Abstract
We have investigated the site-specific folding kinetics of a photoswitchable cross-linked α-helical peptide by using single 13C = 18O isotope labeling together with time-resolved IR spectroscopy. We observe that the folding times differ from site to site by a factor of eight at low temperatures (6°C), whereas at high temperatures (45°C), the spread is considerably smaller. The trivial sum of the site signals coincides with the overall folding signal of the unlabeled peptide, and different sites fold in a noncooperative manner. Moreover, one of the sites exhibits a decrease of hydrogen bonding upon folding, implying that the unfolded state at low temperature is not unstructured. Molecular dynamics simulations at low temperature reveal a stretched-exponential behavior which originates from parallel folding routes that start from a kinetically partitioned unfolded ensemble. Different metastable structures (i.e., traps) in the unfolded ensemble have a different ratio of loop and helical content. Control simulations of the peptide at high temperature, as well as without the cross-linker at low temperature, show faster and simpler (i.e., single-exponential) folding kinetics. The experimental and simulation results together provide strong evidence that the rate-limiting step in formation of a structurally constrained α-helix is the escape from heterogeneous traps rather than the nucleation rate. This conclusion has important implications for an α-helical segment within a protein, rather than an isolated α-helix, because the cross-linker is a structural constraint similar to those present during the folding of a globular protein.
Keywords: cooperativity, infrared spectroscopy, molecular dynamics simulation, peptide folding
In many biomolecular systems, large changes can take place in response to a relatively small perturbation in the environment such as a variation in temperature, denaturant concentration, or the partial pressure of certain gases. Such an “all or nothing” phenomenon is termed a cooperative process. The classical example of cooperativity is the binding affinity of oxygen molecules to the four hemes of hemoglobin (1), which is a factor 100 to 1,000 times larger for the fourth oxygen molecule compared with the first. This leads to a sigmoidal dependence of oxygen binding on oxygen partial pressure with a sharp transition in a relatively small range of the latter. The folding of α-helices, which constitute one of the predominant secondary structures in many proteins, is often described in a similar manner (2): Once an entropically expensive nucleation process has occurred, i.e., a first helical turn with a hydrogen bond is formed, the zipping of additional hydrogen bonds is more likely because it is enthalpically favorable. A thermodynamic (statistical) treatment of the process leads to so-called nucleation-propagation-models (or zipper models), initially introduced by Zimm and Bragg (3) and Lifson and Roig (4). A large number of thermodynamic studies on α-helical peptides has been treated extremely successfully in terms of these models (5–8).
Cooperativity implies that the free energies of the two states of a system are balanced, however, in a way that enthalpic (ΔH) and entropic (−T ΔS) contributions are large and compete against each other, leading to a characteristic sigmoidal transition as a function of the external control parameter. Because, in general, enthalpy and entropy vary in a nonsynchronous way as a function of some order parameter, the resulting free-energy surface ΔG ≡ ΔH − TΔS will be uneven and in most cases will have a pronounced barrier. This is why this definition of cooperativity, which is based on thermodynamic arguments, often also has consequences for the kinetics of the transition between the two states. For example, in the classic case of oxygen binding to hemoglobin, a two-state allosteric model (i.e., the MWC model) can also explain the binding rate that increases with the number of already bound oxygen molecules (1). Cooperativity in protein folding reflects a two-state conformational distribution; its investigation requires a rigorous analysis of the folding transition (9). In the case of the helix–coil transition, high cooperativity would imply that once the rate-limiting nucleation step has occurred somewhere in the sequence, all subsequent helical turns would form at essentially the same time. Then, one common rate would be expected for all sites, corresponding to the nucleation rate (the propagation rate would not be detectable because it is very fast). Indeed, temperature-jump experiments on helix folding have successfully been described by “kinetic-zipper” models (10, 11), in which thermodynamic states of a nucleation-propagation model are linked by rate constants. However, because the cooperativity of isolated α-helices is weak, in particular when they are short, they do not fold in a two-state fashion, but rather with biexponential kinetics resulting from the coupling between nucleation and the only slightly faster diffusive elongation (10, 12).
It has recently been argued that even proteins that appear to be two-state folders can in fact be much more complex when reporting the folding free-energy surface on the level of individual protons by using NMR chemical-shift spectroscopy (13). IR spectroscopy together with site-selective isotope labeling offers the time resolution to perform site-selective folding studies also in a kinetic sense, even for the much faster folding of secondary structure motifs. The amide I′ (C = O stretch) vibrational mode is very sensitive to hydrogen bonding and dipole–dipole coupling among different peptide units (14), and isotope labeling of the carbonyl groups allows one to spectrally single out individual amino acids (12, 15). Gai and coworkers (12) demonstrated by 13C = 16O labeling of groups of four subsequent alanines that the relaxation rates vary by ≈10% throughout the sequence. This result points to a more complex behavior of α-helix folding. Nevertheless, such a small degree of heterogeneity could be still modeled in the framework of nucleation-propagation models, taking into account position-dependent parameters for the different amino acids in the heteropolymer (16). However, as we will show, labeling of groups of several subsequent amino acids might still be too coarse-grained. If the kinetics in various sections of the peptide differ, one might expect variations even on the single amino acid level. Indeed, by using 13C = 18O double labeling of a single amino acid in a small helix bundle protein, Dyer and coworkers (17) obtained considerably different melting curves and considerably different complex folding kinetics for that particular site, compared with the averaged signal.
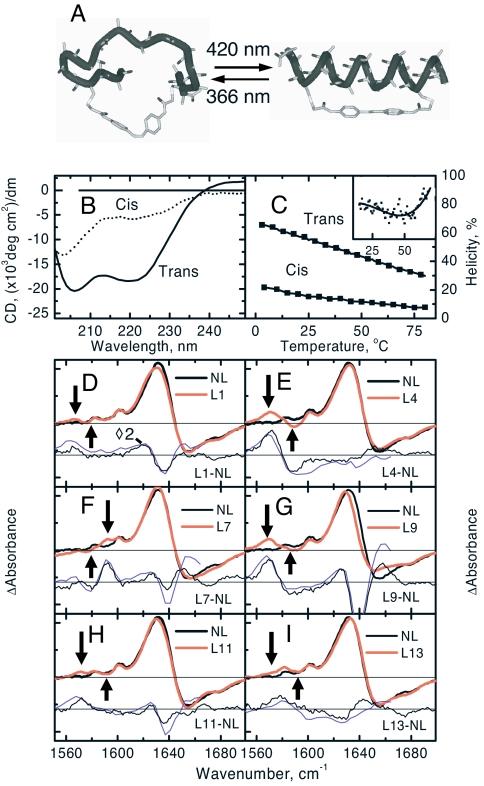
As an alternative approach to study α-helix folding, we recently started to employ an azobenzene moiety as a photoswitchable structural constraint (see Fig. 1A) (18–20). Two cysteines are cross-linked in such a way that the azo-moiety in the trans (cis) conformation stabilizes (destabilizes) the helix, as deduced from CD spectroscopy (Fig. 1 B and C). This allows one to monitor both the folding and the unfolding direction of one and the same molecule at identical temperatures. We observed stretched exponential kinetics in both directions (20), in disagreement with the prediction from nucleation-propagation models that would reveal compressed exponential kinetics in the folding direction (21) (in fact, compressed exponential response has already been obtained in figure 10 of ref. 10, however, remained undiscussed). We therefore speculated in our previous work that the rate-limiting step in folding of our model systems is the escape from misfolded traps, which are consistently obtained in full-atom molecular dynamic (MD) simulations of small peptides (22–29).
Fig. 1.
Photoswitchable peptide and its steady-state spectra at room temperature. (A) Schematic drawing of the photoswitchable peptide in its cis (Left) and trans (Right) conformations. (B) CD-spectra of the peptide in its complete trans and cis states at room temperature. (C) The helicities in the cis and trans conformations at various temperatures. The trans spectrum was obtained in darkness, and the cis-spectrum was obtained under 365-nm illumination together with an estimate that 75% of the molecules are in the cis conformation (based on the UV-vis absorbance difference). (Inset) The first derivative of the trans CD data reveals the inflection point. (D–I) FTIR-difference spectra between the trans (under 436-nm illumination) and cis state (under 365-nm illumination) of each peptide at room temperature. Shown are the difference between not-labeled (NL) and labeled (LX, labeled at residue X) samples taken from the FTIR-spectra (thin black line) and from the late delay-time pump-probe spectra 30 μs after photoswitching (thin blue line). The arrows point to the frequency position from which the site-specific kinetics has been extracted.
Here, we present a comprehensive set of kinetic data of the folding of a photoswitchable α-helix (Ac-AACAK5AAAAK10AAACK15A-NH2) on the single amino acid level by employing 13C = 18O labeling of an alanine-rich peptide, revealing strong variations in the folding kinetics and their temperature dependence along the sequence. The interpretation of the experimental data are corroborated by multiple implicit solvent (30) MD simulations (31) of folding of a similar cross-linked peptide (i.e., Ac-AACAR5AAAAR10AAACR15A-NH2) at low and high temperatures, and their network analysis (22, 29). Nonequilibrium MD simulations of an eight-residue cyclized peptide immersed in dimethyl sulfoxide have been published recently (32, 33) but those studies focused on the contributions to the frequency shift and did not mention pathways and kinetics of (helical) folding. The atomistic detail of the MD simulations and the large sampling of folding events (100 MD runs for a total of 0.4 ms) allow us to directly extract the conformational distribution from the trajectories. The comparison of experimental data and simulation results shows qualitatively similar folding kinetics and temperature dependence. Our analysis provides strong evidence that (i) the azo-cross-linker stabilizes kinetic traps originating from nonnative contacts that are nonexistant in isolated helices and that (ii) the azo-cross-linker finally destroys the already weak cooperativity of isolated α-helices. The analysis of the photoswitching simulations provides insights, at the atomic level of detail, on the folding mechanism and thereby explains the kinetic traces measured experimentally.
Results and Discussion
Noncooperative Helix Folding.
Fig. 1C shows melting curves deduced from the fractional helicity at 222 nm [assuming [Θ]222 = −32,000 deg cm2/dm for the ellipticity of a fully folded helix (34)]. Within the limited temperature range, the sigmoidal dependence of the folding transition is hardly visible (see Fig. 1C Inset). Nevertheless, the helicity in the trans conformation varies slightly more than the relative temperature change (ΔT/T = 24%), hence we conclude that some small degree of cooperativity still remains, in the sense that folding enthalpy and folding entropy compete against each other. A comparison with melting curves from similar but nonlinked peptides (34, 35), which are steeper, indicates that the linker reduces the conformational flexibility of the unfolded ensemble and, hence, diminishes the entropic penalty of folding. It should be noted that the folding rates we observe for the azo-linked peptides (19, 20) are similar to those found in temperature-jump experiments of comparable unlinked peptides (10, 12, 36, 37) (a one-to-one comparison is not possible because the sequences used in temperature-jump experiments are typically a bit longer) and that the amino acid sequence determines the folding rate to a significant extent. That is, at room temperature, the folding rate is 1,200 ns for Ac-EACAR5EAAAR10EAACR15Q-NH2 (19), 700 ns for Ac-AACAR5AAAAR10AAACR15A-NH2 (20), 600 ns for Ac-AACAK5AAAAK10AAACK15A-NH2 (this study), and 2,200 ns for Ac-EMCAR5EMAAR10EMACR15Q-NH2 (data not shown). This variability indicates that interactions among the amino acid side chains and with the cross-linker may act as traps that are, indeed, rate determining.
Stationary FTIR-difference spectra between the two conformations of the photoswitchable helix are shown in Fig. 1 D–I. Upon folding, the unlabeled amide I′ band red-shifts with a positive signal ≈1633 cm−1 and a negative signal with additional substructure ≈1655 cm−1 and 1680 cm−1, reporting on an overall strengthening of hydrogen bonding (14). Additional small bands are observed at lower frequencies that originate from both the ring modes of the azo-moiety (1,602 cm−1 and 1,580 cm−1) (20) and from the isotope labels. By subtracting the FTIR-difference spectrum of the nonlabeled compound (termed NL throughout the text) from that of isotope-labeled compounds (termed LX, where X is the labeling position counted from the N terminus), the contributions of the latter can be isolated. As expected, they are downshifted by ≈65 cm−1 from the main band, exhibiting a dispersive shape with a sharp and distinct positive contribution in either case, whereas the negative contribution is, in general, broader and less clearly identified (see arrows in Fig. 1 D–I). The one notable exception is L7, which shifts to higher frequencies upon folding of the α-helix (Fig. 1F), opposite to all other residues. Because the frequency of the 13C = 18O vibration is related to the strength of hydrogen bonding, the only conclusion can be that hydrogen bonding of this particular site is stronger in the “unfolded” ensemble.
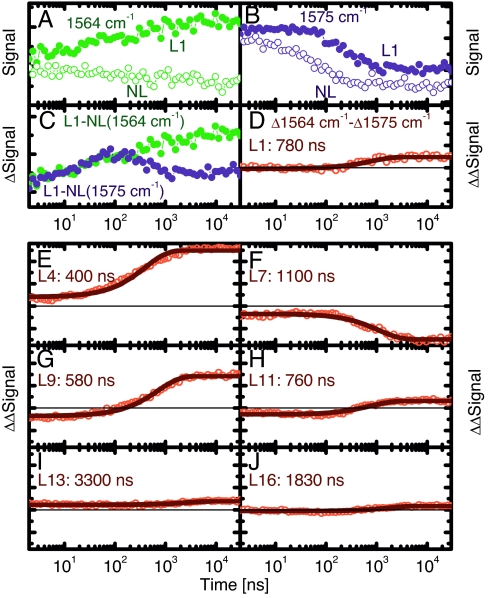
In the time-resolved experiments, helix folding is initiated by a subpicosecond laser pulse isomerizing the azo-photoswitch, and the formation of individual hydrogen bonds is detected in the amide I′ region as a function of time (19, 20). To single out the contribution from the isotope label, difference spectra between nonlabeled and labeled samples had to be taken. Furthermore, to suppress temperature-induced baseline effects, two kinetics were collected for each sample (Fig. 2 A and B; NL, open circles; L1, filled circles) at the frequency positions with the biggest deviations between nonlabeled and labeled sample (left and right from the 13C = 18O vibration, as indicated by arrows in Fig. 1). The result of subtracting the normalized signals from NL and L1 is shown in Fig. 2C (normalization was performed according to concentration obtained from the UV-vis spectrum and excitation power), and the difference between the traces at the two frequencies is shown in Fig. 2D. This “difference of difference signal” constitutes the site-selective folding trace and can be fitted (within signal-to-noise) with a single-exponential function shown as a solid line in Fig. 2D for L1. We note also that the difference–difference signals of the late time transient experiment agree with that from the FTIR-experiment (Fig. 1), validating the correctness of the background subtraction procedure. The site-selective folding traces labeled at all other positions are shown in Fig. 2 E–J. The immediate observation is that the rates scatter quite substantially without clear correlation between neighboring sites (Fig. 3A), thus, each peptide unit behaves independently, or noncooperatively. As a general trend, the signal is larger in the middle of the peptide, and sites 4, 7, and 9 reveal the most dominant signals (however, note again that L7 exhibits kinetics in the opposite direction; Fig. 2F). In line with other studies (e.g., ref. 38), a so-called “end-fraying effect” is observed as evidenced by smaller signals at the C and N termini. As a matter of fact, one would not expect any response from L13 and L16 at all, because they cannot form α-helical hydrogen bonds even in the folded conformation, and indeed, the signals are extremely small. Interestingly, the rates are correlated with the amplitudes of the corresponding signals (Fig. 3A); thus, the larger the driving force, the faster hydrogen bond formation.
Fig. 2.
Site-specific folding signals at 19°C. (A and B) Kinetic traces of the L1 and NL at 1,564 cm−1 (A) and at 1575 cm−1 (B). (C) The difference of the L1 and NL signals at these wavelengths. (D) The difference between the resulting signals together with its single exponential fit (solid line). (E–J) Site-selective folding signals of the other sites. The solid lines are single-exponential fits of the curves with the resulting time constant indicated.
Fig. 3.
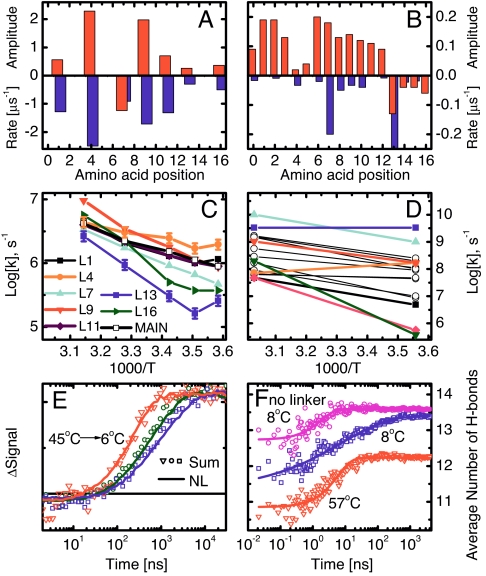
Summary of the experimental (A, C, and E) and MD (B, D, and F) results. (A and B) Amplitudes (upward, red) and corresponding rates (downward, blue) at 19°C (A) and 8°C (B). Note that site 7 in the experiment, and sites 13–16 in the MD simulations, have inverted amplitudes. (C and D) Site-selective folding rates as a function of inverse temperature. In D, black circles are used for residues not measured experimentally. (E) Sum of all site signals (symbols) and amide I′ signal of the nonlabeled peptide (solid lines) at 6°C, 19°C, and 45°C. (F) Average number of hydrogen bonds along the MD simulations: 100 MD runs at 8°C with stretched exponential fit h(t) = 13.6 − 1.6exp(−t/29 ns)0.39 (blue), 50 MD runs at 57°C with single-exponential fit h(t) = 12.2 − 1.4exp(−t/6 ns) (red), and 50 MD runs of peptide without cross-linker at 8°C with single-exponential fit h(t) = 13.6 − 0.9exp(−t/2 ns) (magenta). The standard deviations are almost all <0.3 units. The experimental data in E are normalized, and the background has been removed, whereas the corresponding MD data in F are on their original scale.
Temperature Dependence.
It is useful to plot the rates of folding of individual sites (i.e., the rates of helical hydrogen bond formation of individual backbone carbonyl groups) against reciprocal temperature, as measured by the isotopically labeled peptides (Fig. 3C). The individual sites exhibit considerably different temperature dependencies. The spread of rates is clearly bigger at low temperatures (a factor of approximately eight at 6°C), whereas they approach each other at higher temperatures (a factor of two at 45°C). Interestingly, summing up all site-signals with their relative intensities, the kinetics of the unlabeled band can be reproduced remarkably well (Fig. 3E). This result shows that our set of single-site labeled peptides represents the overall folding kinetics very well and, more importantly, that the averaged kinetics is just a trivial sum of the individual contributions. The average folding time of the NL peptide increases from approximately τ = 240 ns to τ = 1,290 ns when lowering the temperature from 45°C to 6°C and becomes more stretched with a stretching factor that decreases from β = 1.00 to β = 0.71 (fitting it with a function ∝ exp[−(t/τ)β]) (19, 20). The larger spread of rates reveals stretched exponential kinetics at low temperatures, whereas the more uniform values at higher temperatures result in a close to single-exponential response.
Qualitatively speaking, the MD simulations reveal very similar results. Just as in the experiment, the folding rates and amplitudes vary strongly from site to site (Fig. 3B), with a fraying effect toward the ends and a dip in the folding amplitudes in the middle of the helix (albeit not exactly at the same position). Also the temperature dependence observed in the MD simulations is in qualitative agreement with the experimental data (Fig. 3D), despite the larger spread of individual-residue rates, which is likely to originate mainly from the low friction coefficient [see supporting information (SI) Text]. Quantitative agreement is not expected because of the approximations inherent to the force field and implicit solvation model as well as the slightly different amino acid sequence. As an example, the fastest rate observed in MD (L13, blue square symbol in Fig. 3D) is the slowest in the experiment (Fig. 3C), which is in part a consequence of the very small amplitude. Nevertheless, essentially the same overall folding kinetics and temperature dependence emerge from the analysis of the MD simulations in terms of total number of hydrogen bonds involving carbonyl groups (Fig. 3F), which is the MD observable closest to the experimental signal. The folding signal from the MD runs at low temperature shows complex kinetics, clearly deviating from single-exponential behavior.
There are indications for deviations from single-exponential response even on the single-site level in both the experimental (Fig. 2) and the MD data (Fig. S3 and Tables S2 and S3), however, because of limited signal-to-noise, we do not discuss them in detail. Nevertheless, the largest contribution to the nonexponentiality of the overall signal originates from the spread of rates rather than from the nonexponentiality of the individual signals. Moreover, just as in the experiment, faster folding and single-exponential behavior are observed in the MD runs at high temperature (Fig. 3F, red). In particular, the ratio of folding times at low vs. high temperature is close: 29:6 in the simulations of the cross-linked peptide and 1,290:240 as measured experimentally. Finally, we simulated folding of the peptide without the cross-linker (Fig. 3F, magenta, an “experiment” that can only be performed on the computer) and obtain single-exponential response even at low temperature.
Origin of Complex Kinetics Explained by Network Analysis of MD Trajectories.
In the example of oxygen binding to hemoglobin, cooperativity implies that the first step is the rate-limiting step (1). If the same were true for the folding of our photoswitchable α-helix, one common rate would be expected for all sites corresponding to the rate-limiting nucleation step. Such a common rate is not observed; instead, each amino acid site responds individually, reporting on various escape rates from different partially folded or trap states. The averaged signal obtained from the main band is then a nonspecific sum of these rates, and hence, the folding of the cross-linked α-helix is definitely not cooperative in the kinetic sense, although, from thermodynamic considerations (Fig. 1C), one might still deduce some degree of cooperativity. The complexity of the folding process is masked in an averaged signal, as suggested by previous atomistic simulation studies (22, 23), which have demonstrated that projecting the free energy on a single progress variable based on geometry, e.g., number of native contacts or rmsd from native, is not consistent with the complexity of the actual free-energy surface (25). Note that these projections (histogram-based free-energy profiles in Fig. S4) do not reveal any rate-limiting barrier for the photoswitchable α-helix.
The agreement of the MD results with the experiment justifies drawing detailed conclusions from the former. To that end, the folded (i.e., fully α-helical) state, and the most populated metastable states, can be isolated by grouping conformations according to fast relaxation along the MD trajectories, a procedure called kinetic grouping analysis (29). The advantage of this procedure with respect to a simple projection onto one or two progress variables is that structures (i.e., coordinate sets) are grouped into free-energy minima, not according to geometric characteristics, but rather according to the dynamics. The network analysis shows that escape from traps with a mixture of loop and helical content is rate limiting and that there are multiple parallel folding channels originating from a kinetically partitioned unfolded state (Fig. 4). The kinetic grouping analysis, using secondary structure strings (39), reveals that, of the four main folding channels, one starts from within the helical basin, whereas the remaining three start from the following metastable states: two C-terminal α-helical turns formed (Fig. 4B), only N-terminal α-helical turn formed (Fig. 4C), and only one C-terminal α-helical turn formed (Fig. 4D). The unfolded state is kinetically partitioned so that the traps are connected to the one folded state in a star-like manner, and the folded basin acts as a hub (22) for the interchange between misfolded or partially folded states. An example is shown in Fig. 4D where, starting from the trap with only the C-terminal turn formed, the peptide first reaches the fully α-helical state and then unfolds to a structure with broken N-terminal turn (as seen from the decreasing brightness of the yellow coloring of the nodes along this pathway). Importantly, different channels have barriers of different heights (Table S1), with rates ranging from ≈1/(10 ns) for folding from the helical basin (green in Fig. 4A) to ≈1/(1,000 ns) for folding from the structure with only N-terminal turn formed (cyan in Fig. 4A). Therefore, the spread observed in the site-specific rates originates from the heterogeneous degree of formation of individual helical hydrogen bonds in different traps. The interpretation of the experimental data, thanks to the folding mechanism and pathways extracted from the MD simulations, provides strong evidence that the rate-limiting step of helix folding (at low temperature) is exactly the escape rate from a few metastable states (some of them stabilized by nonnative contacts) rather than the commonly assumed nucleation step.
Fig. 4.
The network analysis (22, 29) of the 100 MD runs at 281 K shows parallel folding channels. Each node (i.e., conformation) of the network represents a secondary structure string (39), and a link is a direct transition (within 20 ps) observed in the MD runs. The surface of each node is proportional to its statistical weight, and only the 1,387 nodes with at least 200 snapshots (≈96.7% of total sampling) are shown to avoid overcrowding. (A) The free-energy basins, i.e., native (green nodes) and metastable states [identified by kinetic grouping analysis (29) using a commitment time of 10 ns to group conformations that interconvert rapidly], are shown with different colors, and their characteristics are listed in Table S1. Within each basin, nodes and intrabasin links are shown with the same color, and interbasin links are colored in gray. White diamonds indicate the starting points of 82 of the 100 folding runs, whereas the remaining 18 runs reached directly the most populated node (i.e., fully formed α-helix, large green circle) and are not shown. An enlarged version of the network is shown in Fig. S2. (B–D) Nodes are colored according to the values of the mean first passage time (29) from the most populated node (white squares) of individual metastable states to all other nodes. The time scale goes from 0 (yellow) to >2 μs (blue). The coloring shows that the unfolded state is kinetically partitioned, and the folded (i.e., fully α-helical) state acts as a hub. Visits to unfolded metastable states different from the starting one require a much longer time than reaching the folded state. Representative structures of the folded state and each metastable state are shown by flexible tubes of variable diameter reflecting conformational disorder, with α-helical turns in green, loop segments in gray, N terminus in blue, and cysteine side chains in yellow for emphasizing the position of the linker.
Conclusion
The kinetics and mechanism of folding of a photoswitchable cross-linked α-helix have been investigated by a combined experimental–simulation study. At low temperatures, the hydrogen bond formation rates of different sites scatter by almost one order of magnitude, whereas they approach each other at higher temperatures. The spread of rates is significantly larger than the 10% effect observed for an isolated helix (12) and appears to be too large to be consistent with conventional nucleation-propagation models along the lines of ref. 16. Furthermore, with group L7, we directly observe a nonnative contact in the misfolded ensemble, the existence of which, however, is neglected in nucleation-propagation models. On the other hand, good agreement with an all-atom MD simulation is obtained, which justifies drawing interpretations at atomic resolution from the MD results. Notably, the MD analysis unmasks discrete traps (i.e., nonnative free-energy basins) along parallel folding pathways, which render the overall kinetics nonexponential. The cross-linker actually stabilizes these traps, as observed from the difference in response in MD runs with and without cross-linker (Fig. 3F). However, in contrast to previous studies, where noncooperative folding has been interpreted as barrierless folding (40, 41), we argue here that a completely different scenario, i.e., a few traps in the unfolded state that are separated from the native basin by barriers of different heights, may lead to a similar noncooperative behavior. This scenario (with barriers of different heights) is different from a barrierless landscape, but they share a higher population of conformations with intermediate compactness with respect to the two-state behavior (42).
Isolated α-helices fold in a marginally cooperative manner, as seen by the somewhat steeper melting curve (34, 35) than in Fig. 1C and the only small variation of folding rates along the peptide chain (12). If α-helix folding were cooperative, one could think of it as binary (all or nothing) when studying the folding of tertiary structures in larger proteins. However, addition of an azo-cross-linker as a switchable structural constraint finally destroys the already weak cooperativity of isolated α-helices. Structural constraints of this sort might also exist for a helical segment in a larger protein by the very fact that the helix is connected through the backbone to the rest of the polypeptide chain, and its flexibility is restrained by tertiary contacts with other pieces of the protein. As such, the folding of secondary and tertiary structures cannot be thought of as decoupled. Paradoxically, the folding of the cross-linked α-helix might be closer to the natural situation inside a globular protein than that of an isolated helix.
Materials and Methods
Experimental.
Synthesis of the molecule, Ac-AACAK5AAAAK10AAACK15A-NH2, cross linked by a azo-moiety acting as a photoswitch and with 13C = 18O-labeled alanine at eight different positions, was performed as described (18). IR pump-probe spectroscopy with delays ranging from 10 ps to 40 μs was performed by using two electronically synchronized Ti:S laser systems, one of which was frequency doubled to generate pump pulses at 420 nm and the second pumped an IR-OPA to obtain broadband IR probe pulses (19, 20).
Computational.
MD simulations were performed by using the CHARMM program package (31) using standard procedures and an implicit solvent (30). The force field parameters for the azo-moiety were derived from the PARAM19 for the amide backbone and phenyl ring of Phe as well as from ref. 43 for the dihedral angles of the central N = N bond (Fig. S1). An equilibrium ensemble with the cross-linker in the cis conformation was sampled by two runs of replica exchange MD (44). After instantaneously switching the torsional potential of the central N = N bond to one that strongly favors the trans-configuration, ensembles of nonequilibrium Langevin dynamics runs of 4 μs each were started from the cis equilibrium ensemble at both 330 K and 281 K. Network analysis (22, 29) of the resulting nonequilibrium trajectories was supported by the program WORDOM (45). Because of the implicit solvent model, both the absolute temperatures and rates are somewhat arbitrary. In the comparison with the experiments, we therefore focus on trends, rather than the absolute values.
For a more detailed account of materials and methods, see SI Text.
Supplementary Material
Acknowledgments.
We thank Riccardo Pellarin for suggesting the control runs without cross-linker, Gianluca Interlandi for help in the initial set-up of the MD simulations, Rolf Pfister for the synthesis of the molecules, Francesco Rao for interesting discussions, Ben Schuler for instructive discussions and for the access to the HPLC-equipment, Bernhard Spingler and Philipp Antoni for the access to the CD equipment, and Jan Helbing and Andrea Prunotto for technical assistance. The MD simulations were run on the Matterhorn cluster of the University of Zürich. This work was supported by Swiss National Science Foundation Grants 200020-107492/1 (to P.H.) and 205320-118214 (to A.C.) and by a fellowship of the “Forschungskredit” of the University of Zürich (to E.H.G.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712099105/DCSupplemental.
References
- 1.Eaton WA, Henry ER, Hofrichter J, Mozzarelli A. Is cooperative oxygen binding by hemoglobin really understood? Nat Struct Biol. 1999;6:351–358. doi: 10.1038/7586. [DOI] [PubMed] [Google Scholar]
- 2.Creighton TE. Proteins. New York: Freeman; 1993. [Google Scholar]
- 3.Zimm BH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526–535. [Google Scholar]
- 4.Lifson S, Roig A. On the theory of helix–coil transition in polypeptides. J Chem Phys. 1961;34:1963–1974. [Google Scholar]
- 5.Scholtz JM, Qian H, York EJ, Stewart JM, Baldwin RL. Parameters of helix–coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 1991;31:1463–1470. doi: 10.1002/bip.360311304. [DOI] [PubMed] [Google Scholar]
- 6.Muñoz V, Serrano L. Helix design, prediction and stability. Curr Opin Biotechnol. 1995;6:382–386. doi: 10.1016/0958-1669(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 7.Doig AJ. Recent advances in helix–coil theory. Biophys Chem. 2002;101:281–293. doi: 10.1016/s0301-4622(02)00170-9. [DOI] [PubMed] [Google Scholar]
- 8.Scheraga HA, Vila JA, Ripoll DR. Helix–coil transitions re-visited. Biophys Chem. 2002;101–102:255–265. doi: 10.1016/s0301-4622(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 9.Kaya H, Chan HS. Polymer principles of protein calorimetric two-state cooperativity. Proteins. 2000;40:637–661. doi: 10.1002/1097-0134(20000901)40:4<637::aid-prot80>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 10.Thompson PA, Eaton WA, Hofrichter J. Laser temperature jump study of the helix–coil kinetics of an alanine peptide interpreted with a “kinetic zipper” model. Biochemistry. 1997;36:9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 11.Thompson PA, et al. The helix–coil kinetics of a heteropeptide. J Phys Chem B. 2000;104:378–389. [Google Scholar]
- 12.Huang C-Y, et al. Helix formation via conformation diffusion search. Proc Natl Acad Sci USA. 2002;99:2788–2793. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadqi M, Fushman D, Muñoz V. Atom-by-atom analysis of global downhill protein folding. Nature. 2006;442:317–321. doi: 10.1038/nature04859. [DOI] [PubMed] [Google Scholar]
- 14.Krimm S, Bandekar J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv Protein Chem. 1986;38:181–364. doi: 10.1016/s0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- 15.Silva RAGD, Kubelka J, Bour P, Decatur SM, Keiderling T. A site-specific conformational determination in thermal unfolding studies of helical peptides using vibrational circular dichroism with isotopic substitution. Proc Natl Acad Sci USA. 2002;97:8318–8323. doi: 10.1073/pnas.140161997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doshi U, Muñoz V. The principles of α-helix formation: Explaining complex kinetics with nucleation-elongation theory. J Phys Chem B. 2004;108:8497–8506. [Google Scholar]
- 17.Brewer SH, Song B, Raleigh DP, Dyer RB. Residue specific resolution of protein folding dynamics using isotope-edited infrared temperature jump spectroscopy. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 18.Kumita JR, Smart OS, Woolley GA. Photo-control of helix content in a short peptide. Proc Natl Acad Sci USA. 2000;97:3803–3808. doi: 10.1073/pnas.97.8.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bredenbeck J, Helbing J, Kumita JR, Woolley GA, Hamm P. α-Helix formation in a photoswitchable peptide tracked from picoseconds to microseconds by time resolved IR spectroscopy. Proc Natl Acad Sci USA. 2005;102:2379–2384. doi: 10.1073/pnas.0406948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihalainen JA, et al. Folding and unfolding of a photoswitchable peptide. Proc Natl Acad Sci USA. 2007;104:5383–5388. doi: 10.1073/pnas.0607748104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamm P, Helbing J, Bredenbeck J. Stretched versus compressed exponential kinetics in α-helix folding. Chem Phys. 2006;323:54–65. [Google Scholar]
- 22.Rao F, Caflisch A. The protein folding network. J Mol Biol. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 23.Krivov SV, Karplus M. Hidden complexity of free energy surfaces for peptide (protein) folding. Proc Natl Acad Sci USA. 2004;101:14766–14770. doi: 10.1073/pnas.0406234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hummer G, Garcia AE, Garde S. Helix nucleation kinetics from molecular simulations in explicit solvent. Proteins. 2001;42:77–84. [PubMed] [Google Scholar]
- 25.Caflisch A. Network and graph analyses of folding free energy surfaces. Curr Opin Struct Biol. 2006;16:71–78. doi: 10.1016/j.sbi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Chekmarev SF, Krivov SV, Karplus M. Folding of ubiquitin: A simple model describes the strange kinetics. J Phys Chem B. 2006;110:8865–8869. doi: 10.1021/jp056799o. [DOI] [PubMed] [Google Scholar]
- 27.Chowdhury S, Zhang W, Wu C, Xiong G, Duan Y. Breaking non-native hydrophobic clusters is the rate limiting step in the folding of an alanine-based peptide. Biopolymers. 2003;68:63–75. doi: 10.1002/bip.10216. [DOI] [PubMed] [Google Scholar]
- 28.Makowska J, et al. Polyproline II conformation is one of many local conformational states and is not an overall conformation of unfolded peptides and proteins. Proc Natl Acad Sci USA. 2006;103:1744–1749. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muff S, Caflisch A. Kinetic analysis of molecular dynamics simulations reveals changes in the denatured state and switch of folding pathways upon single-point mutation of a β-sheet miniprotein. Proteins Struct Funct Bioinf. 2008;70:1185–1195. doi: 10.1002/prot.21565. [DOI] [PubMed] [Google Scholar]
- 30.Ferrara P, Apostolakis J, Caflisch A. Evaluation of a fast implicit solvent model for molecular dynamics simulations. Proteins Struct Funct Bioinf. 2002;46:24–33. doi: 10.1002/prot.10001. [DOI] [PubMed] [Google Scholar]
- 31.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 32.Nguyen PH, Stock G. Nonequilibrium molecular dynamics simulation of a photoswitchable peptide. Chem Phys. 2006;323:36–44. [Google Scholar]
- 33.Nguyen PH, Gorbunov RD, Stock G. Photoinduced conformational dynamics of a photoswitchable peptide: A nonequilibrium molecular dynamics simulation study. Biophys J. 2006;91:1224–1234. doi: 10.1529/biophysj.106.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alaninen-based peptides. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C-Y, Klemke JW, Getahun Z, DeGrado WF, Gai F. Temperature-dependent helix–coil transition of an alanine based peptide. J Am Chem Soc. 2001;123:9235–9238. doi: 10.1021/ja0158814. [DOI] [PubMed] [Google Scholar]
- 36.Werner JH, Dyer RB, Fesinmeyer RM, Andersen NH. Dynamics of the primary processes of protein folding: Helix nucleation. J Phys Chem B. 2002;106:487–494. [Google Scholar]
- 37.Gooding EA, et al. The effects of individual amino acids on the fast folding dynamics of α-helical peptides. Chem Commun. 2005:5985–5987. doi: 10.1039/b511072f. [DOI] [PubMed] [Google Scholar]
- 38.Rohl CA, Baldwin RL. Deciphering rules of helix stability in peptides. Methods Enzymol. 1998;295:1–26. doi: 10.1016/s0076-6879(98)95032-7. [DOI] [PubMed] [Google Scholar]
- 39.Andersen CAF, Palmer AG, Brunak S, Rost B. Continuum secondary structure captures protein flexibility. Structure (London) 2002;10:174–184. doi: 10.1016/s0969-2126(02)00700-1. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Mira MM, Sadqi M, Fischer N, Sanchez-Ruiz JM, Muñoz V. Experimental identification of downhill protein folding. Science. 2002;298:2191–2195. doi: 10.1126/science.1077809. [DOI] [PubMed] [Google Scholar]
- 41.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193–197. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 42.Knott M, Chan HS. Criteria for downhill protein folding: Calorimetry, chevron plot, kinetic relaxation, and single-molecule radius of gyration in chain models with subdued degrees of cooperativity. Proteins. 2006;65:373–391. doi: 10.1002/prot.21066. [DOI] [PubMed] [Google Scholar]
- 43.Carstens H. Munich: Ludwig Maxmilians University; 2004. Conformation dynamics of light-stable peptide: Molecular dynamics simulations and data-driven model-building. PhD thesis. [Google Scholar]
- 44.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 45.Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A. WORDOM: A program for efficient analysis of molecular dynamics simulations. Bioinformatics. 2007;23:2625–2627. doi: 10.1093/bioinformatics/btm378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.