterations of nuclear architecture. Biochemical evidence indicates that the protein composition of the nuclear matrix is modified in tumor cells (1–7). The nuclear matrix is a scaffold that provides a means for localizing genes and regulatory factors throughout the nuclear space (8–11). Consequently, analysis of targeting signals that direct regulatory factors to nuclear matrix-associated subnuclear sites (12) may provide insight into nuclear structure–function relationships that are compromised in cancer.
The acute myelogenous leukemia (AML) transcription factor AML11 is a key regulator of hematopoiesis (13, 14). Numerous cytogenetic abnormalities that involve genes encoding AML1 or its partner core binding factor β have been identified in AML and acute lymphocytic leukemia (15–18). The frequent 8;21 translocation produces a chimeric protein (AML1/ETO) in which the C terminus of AML1 is replaced by the unrelated ETO (MTG8) protein (19–23). The 8;21 translocation occurs in approximately 15% of AML in adult patients (24–26).
The normal form of the AML1 protein has an N-terminal region containing a runt homology DNA-binding domain (27) and a multifunctional C-terminal region that supports transactivation, repression, and intranuclear targeting (28–34). Alternative splicing of the AML1 gene can generate several protein isoforms. The predominant isoform AML1B (480 aa) contains a 31-aa nuclear matrix-targeting signal (NMTS) in the C terminus. This targeting signal is necessary and sufficient to direct the factor to nuclear matrix-associated subnuclear sites that support transcription (30). Many of these AML1 sites are localized together with the hyperphosphorylated active form of RNA polymerase IIo, and this association requires the presence of the runt homology domain of AML1B (35).
The AML1/ETO fusion product is expressed from the AML promoter and contains the DNA-binding domain of AML1 and all but the first 30 aa of the ETO protein (19, 20, 36). However, this fusion protein lacks the AML1 multifunctional C terminus, including its subnuclear targeting signal. The ETO protein that replaces it is a highly conserved gene regulatory factor related to nervy, a Drosophila homeotic target gene product (20, 37–41). The AML1/ETO fusion protein acts as a dominant-negative factor in mice (42) and in transient assays is a dominant-negative regulator of AML1B-mediated transcriptional activation (43). Additionally, AML1/ETO blocks myeloid differentiation (40, 44). AML1/ETO may prevent apoptosis by activating the Bcl2 promoter (45) and causes cellular transformation of NIH 3T3 cells (46). Thus, the t(8;21) lesion compromises normal regulatory properties of the AML1 protein by relinquishing AML1-encoded functions and acquiring ETO-related characteristics.
Targeting of gene regulatory factors to specific subnuclear sites may be critical for accurate control of gene expression. Consequently, AML1/ETO provides a paradigm for experimentally addressing whether loss of the AML1B NMTS compromises the subnuclear trafficking of AML factors. Here we show that the AML1/ETO protein is directed to intranuclear sites that do not overlap AML1 domains. Instead, the subnuclear targeting of the fusion protein is controlled by an ETO intranuclear targeting signal. Thus, our findings indicate that the misrouting of gene regulatory factors as a consequence of gene rearrangement is an important characteristic of translocation-associated leukemias.
Materials and Methods
Cell Culture.
Saos-2 cells were grown in McCoy's 5A Medium (GIBCO/BRL) supplemented with 15% FBS.
Transient Transfections.
Saos-2 cells were plated on 0.5% gelatin coated coverslips (Fisherbrand, no. 12–545-101, 22cir-1; Fisher) in 6-well tissue culture trays at a density of 0.2 × 106 cells/well. Cells were grown approximately 18 h after plating on coverslips in McCoy's 5A + 15% FBS. Cell density was approximately 50% at the time of transfection. Saos-2 cells were transfected by using either calcium phosphate (47) or Superfect as described by the manufacturer (Qiagen, Chatsworth, CA) with identical results. It was essential to achieve low levels of expression per cell to analyze colocalization. Optimization of the Superfect procedure included using 250 ng of expression vector and 5 μl of Superfect reagent per well. Cells were processed for immunofluorescence 18 h after transfection, as described below (except when noted otherwise). Epitope-tagged DNA constructs used in these experiments are described below.
Plasmids.
Plasmids hemagglutinin epitope (HA)-AML1B, HA-AML1/ETO, and Flag-ETO were constructed as described (30, 48, 49). Plasmid enhanced green fluorescent protein (EGFP)-AML1B was constructed by fusing the AML1B coding sequence (amino acids 27–480) in frame with an EGFP epitope tag cloned into pcDNA3.
Cell Extraction and Fixation.
Cells were processed for whole-cell or nuclear matrix–intermediate filament (NM-IF), as described previously (50).
Immunofluorescence.
PBS-A (PBS containing 0.5% BSA) was used to block nonspecific antibody binding, as the wash solution, and for antibody dilution, unless otherwise indicated. Antisera were as follows: a rabbit polyclonal antiserum to the HA epitope was diluted 1:1,000 (Santa Cruz Biotechnology, no. sc805); a mouse monoclonal antibody to the Flag-epitope was diluted 1:1,000 (Kodak, no. IB13010 or Sigma, no. F3165); a mouse monoclonal antibody to human promyelocytic leukemia was diluted 1:1,000 (Santa Cruz Biotechnology, no. SC966); and a rabbit polyclonal to ETO protein was diluted 1:500. Diluted antibody was added as a 50-μl drop to coverslips in wells, covered lightly with Parafilm, and incubated for 1 h at 37°C or overnight at 4°C. Coverslips were rinsed four times with PBS-A, and secondary antibody was added. Secondary antibody was goat anti-rabbit IgG conjugated to fluorescein or Texas red or Donkey anti-mouse IgG conjugated to fluorescein or Texas red (Jackson ImmunoResearch), diluted 1:500, added to coverslips, and incubated 1 h at 37°C. After incubation, coverslips were rinsed four times with PBS-A; one time with PBS-A containing 0.1% Triton X-100 and 0.05 μg/ml of the DNA counterstain 4′,6-diamidino-2-phenylindole (DAPI); one time with PBS-A containing 0.1% Triton X-100; and twice in PBS. Coverslips were mounted in Vectashield (Vector Laboratories) as an antifade mounting medium.
Microscopy.
A Zeiss Axioplan 2 microscope equipped with epifluorescence filters and a charge-coupled device camera interfaced with the MetaMorph Imaging System (Universal Imaging, Media, PA) was used.
Results
Subnuclear Targeting of the t(8;21) AML1/ETO Translocation Fusion Protein.
There are at least three signals in the AML1B transcription factor that determine its subcellular location. The nuclear localization signal of AML1 is required for nuclear import (28), whereas its NMTS directs it to the appropriate sites on the nuclear matrix (30). The DNA binding domain facilitates linkage with transcriptionally active subnuclear foci (35).
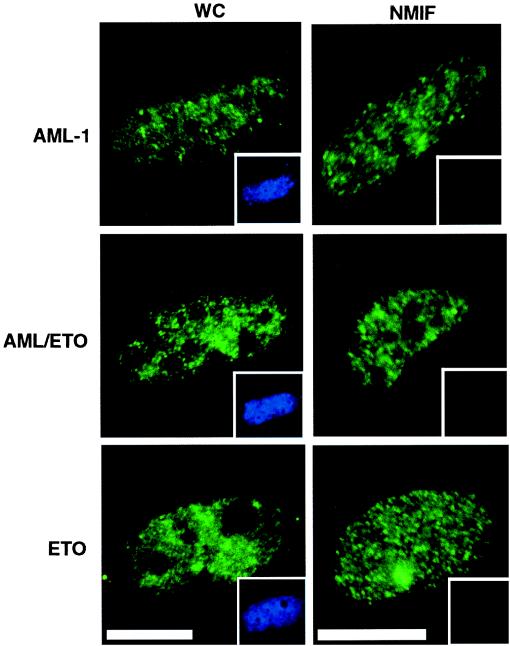
The AML1/ETO translocation results in the removal of the AML NMTS from the DNA-binding domain of the AML1 protein. The loss of the NMTS should result in aberrant intranuclear targeting of AML1. To determine the location of the AML1/ETO fusion protein, we examined whole-cell and NM-IF preparations of Saos-2 cells transfected with epitope-tagged AML1B and AML1/ETO expression constructs. Expressed proteins were detected with epitope-specific antibodies and visualized by using an epifluorescence microscope interfaced with a digital imaging system. We observed that both AML1B and AML1/ETO proteins were present in whole-cell preparations (Fig. 1). As reported previously, AML1B was retained in the NM-IF fraction. However, the AML1/ETO fusion protein, which lacks the AML1B NMTS, was nevertheless associated with the NM-IF (Fig. 1). Thus, the ETO moiety may contain a compensatory subnuclear targeting signal that supports nuclear matrix association and substitutes for the AML1B NMTS. To test this possibility directly, we analyzed the subnuclear distribution of the ETO protein. The results (Fig. 1) show that ETO indeed associates with the nuclear matrix and suggest that the ETO protein contains an independent subnuclear targeting signal. The presence of this signal in the AML1/ETO fusion protein apparently serves to direct the chimeric protein to the nuclear matrix in the absence of the AML NMTS.
Figure 1.
The chimeric AML/ETO fusion protein is directed to the nuclear matrix in situ in the absence of the AML1B NMTS. Immunofluorescence localization of AML1B (Top), AML/ETO (Middle), and ETO (Bottom) proteins expressed in Saos-2 cells was examined in whole-cell (Left) and in situ nuclear-matrix (NMIF, Right) preparations by using specific antibodies to detect the epitope-tagged proteins and visualized by using a FITC secondary antibody. 4′,6-diamindino-2-phenylindole (DAPI) staining was used to evaluate DNA content and is shown (Inset) for each image. (Bar = 10 μm.)
AML1 and the AML1/ETO Fusion Protein Are Targeted to Different Subnuclear Locations.
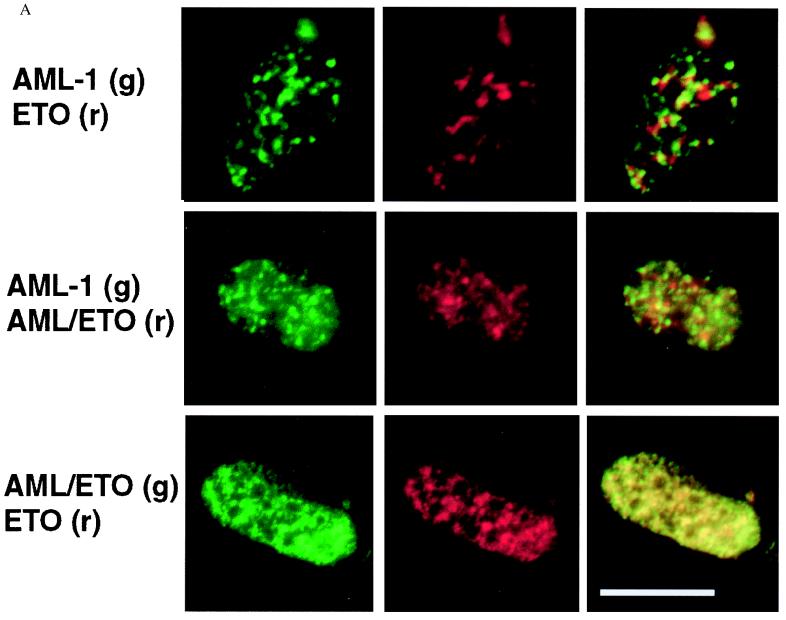
To assess whether loss of the AML1 NMTS and/or fusion of ETO sequences results in an altered spatial distribution of AML1B, we next compared the nuclear matrix-associated foci containing AML1B with those containing the translocation gene product AML1/ETO. Saos-2 cells were cotransfected with equal concentrations of AML1B and AML1/ETO epitope-tagged expression constructs and extracted to obtain the NM-IF fraction. The epitope-tagged proteins were visualized by using digital fluorescence microscopy or laser-scanning confocal microscopy. AML1B and AML1/ETO are both observed as punctate nuclear signals, one red and one green, that are associated with the nuclear matrix. There is only a minor yellow signal, indicating very little overlap of the fluorescent signals of the two expressed proteins (Fig. 2). Thus the AML1/ETO fusion protein is targeted to subnuclear sites within the nuclear matrix different from those of the full-length AML1B protein. We conclude that the substitution for the AML1B C terminus results in misrouting of the AML gene regulatory factor within the nucleus.
Figure 2.
AML1B and AML/ETO proteins are targeted to distinct subnuclear locations. Immunofluorescence localization of coexpressed proteins in Saos-2 cells was examined in in situ nuclear-matrix preparations by using digital fluorescence microscopy (A) or laser-scanning confocal microscopy (B). AML and ETO coexpression (Top), AML and AML/ETO coexpression (Middle), and AML/ETO and ETO coexpression (Bottom). Epitope-tagged proteins were detected with specific antibodies and visualized by using a Texas red-conjugated secondary antibody seen as a red fluorescent signal (r, red, Center), and either a FITC secondary antibody or an EGFP epitope tag, seen as a green fluorescent signal (g, green, Left). Colocalization of red and green immunofluorescence signals is observed as yellow staining in the merged images (Right).
We then analyzed the subnuclear locations of coexpressed AML1B and ETO proteins (Fig. 2). Similar to the results for AML1B and AML1/ETO, minimal overlap is observed with these expressed proteins, and most sites are detected as distinct green or red immunofluorescence signals, respectively. The results show that the majority of AML1B and ETO proteins do not colocalize. Hence, AML1B and ETO are directed to distinct subnuclear compartments, which may reflect differences in the regulatory activities of these factors.
To address the specificity of subnuclear targeting, we compared the subnuclear distribution of AML1B proteins containing different epitope tags. We cotransfected HA-AML1B and EGFP AML1B into Saos-2 cells and examined the localization of the expressed proteins, as described above. The data show that the majority of the expressed proteins are targeted to the same nuclear matrix-associated foci, because they are detected mostly as colocalizing yellow immunofluorescence signals (Fig. 3). We conclude that coexpression of AML1B with two different epitope tags (HA and EGFP) results in targeting to the same subnuclear compartment. Thus, epitope tagging does not interfere with specificity of intranuclear targeting.
Figure 3.
EGFP or the HA epitope tag does not interfere with specificity of intranuclear targeting. Coexpression of the AML1B protein tagged with either EGFP or the HA epitope was examined in Saos-2 cells by using both digital fluorescence microscopy (A) and laser-scanning confocal microscopy (B). EGFP-AML1B (Left), HA-AML1B (Center), and merged images (Right) are shown. (Bar = 10 μm.)
We evaluated whether the ETO portion of AML1/ETO redirects the fusion protein to the ETO containing foci associated with the nuclear matrix. ETO and AML1/ETO were cotransfected into Saos-2 cells, and the intranuclear localization of both proteins was analyzed. In contrast to the other pairs of proteins examined, we find that the majority of ETO and AML1/ETO signals overlap, as reflected by mostly yellow immunofluorescence signal in the merged image (Fig. 2). Therefore, AML1/ETO and ETO are targeted to the same subnuclear compartment of the nuclear matrix. Taken together, our results show that the fusion of ETO with AML1 results in misdirected targeting of the AML DNA-binding domain to subnuclear foci that are determined by the ETO protein.
Discussion
Our results link the 8;21 chromosomal translocation in AML with modifications in the intranuclear trafficking of a key hematopoietic regulatory factor, AML1. Our principal finding is that substitution of the multifunctional C terminus of AML1 with the chromosome 8-derived ETO protein precludes targeting of the factor to AML1 subnuclear domains. Instead, the AML1/ETO fusion protein is redirected to different nuclear foci. We have also demonstrated that AML1/ETO and ETO can localize at the same intranuclear sites. Thus, our results show that AML1 does not affect ETO trafficking and suggest that ETO contains a targeting signal that specifies a distinct intranuclear address.
The leukemia-related AML1 and promyelocytic leukemia proteins have previously been identified as nuclear matrix-associated factors, consistent with recent findings by others (23, 34). Here we report that AML1/ETO and ETO are also nuclear matrix associated. This association of multiple distinct factors is in agreement with the concept that the nuclear matrix supports the dynamic organization of functionally specialized foci that are spatially distinct. Taken together, our findings suggest that nuclear matrix-related mechanisms for spatial targeting and subnuclear organization of regulatory factors are aberrant in leukemia cells.
Regulation of the proximity of genes and cognate transcription factors may be mediated by specific targeting signals. Experimental support for this trafficking mechanism is provided by the recent identification of a sequence- and structure-specific NMTS in the AML1 protein (30, 51). The NMTS directs this hematopoietic transcription factor to nuclear matrix-associated foci that support gene expression (35). Other classes of gene regulatory factors, including the androgen and glucocorticoid receptors (52, 53), Pit-1 (54), and YY-1 (50, 55), have also been shown to contain NMTSs that differ from the AML1 NMTS. This diversity of trafficking signals may provide the requisite specificity to direct factors to independent subnuclear locations.
An important ramification of this model is that molecular alterations may cause misrouting of transcription factors resulting in compromised gene expression and development of disease. The t(8;21) fusion protein provides a striking demonstration of the functional consequences of altered subnuclear targeting. Our findings show that this translocation rearranges the normal targeting signals of AML1 and ETO and misroutes the fusion protein away from AML1 foci. This misdirected targeting may directly contribute to the deregulation of AML1 responsive genes observed in t(8;21)-related AMLs. In a broader perspective, deregulation of transcription factor targeting may be fundamental to aberrations in gene expression characteristic of many leukemias and other tumors involving chromosomal translocations.
Acknowledgments
The authors thank Dr. Jeffrey Nickerson for his contributions to developing the in situ immunofluorescence analysis, Dr. Sheldon Penman for insightful discussions, and Elizabeth Bronstein for editorial assistance with the preparation of this manuscript. Elizabeth Buffone provided expert assistance with cell culture. This work was supported by grants from the National Institutes of Health (AR45688 and DK50222).
Abbreviations
- AML
acute myelogenous leukemia
- ETO
eight
- twenty one protein
NMTS, nuclear matrix-targeting signal
- NM-IF
nuclear matrix–intermediate filament
- HA
hemagglutinin
- EGFP
enhanced green fluorescent protein
References
- 1.Fey E G, Penman S. Proc Natl Acad Sci USA. 1988;85:121–125. doi: 10.1073/pnas.85.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bidwell J P, Fey E G, van Wijnen A J, Penman S, Stein J L, Lian J B, Stein G S. Cancer Res. 1994;54:28–32. [PubMed] [Google Scholar]
- 3.Getzenberg R H, Pienta K J, Huang E Y, Coffey D S. Cancer Res. 1991;51:6514–6520. [PubMed] [Google Scholar]
- 4.Stuurman N, van Driel R, de Jong L, Meijne A M, Van Renswoude J. Exp Cell Res. 1989;180:460–466. doi: 10.1016/0014-4827(89)90072-4. [DOI] [PubMed] [Google Scholar]
- 5.Nickerson J A. J Cell Biochem. 1998;70:172–180. [PubMed] [Google Scholar]
- 6.Holth L T, Chadee D N, Spencer V A, Samuel S K, Safneck J R, Davie J R. Int J Oncol. 1998;13:827–837. doi: 10.3892/ijo.13.4.827. [DOI] [PubMed] [Google Scholar]
- 7.Samuel S K, Minish T M, Davie J R. J Cell Biochem. 1997;66:9–15. [PubMed] [Google Scholar]
- 8.Nickerson J A, He D C, Krochmalnic G, Penman S. Proc Natl Acad Sci USA. 1990;87:2259–2263. doi: 10.1073/pnas.87.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei X, Somanathan S, Samarabandu J, Berezney R. J Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei X, Samarabandu J, Devdhar R S, Siegel A J, Acharya R, Berezney R. Science. 1998;281:1502–1505. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- 11.Ward W S, Coffey D S. Biochem Biophys Res Commun. 1990;173:20–25. doi: 10.1016/s0006-291x(05)81015-0. [DOI] [PubMed] [Google Scholar]
- 12.Stein, G. S., van Wijnen, A. J., Stein, J. L., Lian, J. B., McNeil, S. & Pockwinse, S. M. (1999) J. Cell. Biochem., in press. [DOI] [PubMed]
- 13.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Speck N A, Stacy T. Crit Rev Eukaryotic Gene Expression. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 16.Meyers S, Hiebert S W. Crit Rev Eukary Gene Expr. 1995;5:365–383. doi: 10.1615/critreveukargeneexpr.v5.i3-4.70. [DOI] [PubMed] [Google Scholar]
- 17.Sawyers C L. Lancet. 1997;349:196–200. doi: 10.1016/S0140-6736(96)07535-6. [DOI] [PubMed] [Google Scholar]
- 18.Rowley J D. Cancer Res. 1984;44:3159–3168. [PubMed] [Google Scholar]
- 19.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 20.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozu T, Komori A, Sueoka E, Fujiki H, Kaneko Y, Matsui T, Uehara T, Seino Y, Ishii M. Leukemia. 1997;11:S3. , 297–298. [PubMed] [Google Scholar]
- 23.Le X F, Claxton D, Kornblau S, Fan Y H, Mu Z M, Chang K S. Eur J Haematol. 1998;60:217–225. doi: 10.1111/j.1600-0609.1998.tb01027.x. [DOI] [PubMed] [Google Scholar]
- 24.Koeffler H P. Ann Intern Med. 1987;107:748–758. doi: 10.7326/0003-4819-107-5-748. [DOI] [PubMed] [Google Scholar]
- 25.Schiffer C A, Lee E J, Tomiyasu T, Wiernik P H, Testa J R. Blood. 1989;73:263–270. [PubMed] [Google Scholar]
- 26.Tashiro S, Kyo T, Tanaka K, Oguma N, Hashimoto T, Dohy H, Kamada N. Cancer. 1992;70:2809–2815. doi: 10.1002/1097-0142(19921215)70:12<2809::aid-cncr2820701214>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Daga A, Tighe J E, Calabi F. Nature (London) 1992;356:484. doi: 10.1038/356484b0. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Maruyama M, Satake M, Bae S C, Ogawa E, Kagoshima H, Shigesada K, Ito Y. Mol Cell Biol. 1995;15:1651–1661. doi: 10.1128/mcb.15.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyers S, Downing J R, Hiebert S W. Mol Cell Biol. 1993;13:6336–6345. doi: 10.1128/mcb.13.10.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, et al. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanno T, Kanno Y, Chen L F, Ogawa E, Kim W Y, Ito Y. Mol Cell Biol. 1998;18:2444–2454. doi: 10.1128/mcb.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrovick M S, Hiebert S W, Friedman A D, Hetherington C J, Tenen D G, Zhang D E. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruhn L, Munnerlyn A, Grosschedl R. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 34.Chen L F, Ito K, Murakami Y, Ito Y. Mol Cell Biol. 1998;18:4165–4176. doi: 10.1128/mcb.18.7.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng C, McNeil S, Pockwinse S, Nickerson J A, Shopland L, Lawrence J B, Penman S, Hiebert S W, Lian J B, van Wijnen A J, et al. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozu T, Miyoshi H, Shimizu K, Maseki N, Kaneko Y, Asou H, Kamada N, Ohki M. Blood. 1993;82:1270–1276. [PubMed] [Google Scholar]
- 37.Feinstein P G, Kornfeld K, Hogness D S, Mann R S. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erickson P F, Dessev G, Lasher R S, Philips G, Robinson M, Drabkin H A. Blood. 1996;88:1813–1823. [PubMed] [Google Scholar]
- 39.Davis J N, Williams B J, Herron J T, Galiano F J, Meyers S. Oncogene. 1999;18:1375–1383. doi: 10.1038/sj.onc.1202412. [DOI] [PubMed] [Google Scholar]
- 40.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Hoshino T, Redner R L, Kajigaya S, Liu J M. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuda T, Cai Z, Yang S, Lenny N, Lyu C J, van Deursen J M, Harada H, Downing J R. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 43.Frank R, Zhang J, Uchida H, Meyers S, Hiebert S W, Nimer S D. Oncogene. 1995;11:2667–2674. [PubMed] [Google Scholar]
- 44.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klampfer L, Zhang J, Zelenetz A O, Uchida H, Nimer S D. Proc Natl Acad Sci USA. 1996;93:14059–14064. doi: 10.1073/pnas.93.24.14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frank R C, Sun X, Berguido F J, Jakubowiak A, Nimer S D. Oncogene. 1999;18:1701–1710. doi: 10.1038/sj.onc.1202459. [DOI] [PubMed] [Google Scholar]
- 47.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 48.Meyers S, Lenny N, Hiebert S W. Mol Cell Biol. 1995;15:1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Rosenfeld M G, et al. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McNeil S, Guo B, Stein J L, Lian J B, Bushmeyer S, Seto E, Atchison M L, Penman S, van Wijnen A J, Stein G S. J Cell Biochem. 1998;68:500–510. [PubMed] [Google Scholar]
- 51.Tang L, Guo B, Javed A, Choi J Y, Hiebert S, Lian J B, van Wijnen A J, Stein J L, Stein G S, Zhou G W. J Biol Chem. 1999;274:33580–33586. doi: 10.1074/jbc.274.47.33580. [DOI] [PubMed] [Google Scholar]
- 52.Tang Y, Getzenberg R H, Vietmeier B N, Stallcup M R, Eggert M, Renkawitz R, DeFranco D B. Mol Endocrinol. 1998;12:1420–1431. doi: 10.1210/mend.12.9.0169. [DOI] [PubMed] [Google Scholar]
- 53.van Steensel B, Jenster G, Damm K, Brinkmann A O, van Driel R. J Cell Biochem. 1995;57:465–478. doi: 10.1002/jcb.240570312. [DOI] [PubMed] [Google Scholar]
- 54.Mancini M G, Liu B, Sharp Z D, Mancini M A. J Cell Biochem. 1999;72:322–338. [PubMed] [Google Scholar]
- 55.Bushmeyer S M, Atchison M L. J Cell Biochem. 1998;68:484–499. [PubMed] [Google Scholar]