Summary
Many tumor cells have elevated levels of hydrolytic and proteolytic enzymes, presumably to aid in key processes such as angiogenesis, cancer cell invasion, and metastasis. Since the activities of these enzymes are often regulated by post-translational mechanisms, their functional roles in cancer progression are difficult to study using traditional genomic and proteomic methods. Thus, methods that allow for the direct monitoring of enzyme activity in a physiologically relevant environment are required to better understand the roles of specific players in the complex process of tumorigenesis. This review highlights advances in the field of activity-based proteomics, which uses small molecules known as activity-based probes (ABPs) that covalently bind to the catalytic site of target enzymes. We discuss the application of ABPs to cancer biology, specifically to the discovery of tumor biomarkers, the screening of enzyme inhibitors, and the imaging of enzymes implicated in cancer.
Introduction
During the development of many types of cancer, a complex series of events occurs, including the formation of new tumor blood vessels (angiogenesis), escape of tumor cells from the primary tumor, cell migration and invasion of adjacent tissues and blood vessels, and the establishment of new tumor colonies at distant sites (metastasis) [1]. A crucial step in all of these processes is the degradation and remodeling of the extracellular matrix (ECM). For cancer cells to invade and metastasize to a new organ, the cells must produce hydrolytic enzymes that break down the proteins of the ECM to permit the passage of tumor cells to the blood and lymphatic vessels [2–4]. Hydrolytic enzymes are also produced when new tumor blood vessels remodel and migrate through the ECM [1–3]. Extracellular and cell-surface enzymes from the matrix metalloproteinase and the serine hydrolase families, as well as intracellular enzymes from the cysteine cathepsin protease family, are associated with the degradation of the ECM [2–6]. Many of these hydrolytic and proteolytic enzymes show great promise as tumor biomarkers for the diagnosis and prognosis of human cancers [3].
Numerous proteomic methods have been developed in the past twenty years to aid us in our understanding of enzyme function in biological processes and human disease states, including cancer [7–9]. However, many of these techniques, such as two-dimensional gel electrophoresis [8] or isotope-coded affinity-tagging [7], only focus on measuring changes in protein abundance. Since most enzymes are expressed as inactive zymogens or reside in complex with their endogenous inhibitors, protein abundance does not necessarily correlate with activity. Other methods, such as protein microarrays [9], can provide information about an enzyme’s activity state but generally require recombinantly expressed proteins that are monitored in isolation. Therefore, these technologies do not provide us with a functional understanding of native proteins in the physiologically relevant environment of a cell or whole organism.
To address the limitations in classical proteomics methods, a new research field termed activity-based proteomics or chemical proteomics has been established. Activity-based proteomics uses small molecules known as activity-based probes (ABPs) that covalently bind to the catalytic site of specific target enzymes in complex proteomes derived from cells, tissues, and in some cases, whole organisms. Since many of the regulatory mechanisms for enzyme activity alter a protein’s active site (i.e. endogenous inhibitors sterically block the active site; zymogens have misaligned catalytic residues), ABPs can be designed to react only with the functionally active form of target enzymes. Modification of protein targets by these probes thus provides an indirect measure of enzyme activity and also allows for their purification and identification by mass spectrometry (MS). Several recent reviews have outlined the design of ABPs and their biological applications [10–16]. In this review, we will focus on the use of small molecule probes in cancer biology. Particular attention will be given to the application of ABPs to the discovery of novel cancer biomarkers, the screening of potential enzyme inhibitors, and the imaging of enzymes involved in tumorigenesis.
The structure of an activity-based probe
A typical ABP consists of three basic elements: (1) a reactive functional group (also termed a warhead) that covalently reacts with the enzyme’s active site, (2) a linker region that confers specificity, directs binding to the target, and prevents steric congestion, and (3) a reporter tag used for the identification, purification, or direct visualization of the probe-labeled proteins (Figure 1). The warhead is the most crucial component of an ABP – it must be reactive enough to covalently bind to the desired active site nucleophile but not too reactive to lead to nonspecific modification of other abundant molecules in the proteome (i.e. glutathione or free amines). This warhead is often derived from the electrophilic group of an irreversible enzyme inhibitor, such as an epoxide or fluorophosphonate. For enzymes that do not covalently bind their substrates, such as metalloproteases, ABPs have been developed that contain a chelator moiety (for noncovalent binding to the conserved metal atom in the active site) and a photoinducible chemical crosslinker (for covalent binding to the enzyme active site upon ultraviolet irradiation) [17]. The linker region of an ABP can influence the specificity of the probe. Depending on the type of linker used, ABPs can be engineered to bind to a specific enzyme, class of enzymes, or a broad range of enzymes with related mechanisms. Finally, the tag facilitates the detection of probe-labeled enzyme targets, either for affinity purification, gel-based screening assays, or imaging. Radioactive or fluorophoric tags are often used for gel analysis or imaging, whereas biotinylated tags enable the enrichment, purification, and MS identification of ABP-tagged enzymes.
Figure 1.
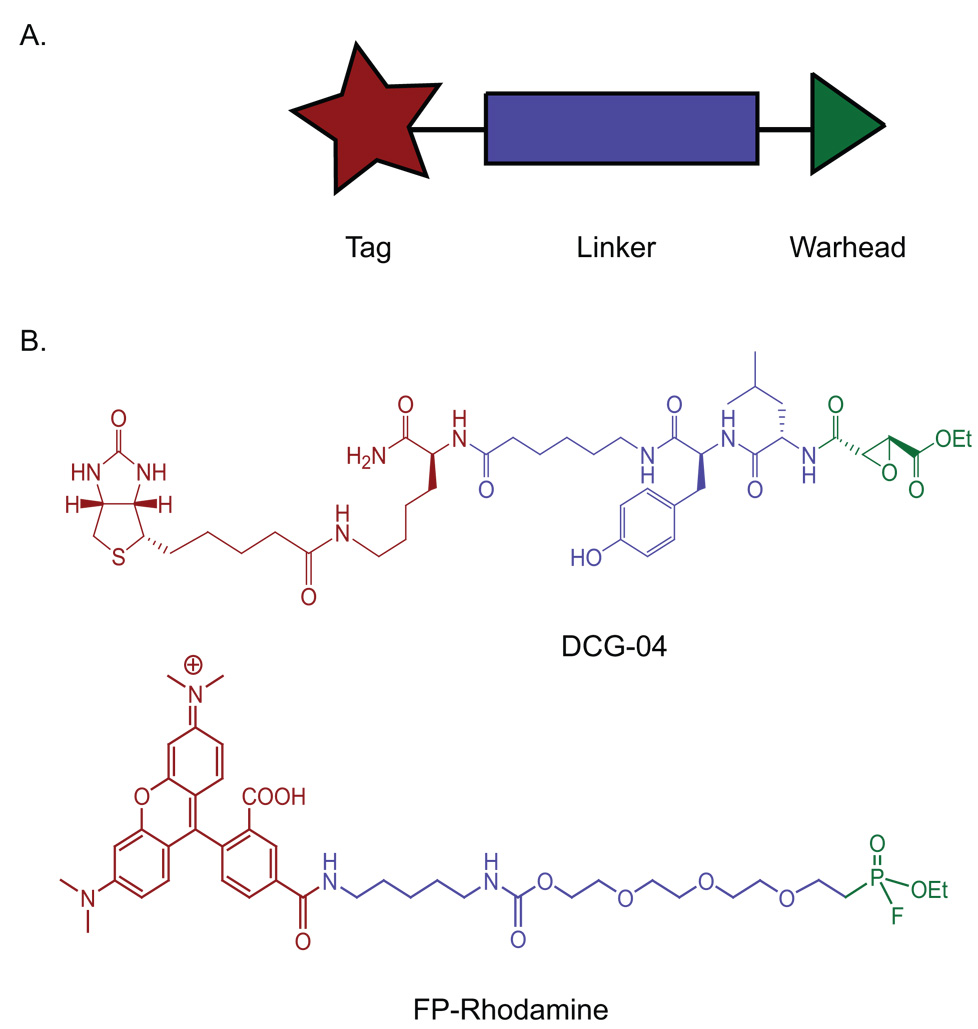
General and specific structures of activity-based probes (ABPs) (A) General structure of an ABP. (B) DCG-04 is an ABP that targets cysteine proteases and contains a biotin tag (red), a dipeptide-containing linker (blue), and an epoxide as a warhead (green). FP-rhodamine is an ABP that targets the serine hydrolase superfamily of enzymes and contains a rhodamine fluorophore (red), a poly(ethylene glycol) linker (blue), and a fluorophosphonate as a warhead (green).
Profiling and discovery of enzymes involved in cancer
Several ABPs have been developed to target enzymes implicated in cancer progression and tumorigenesis, including metalloproteases, cysteine cathepsins, and esterases [11,14,16]. These ABPs have been used to profile human tumors and tumor cell lines and identify novel enzyme activities for the diagnosis and treatment of cancer (Table 1). In a typical experiment, normal and disease proteomes are labeled with an ABP and the proteins are separated and analyzed by gel electrophoresis (Figure 2). Enzymes that differ in their activity levels can then be identified using selective antibody pulldown or MS methods. In one application of ABPs to cancer biomarker discovery, FP-rhodamine, an ABP that targets the serine hydrolase superfamily of enzymes, was used to profile the activities of these enzymes in a set of human breast and melanoma cancer cell lines [18]. This study confirmed that highly invasive cancer cells from several different tumor types upregulate a distinct set of serine hydrolase activities, including the protease urokinase and a novel integral membrane hydrolase, KIAA1363. Although urokinase was known to be involved in tumor progression, KIAA1363 had never been implicated in cancer and therefore represents a potentially novel cancer biomarker [18]. In a related study, a panel of primary human breast cancer tissues was probed with the biotin-labeled version of FP-rhodamine [19]. Probe-labeled proteins were enriched using avidin-conjugated beads, digested by trypsin, and subjected to semi-quantitative MS analysis. A set of enzymes, including KIAA1363, with elevated activities in the most aggressive tumor tissues were identified as potential breast cancer biomarkers. Recently, both FP-rhodamine and FP-biotin were used to identify enzymes that are involved in cancer cell intravasation, the process by which tumor cells enter into the vasculature [20]. The activity level of the serine protease urokinase-type plasminogen activator (uPA) was substantially elevated in the high intravasating (HT-hi/diss) variants of the human fibrosarcoma cell line HT-1080. Inhibition of uPA activity significantly reduced the rate of intravasation and metastasis of HT-hi/diss cells, suggesting that active uPA is a key determinant of these processes [20].
Table 1.
Enzyme classes characterized using ABPs.
| ABP | ABP structure | Proteome | Enzyme activity | References |
|---|---|---|---|---|
| FP-rhodamine | 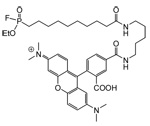 |
Human breast melanoma cell lines Human ER(+) and ER(−) breast tissue samples HT-hi/diss and HT-low/diss human fibrosarcoma cell lines MDA-MB-231 breast cancer cells before and after passage in mice |
Serine hydrolases | 18, 19, 20, 29 |
| FP-biotin | 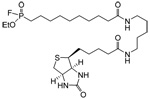 |
Human ER(+) and ER(−) breast cancer tissue samples HT-hi/diss and HT-low/diss human fibrosarcoma cell lines |
Serine hydrolases | 19, 20 |
| HxBP-Rh | 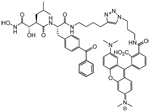 |
Human melanoma cell lines Human breast cancer cell lines |
Metalloproteases | 17, 22 |
| SAHA-BPyne | 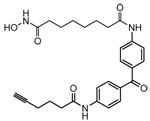 |
Human melanoma and breast cancer cell lines | Class I/II HDACs | 24 |
| HAUb-VME | Cervical carcinoma biopsies tissue samples and cell lines Human tumor cell lines |
USPs | 25, 27 | |
| DCG-04 | 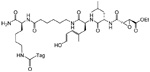 |
PyMT/ctsb+/+, PyMT/ctsb−/−, and PyMT/ctsb+/− murine tumor cells RIP1-TAG2 murine normal and tumor cells | Cysteine cathepsins | 28, 42, 43 |
| Phenyl-SE | MDA-MB-231 breast cancer cells before and after passage in mice Human breast cancer cell lines |
Multiple enzymes, including thiolases, sugar kinases, and glutathione S-transferases | 29, 30, 31, 32 | |
| GB137 | 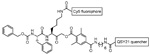 |
Human and mouse tumor cell lines MDA-MB-231 xenografted breast cancer cell lines from mice |
Cysteine cathepsins | 49 |
Figure 2.
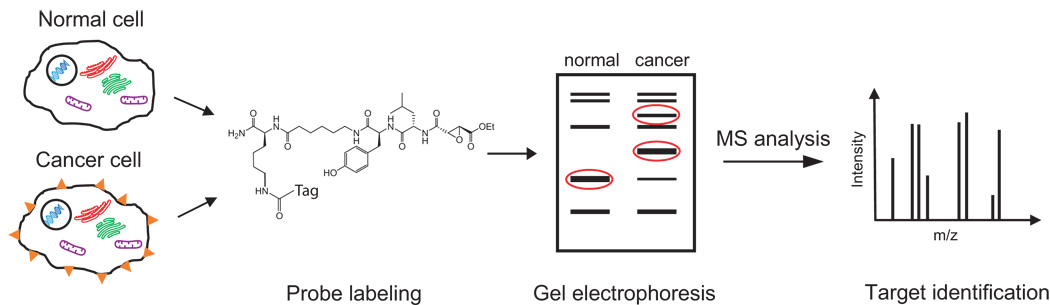
Tumor biomarker discovery using ABPs. Enzymes from cancer cells and normal cells are reacted with a biotin-containing ABP and are then separated and analyzed by gel electrophoresis. Probe-labeled enzymes are visualized, and enzymes with altered activities in normal and cancerous cells are identified. These potential tumor biomarkers can then be identified by MS analysis.
Metalloproteases are another family of enzymes that play key roles in cancer progression events such as angiogenesis and metastasis [4]. Several metalloprotease genes are overexpressed in metastatic cancers, and inhibitors of these enzymes reduce tumor angiogenesis in animal models of cancer [21]. Most enzymes from the metalloprotease family use a zinc-activated water molecule for catalysis and do not covalently bind to their substrates, thus complicating probe design. However, ABPs for these enzymes have been constructed that contain a zinc-chelating moiety and a photocrosslinking group that allows for covalent labeling of the target enzymes. A library of metalloprotease probes was used to profile the activities of metalloproteases in both breast carcinoma and melanoma cell lines [17,22]. Neprilysin, alanyl aminopeptidase, and ADAM10 activities were found to be elevated in invasive cells. Although neprilysin has historically been considered a negative regulator of tumorigenesis, its high activity in invasive melanoma cells suggests that this enzyme may contribute to cancer progression.
Histone deacetylases (HDACs) are enzymes that remove acetyl groups from lysine residues on histone tails and therefore are important regulators of gene expression. These enzymes have also been implicated in tumor growth and development [23]. HDACs hydrolyze amide bonds using an active-site zinc cation; therefore, ABPs that target these enzymes are similar to probes that target metalloproteases. An HDAC-selective ABP has been designed based on a hydroxamic acid zinc-chelating moiety and a photoactivatible benzophenone group [24]. This probe was used to analyze HDAC activity in melanoma and ovarian cacncer cell proteomes [24]. Differences in the composition and activity of HDACs were found among cancer cells with distinct biological properties, indicating that the members of the HDAC enzyme family may have a variety of functional roles in cancer.
The ubiquitin-specific proteases (USPs) are a large family of proteolytic enzymes that regulate the production and recycling of ubiquitin and are involved in cell growth and differentiation [25,26]. ABPs containing a warhead conjugated to the full-length ubiquitin protein are highly selective probes of the USPs. A number of these probes were used to identify unique and tumor-specific activities in a variety of human tumor cell lines [27]. One specific USP, UCH-L1, was highly active in numerous malignant tumor cell lines. UCH-L1 activity was also found to be upregulated in normal B cells after in vitro Epstein-Barr virus infection. This upregulation correlated with a transition from slow to rapid proliferation of the cells, implicating UCH-L1 in this adaptation. USP-specific ABPs have been used to profile USP activity in human cervical cancer biopsies [25]. The activities of two USPs, UCH-L3 and UCH37, were elevated in tumor tissue when compared to normal tissue. Additionally, the activities of four USPs were upregulated in primary keratinocytes upon infection with human papilloma virus oncogenes, providing further evidence that the USPs are involved in growth transformation [25].
ABPs have also been applied to functionally characterize enzyme activities in mouse models of cancer. The biotinylated ABP DCG-04 that targets the papain family of cysteine proteases was used to evaluate cysteine cathepsin activity in mammary tumor cells from PyMT;ctsb−/− mice, a mouse mammary cancer model deficient in cathepsin B [28]. Although cathepsin B is the most active cysteine cathepsin on the surface of PyMT;ctsb+/+ mammary cells, tumor cells lacking this protease (from PyMT;ctsb−/− mice) show an upregulation of active cathepsin X on their cell surfaces. Cathepsin X activity partially compensates for the deficiency of cathepsin B in these tumor cells. Data from these experiments suggest that proteases can dynamically compensate for each other, thus complicating the analysis of knock-out data.
In an effort to more fully characterize the enzyme activity profiles of xenografted mouse tumors, ABPs such as FP-rhodamine have been used to characterize enzyme activities in MDA-MB-231 breast cancer cells both before and after growth as tumors in the mammary fat pad of immune-deficient mice [29]. Many serine hydrolase activities, such as uPA and tissue plasminogen activator (tPA), were highly elevated in the in vivo-derived lines of MDA-MB-231 and correlated with increased tumor growth rates and metastasis upon reintroduction into mice.
Most of the ABPs developed to date are specific for an individual class of enzymes; however, broad-spectrum probes have also been designed to target enzymes for which no covalent mechanism-based inhibitors exist [30–32]. Adam and coworkers synthesized a variety of ABPs based on the reactive sulfonate ester (SE) warhead. These probes were found to covalently modify a wide range of mechanistically distinct enzymes including sugar kinases and thiolases [30]. Profiling estrogen receptor-positive (ER+) and –negative (ER−) breast cancer cells with SE-containing ABPs identified omega glutathione S-transferase (GSTO 1-1) as having elevated activity in ER− cells [30]. GSTO 1-1 had no previous association with invasive breast cancer and therefore represents a potentially new cancer biomarker. SE-based ABPs have also been used to evaluate enzyme activities in live human breast cancer cells [32]. These efforts led to the identification of a novel enoyl-CoA hydratase whose activity was highly upregulated in the ER+ cell line T-47D.
Enzyme inhibitor discovery and verification
In traditional drug discovery, libraries of small molecules are screened in vitro against purified, often recombinant, protein targets to identify inhibitors. However, in vitro assays provide only limited information regarding the in vivo potency and selectivity of an inhibitor for a related series of enzymes. Since ABPs bind to the active sites of their enzyme targets, these probes have been used to develop small molecule inhibitor screens that resolve many of the shortcomings that plague standard in vitro inhibitor assays [11,14,33]. In an ABP-based screen, whole cells, cell lysates, or even whole organisms are treated with a range of concentrations of a potential inhibitor (Figure 3). Total tissue or cell extracts are then reacted with an ABP and subjected to gel electrophoresis to separate the labeled enzymes. Small molecule inhibitor binding to a target is then measured as a decrease in enzyme labeling by the ABP. The resulting percent competition values can be measured by quantification of labeled proteins and used to generate IC50 values of the small molecule for each of the primary targets of the ABP. In contrast to standard inhibitor assays, ABP-based assays can be performed in complex proteome mixtures (including cells and whole organisms) containing multiple related enzymes, thus allowing for the evaluation of both potency and selectivity in a native cellular environment. These assays eliminate the time-consuming expression and purification of drug targets and can be used to identify inhibitors for enzymes that lack known substrates. Finally, when used in vivo, ABP-based drug screens can be used to obtain information regarding potency, selectivity, and biodistribution of an inhibitor in the context of a whole organism.
Figure 3.
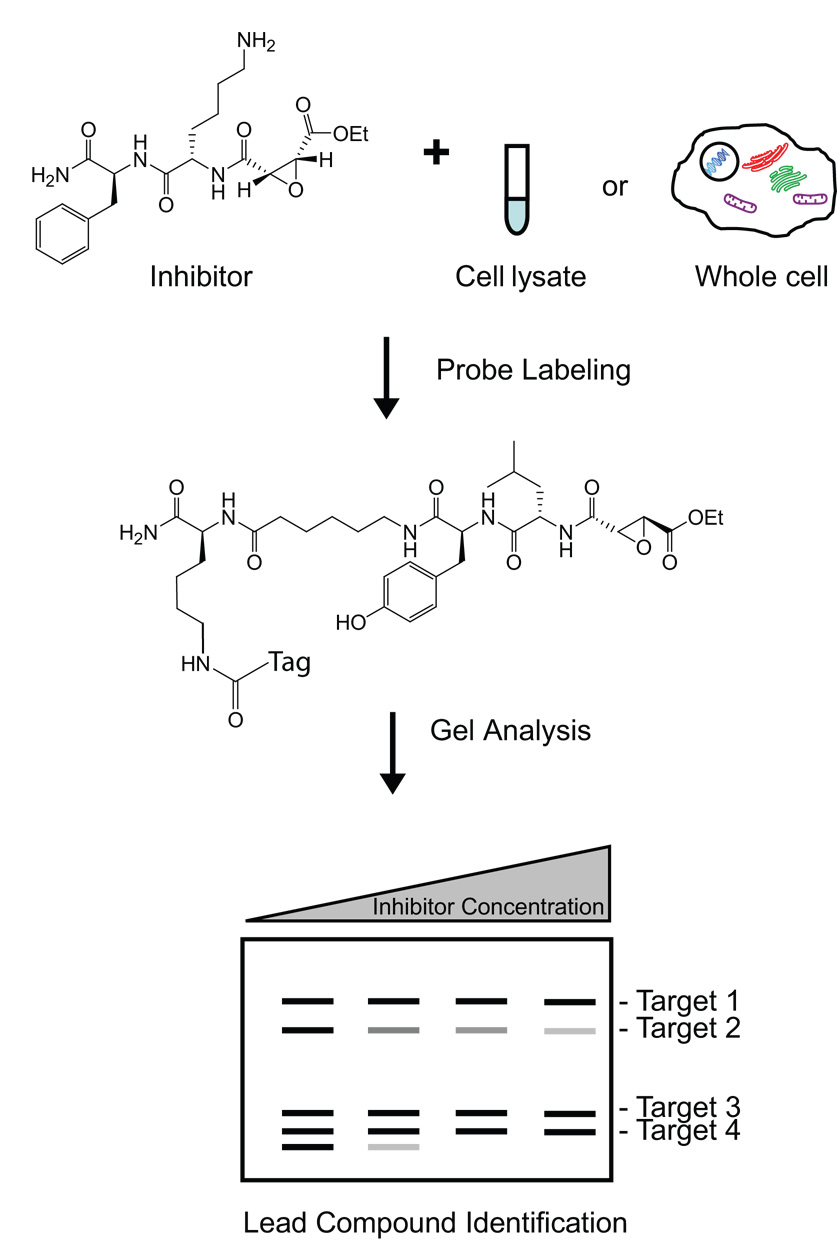
Enzyme inhibitor discovery using an ABP-based assay. Cell lysates or whole cells are treated with a range of concentrations of an inhibitor. These samples are then reacted with an ABP and subjected to gel electrophoresis to separate active enzymes. A decrease in residual activity corresponds to more potent inhibition by the inhibitor. Additionally, the selectivity of the inhibitor for one or multiple enzymes can be determined using this assay.
In one example of an ABP-based competition study, the potency and selectivity of a series of cysteine protease inhibitors was monitored in rat liver extracts [33]. This screen identified a small molecule that selectively targeted cathepsin B activity. Since cathepsin B is suspected of facilitating tumor invasion, this compound could potentially be used as a lead target for cancer therapy. ABP-based assays using the serine hydrolase probe FP-rhodamine have also been applied to the discovery of novel, selective inhibitors of KIAA1363, a poorly characterized enzyme with highly elevated activity in invasive cancer cells [34,35]. This screen yielded valuable lead compounds that facilitated further study of the function of KIAA1363 in the metabolism of lipids. Highly selective inhibitors of the caspases, cysteine proteases involved in apoptosis that are often dysregulated in cancer, have also been identified using an ABP-based competition assay [36].
In addition to the discovery of new inhibitors for cancer-related enzymes, ABPs have been applied to the characterization of existing drugs. Proteasome-directed ABPs have been used to evaluate the specificity of bortezomib, a clinically approved proteasome inhibitor for the treatment of multiple myeloma [37,38]. Myeloma cells were cultured in the presence or absence of bortezomib, incubated with a cell-permeable, proteasome-specific ABP, lysed, and then analyzed by gel electrophoresis. Results from these experiments revealed that only the activities of the β1/β1i and β5/β5i subunits of the proteasome were inhibited by bortezomib [37,38]. The cancer drug, paclitaxel, used to treat breast and ovarian cancers, has also been tested for its efficacy in a cell-based assay using a wortmannin-containing ABP that targets polo-like kinase 1 (PLK1), an enzyme with highly elevated activity in the M phase of the cell cycle [39]. Since paclitaxel arrests the cell cycle at M phase, the elevated activity levels of PLK1 in Jurkat cells treated with this drug were monitored using the wortmannin-containing ABP [39]. This study demonstrated that PLK1 activity could potentially serve as a biomarker for paclitaxel efficacy in cancer treatment.
ABPs have also been employed for the in vivo evaluation of inhibitors that target enzymes involved in cancer. Kraus and colleagues used a proteasome-directed ABP to identify the active human proteasomal subunits targeted by bortezomib in patients receiving this drug [40]. Blood cells obtained from a patient receiving bortezomib monotherapy for multiple myeloma were treated with a proteasome-specific ABP. Bortezomib treatment was shown to reversibly eliminate both β1 and β5 proteasomal activities and reduce β2 proteasomal activity in human blood cells. Since proteasomal subunits in cancer cells are known to have variable activity, the preferences of bortezomib for certain subunits may explain the differences in patient sensitivity to this cancer drug [40]. The in vivo specificity and biodistribution of another proteasome inhibitor, MG262, was monitored in murine tissues using a fluorescently-labeled proteasome-specific ABP [41].
The cathepsin-specific ABP DCG-04 has been applied to the evaluation of cathepsin inhibitors in RIP1-TAG2 transgenic mice, a mouse model of pancreatic cancer [42,43]. In these studies, inhibitors were injected into mice, and normal and tumor tissue samples were collected and analyzed for residual cathepsin activity. The inhibition of these proteases by the cathepsin-specific inhibitor JPM-OEt resulted in a reduction in invasion, angiogenesis, and tumor growth [43]. Importantly, fluorescently-labeled DCG-04 enabled the biochemical identification and monitoring of the cysteine cathepsins during tumorigenesis in these mice. In a follow-up study, a panel of cathepsin inhibitors was evaluated in these mice using radiolabeled DCG-04 [42]. Inhibitors were optimized for selectivity and potency based on these results and then were reprofiled in vivo. A set of inhibitors was identified that blocked cysteine cathepsin activity in tumor tissues and can be used as lead compounds for cancer therapy. These studies demonstrate that ABPs can be a valuable tool for the analysis of drug specificity and pharmacodynamic properties in vivo.
The principles of ABP competition assays were also used by Evans and coworkers in their screen of a small library of inhibitors against the invasive human breast cancer cell line MDA-MB-231 [44]. These inhibitors contain an electrophilic spiroepoxide that covalently modifies enzyme targets and an alkyne for the visualization and identification of target proteins using click chemistry to rhodamine-conjugated azides. One compound from the inhibitor library, MJE3, prevented cell proliferation and was used as an ABP to identify its protein target phosphoglycerate mutase B (PGAM1) by MS analysis. These experiments have identified a novel small molecule inhibitor of PGAM1 and implicate PGAM1 in cancer cell proliferation.
Imaging enzyme activity in tumors
One of the major challenges in cancer diagnosis is the early detection of small primary tumors [45]. Since many enzyme activities are upregulated in tumor cells, probes that report on enzymatic activity represent valuable tools for early diagnostic imaging strategies [45–47]. Current methods for imaging enzymes mainly rely on antibody labeling or on substrates that become fluorescent after enzyme cleavage [13,47]. Although antibodies are specific for their enzyme targets, they are not cell permeable and do not give information about enzyme activity. Fluorescent substrates are useful for the activity-based imaging of proteases; however, these compounds often suffer from a lack of specificity, leading to cleavage by multiple classes of proteases [13,47]. Furthermore, there is no way to determine which protease is responsible for substrate processing in vivo using fluorescent substrate reporters. In contrast, ABPs covalently bind to active enzymes, thus permitting assignment of imaging signals to specific enzymes [13,14]. In fact, a number of ABPs that target cysteine proteases have been used to image enzyme activity in tumor cells both in vitro and in vivo [43,48,49].
A fluorescently-tagged DCG-04 analog has been used to image cysteine cathepsin activity during tumorigenesis in RIP1-TAG2 transgenic mice [43]. In this study, the ABP was administered systemically by intravenous injection into a mouse. After allowing the probe to circulate for several hours, pancreatic tumor tissue was collected and imaged using fluorescent microscopy. Cathepsin activity was found to be elevated in tumors and at the invasive edges of islet carcinomas. After imaging, tumor tissues were lysed and analyzed by gel electrophoresis, providing an activity profile that could be used to identify and quantify the levels of probe-modified cathepsins that produced the fluorescent signals. Additionally, this ABP was applied to the imaging of cathepsin activity in a mouse model of cervical carcinogenesis (K14-HPV/E2 mice) [43]. Cathepsin activity levels were also elevated in cervical tumor tissues, further confirming that cysteine cathepsin activity can serve as a useful cancer biomarker and that cathepsin-specific ABPs have potential value as imaging agents to monitor tumor progression in whole animals.
In a recent advance, a cathepsin-specific ABP that becomes fluorescent only upon binding to its enzyme target has been developed [48]. Since tagged ABPs used in imaging are constitutively fluorescent, they generate a high nonspecific fluorescent background when used in living cells. The newly designed quenched ABP (qABP) makes use of the acyloxy leaving group found on the acyloxymethyl ketone warhead. By attaching a fluorescent quencher, the probe is rendered non-fluorescent when free in solution. Covalent modification of cysteine cathepsins by this probe liberates the quencher moiety and the probe becomes fluorescent. This cell-permeable, quenched probe has been used to image cathepsin activity levels in both the murine fibroblast cell line NIH-3T3 and the human MCF-10A breast cancer cell line [48]. Cells treated with the qABP showed distinct punctuate fluorescent staining of lysosomal compartments, whereas the unquenched control probe produced bright, nonspecific intracellular fluorescence that required extensive washing to reveal specific target labeling. Importantly, the fluorescent signals could be specifically blocked by pre-treatment of cells with a general cysteine protease inhibitor.
In a follow-up study, Blum et al. used a related series of quenched and nonquenched ABPs to non-invasively image cathepsin activity in a xenografted mouse model of breast cancer [49]. Near-infrared-labeled versions of the cysteine cathepsin probes produced spatially resolvable fluorescence in the tumor tissues of live mice that correlated with the levels of active cathepsins in those tissues. Both quenched and nonquenched ABPs were able to selectively label tumor tissue and had similar signal-to-background ratios; however, the quenched probe (GB137) achieved its maximum signal-to-background ratio much more rapidly than the nonquenched probe. Ex vivo analysis of tumor tissues from these mice further confirmed that the signals observed in the live animals were due to specific probe labeling of active cathepsins. In addition, the authors demonstrated that non-invasive imaging methods could be used to monitor changes in cysteine cathepsin activity in animals treated with small molecule protease inhibitors. Thus, ABPs represent potentially valuable tools for the evaluation of clinical efficacy of small molecule cancer chemotherapeutics.
Conclusion and future directions
Over the past several years, the field of activity-based proteomics has produced a wealth of new technologies for the direct biological study of enzymes. ABPs that target numerous diverse enzyme classes have been synthesized, and these probes have been applied to many biologically and pathologically relevant fields. Additionally, a number of new tools, including gel-free screening systems and quenched ABPs, have been developed that allow rapid identification and visualization of probe-labeled enzymes in vitro and in vivo. ABPs have also been applied to the identification and evaluation of potential enzyme inhibitors in the physiologically relevant environments of a complex proteome, cell, or even whole animal. However, challenges in the field of activity-based proteomics still remain to be addressed. In order to identify new probe scaffolds that allow for greater proteomic coverage by ABPs, structurally diverse probe libraries need to be developed. Furthermore, advances in gel-free analysis systems will be required to profile proteins with low activities or abundances and to rapidly identify large numbers of proteins targeted by ABPs. Perhaps the most important challenge facing activity-based proteomics is the need to combine the data from ABP-based assays with relevant biological experiments to gain a more complete understanding of enzyme function in cancer and other biological processes and diseases. With time and the rapid advance in technology, we are likely to see a sharp increase in the number and types of applications of ABPs to the study of cancer biology.
Figure 4.
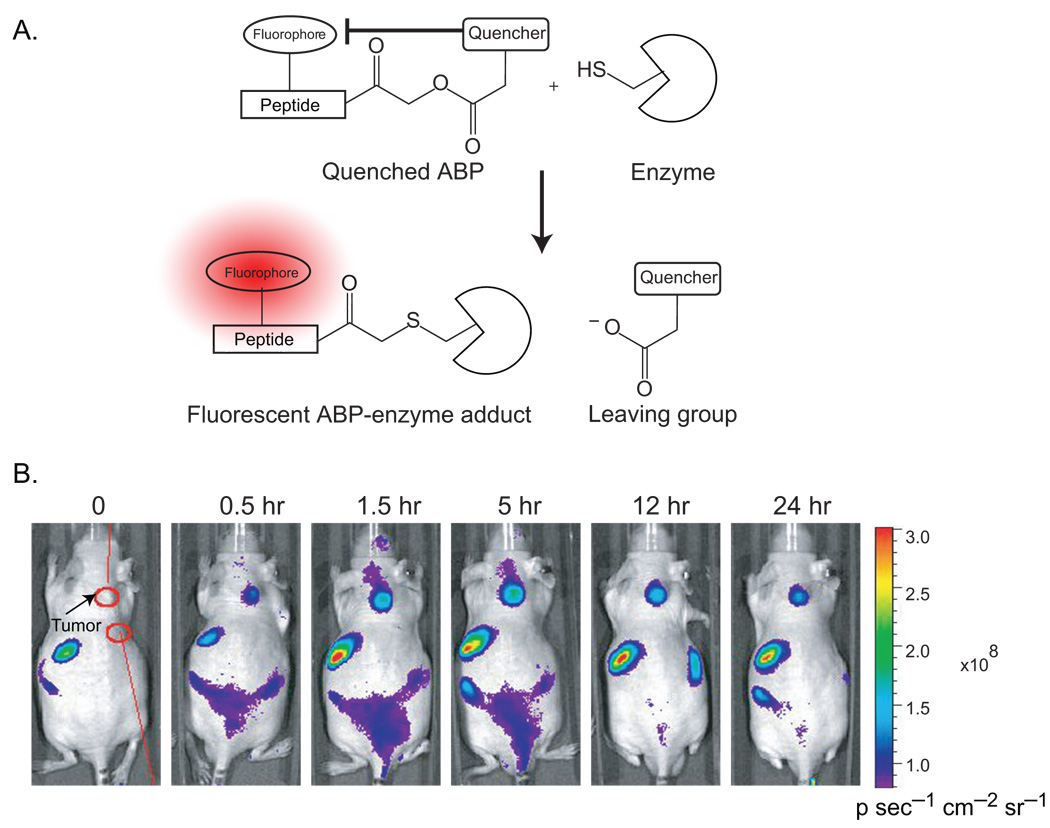
Quenched ABPs for the non-invasive imaging of tumors in vivo. (A) Covalent labeling of a cysteine protease target by a quenched ABP (GB137). Activity-based labeling of the target enzyme results in the loss of the quenching group and subsequent generation of a fluorescently-labeled enzyme. (B) Optical imaging of MDA-MB-231 breast cancer xenograft tumors in nude mice using a quenched cysteine cathepsin-specific ABP. The quenched probe was injected intravenously and fluorescent images of the mice were taken at various time points after injection. Images taken from reference 49.
Acknowledgements
We thank the members of the Bogyo lab for helpful discussions and manuscript comments. This work was supported by funding from a NIH National Technology Center for Networks and Pathways (NTCNP) grant U54 RR020843.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Lee M, Fridman R, Mobashery S. Extracellular proteases as targets for treatment of cancer metastases. Chem Soc Rev. 2004;33:401–409. doi: 10.1039/b209224g. [DOI] [PubMed] [Google Scholar]
- 3.Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 4.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 5.Patten LC, Berger DH. Role of proteases in pancreatic carcinoma. World J Surg. 2005;29:258–263. doi: 10.1007/s00268-004-7816-3. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, Bilgin M, Snyder M. Proteomics. Annu Rev Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- 9.MacBeath G. Protein microarrays and proteomics. Nat Genet. 2002;32 Suppl:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 10.Sadaghiani AM, Verhelst SHL, Bogyo M. Tagging and detection strategies for activity-based proteomics. Current Opinion in Chemical Biology. 2007;11:20–28. doi: 10.1016/j.cbpa.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 11. Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chemical Reviews. 2006;106:3279–3301. doi: 10.1021/cr050288g. **This is a recent comprehensive review of activity-based proteomics that focuses on the design and biological applications of ABPs.
- 12.Berger AB, Vitorino PM, Bogyo M. Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am J Pharmacogenomics. 2004;4:371–381. doi: 10.2165/00129785-200404060-00004. [DOI] [PubMed] [Google Scholar]
- 13.Baruch A, Jeffery DA, Bogyo M. Enzyme activity--it's all about image. Trends Cell Biol. 2004;14:29–35. doi: 10.1016/j.tcb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Fonovic M, Bogyo M. Activity based probes for proteases: applications to biomarker discovery, molecular imaging and drug screening. Curr Pharm Des. 2007;13:253–261. doi: 10.2174/138161207779313623. [DOI] [PubMed] [Google Scholar]
- 15.Phillips CI, Bogyo M. Proteomics meets microbiology: technical advances in the global mapping of protein expression and function. Cell Microbiol. 2005;7:1061–1076. doi: 10.1111/j.1462-5822.2005.00554.x. [DOI] [PubMed] [Google Scholar]
- 16.Schmidinger H, Hermetter A, Birner-Gruenberger R. Activity-based proteomics: enzymatic activity profiling in complex proteomes. Amino Acids. 2006;30:333–350. doi: 10.1007/s00726-006-0305-2. [DOI] [PubMed] [Google Scholar]
- 17. Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. *This is the first reported ABP that targets metalloproteases using a photocrosslinking group. The authors apply their ABP to profile metalloprotease activities in human melanoma cell line proteomes.
- 18.Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc Natl Acad Sci U S A. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. *This work describes a functional proteomics strategy that uses ABPs to generate enzyme activity signatures and then analyzes these signatures using 2D liquid chromatography-mass spectrometry.
- 20.Madsen MA, Deryugina EI, Niessen S, Cravatt BF, Quigley JP. Activity-based protein profiling implicates urokinase activation as a key step in human fibrosarcoma intravasation. J Biol Chem. 2006;281:15997–16005. doi: 10.1074/jbc.M601223200. [DOI] [PubMed] [Google Scholar]
- 21.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nature Reviews Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 22.Sieber SA, Niessen S, Hoover HS, Cravatt BF. Proteomic profiling of metalloprotease activities with cocktails of active-site probes. Nat Chem Biol. 2006;2:274–281. doi: 10.1038/nchembio781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 24.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolen U, Kobzeva V, Gasparjan N, Ovaa H, Winberg G, Kisseljov F, Masucci MG. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45:260–269. doi: 10.1002/mc.20177. [DOI] [PubMed] [Google Scholar]
- 26.Ovaa H. Active-site directed probes to report enzymatic action in the ubiquitin proteasome system. Nat Rev Cancer. 2007;7:613–620. doi: 10.1038/nrc2128. [DOI] [PubMed] [Google Scholar]
- 27.Ovaa H, Kessler BM, Rolen U, Galardy PJ, Ploegh HL, Masucci MG. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc Natl Acad Sci U S A. 2004;101:2253–2258. doi: 10.1073/pnas.0308411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vasiljeva O, Papazoglou A, Kruger A, Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt K, Bogyo M, et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. *In this study, cathepsin-specific ABPs were used to detect cathepsin activities in PyMT mice deficient in cathepsin B. Tumor cells from these mice had an upregulation of cathepsin X on their cell surface, suggesting that proteases can dynamically compensate for each other.
- 29.Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, 3rd, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci U S A. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adam GC, Sorensen EJ, Cravatt BF. Proteomic profiling of mechanistically distinct enzyme classes using a common chemotype. Nat Biotechnol. 2002;20:805–809. doi: 10.1038/nbt714. [DOI] [PubMed] [Google Scholar]
- 31.Adam GC, Sorensen EJ, Cravatt BF. Trifunctional chemical probes for the consolidated detection and identification of enzyme activities from complex proteomes. Mol Cell Proteomics. 2002;1:828–835. doi: 10.1074/mcp.t200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 32.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 33. Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, Bogyo M. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. *One of the first applications of ABPs to enzyme inhibitor discovery. The authors identified a selective inhibitor of cathepsin B which can be used as a lead target for cancer therapeutics.
- 34.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 35. Leung D, Hardouin C, Boger DL, Cravatt BF. Discovering potent and selective reversible inhibitors of enzymes in complex proteomes. Nat Biotechnol. 2003;21:687–691. doi: 10.1038/nbt826. *This paper reports an ABP-based inhibitor screen that is able to identify reversible enzyme inhibitors. Using this screen, the authors discovered a novel reversible inhibitor of KIAA1363, an enzyme with highly elevated activity in invasive cancer cells.
- 36.Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23:509–521. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Berkers CR, Verdoes M, Lichtman E, Fiebiger E, Kessler BM, Anderson KC, Ploegh HL, Ovaa H, Galardy PJ. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nat Methods. 2005;2:357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 38.Altun M, Galardy PJ, Shringarpure R, Hideshima T, LeBlanc R, Anderson KC, Ploegh HL, Kessler BM. Effects of PS-341 on the activity and composition of proteasomes in multiple myeloma cells. Cancer Res. 2005;65:7896–7901. doi: 10.1158/0008-5472.CAN-05-0506. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Shreder KR, Gai W, Corral S, Ferris DK, Rosenblum JS. Wortmannin, a widely used phosphoinositide 3-kinase inhibitor, also potently inhibits mammalian polo-like kinase. Chem Biol. 2005;12:99–107. doi: 10.1016/j.chembiol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 40. Kraus M, Ruckrich T, Reich M, Gogel J, Beck A, Kammer W, Berkers CR, Burg D, Overkleeft H, Ovaa H, et al. Activity patterns of proteasome subunits reflect bortezomib sensitivity of hematologic malignancies and are variable in primary human leukemia cells. Leukemia. 2007;21:84–92. doi: 10.1038/sj.leu.2404414. *The authors of this study used a proteasome-directed ABP to identify the active human proteasomal subunits targeted by bortezomib in patients receiving this drug. This is one of the first studies using ABPs to study enzyme activities of human patients receiving cancer treatment.
- 41.Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, van den Nieuwendijk AM, Hofmann T, Berkers CR, van Leeuwen FW, et al. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem Biol. 2006;13:1217–1226. doi: 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 42. Sadaghiani AM, Verhelst SH, Gocheva V, Hill K, Majerova E, Stinson S, Joyce JA, Bogyo M. Design, synthesis, and evaluation of in vivo potency and selectivity of epoxysuccinyl-based inhibitors of papain-family cysteine proteases. Chem Biol. 2007;14:499–511. doi: 10.1016/j.chembiol.2007.03.010. *In this paper, a set of cathepsin-specific inhibitors were identified using an ABP-based in vivo screen.
- 43. Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. *This work was the first example of the use of fluorescent ABPs for direct imaging of cathepsin activity in vivo.
- 44.Evans MJ, Saghatelian A, Sorensen EJ, Cravatt BF. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat Biotechnol. 2005;23:1303–1307. doi: 10.1038/nbt1149. [DOI] [PubMed] [Google Scholar]
- 45.Weissleder R, Tung CH, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 46.Mahmood U, Weissleder R. Near-infrared optical imaging of proteases in cancer. Mol Cancer Ther. 2003;2:489–496. [PubMed] [Google Scholar]
- 47.Sloane BF, Sameni M, Podgorski I, Cavallo-Medved D, Moin K. Functional imaging of tumor proteolysis. Annu Rev Pharmacol Toxicol. 2006;46:301–315. doi: 10.1146/annurev.pharmtox.45.120403.095853. [DOI] [PubMed] [Google Scholar]
- 48. Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1:203–209. doi: 10.1038/nchembio728. *The first example of a fluorescently-quenched ABP that can be used for real-time imaging of cathepsin protease activity in live cells.
- 49. Blum G, von Degenfeld G, Merchant MJ, Blau HM, Bogyo M. Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes. Nat Chem Biol. 2007;3:668–677. doi: 10.1038/nchembio.2007.26. *This study reports the first use of a fluorescently-quenched ABP for in vivo tumor imaging. Mice treated with a cathepsin-specific, fluorescently-quenched ABP showed tumor-specific labeling with very low background fluorescence.


