Abstract
Engineering proteins for higher thermal stability is an important and difficult challenge. We describe a bioinformatic method incorporating sequence alignments to redesign proteins to be more stable through optimization of local structural entropy. Using this method, improved configurational entropy (ICE), we were able to design more stable variants of a mesophilic adenylate kinase with only the sequence information of one psychrophilic homologue. The redesigned proteins display considerable increases in their thermal stabilities while still retaining catalytic activity. ICE does not require a three-dimensional structure or a large number of homologous sequences, indicating a broad applicability of this method. Our results also highlight the importance of entropy in the stability of protein structures.
Keywords: adenylate kinase, improved configurational entropy, local structural entropy, protein engineering, protein stability
Thermally stable proteins have a variety of practical applications in research and industrial settings (1, 2). Chemical reactions are intrinsically faster at higher temperatures, and using enzymes that are stable at higher temperatures would lead to more efficient industrial processes (3). In the laboratory, thermally stable proteins are also easier to store and handle, and can be used in high-temperature reactions such as PCR.
Numerous studies have been performed to find the principles of protein thermal stability and apply them to engineering thermal stability (4, 5). These include comparative studies using thermophilic and mesophilic homologues (6, 7). Although various individual structural features have been identified from those studies, they are used unpredictably to different extents in different proteins. Another strategy to improve protein thermal stability is directed evolution (8, 9). Despite many successes and recent technology development, directed evolution still has some practical limitations and thus can be labor-intensive and expensive. Efforts have also been made to computationally design thermally stable proteins, some requiring the three-dimensional structure of the target to be known (10, 11). The “consensus approach” is a sequence-based method for protein thermal stabilization that seeks to find commonality within a protein family (12). Although this method does not need structural information, it requires many homologous sequences for extensive sequence analysis and often involves arbitrary constraints in its algorithm (13).
We have developed a bioinformatic approach, which we call improved configurational entropy (ICE), to design more stable proteins using as few as two protein sequences. The essence of ICE is to optimize an empirical descriptor of the local structural entropy (LSE) (14) in the protein sequence guided by simple sequence alignment. This descriptor is based on structural information derived from the Protein Data Bank (15). A method for computing the LSE of a protein has been described previously (14). This approach evaluated the likelihood of each protein segment being in one of eight secondary structure configurations. The idea is that protein segments, which can exist equally well in many configurations, have higher entropy than those that exist primarily in one or a few secondary structure states. LSE values for all possible protein segments of length four were previously calculated based on their structural diversity found in the Protein Data Bank (14, 15). Although the absolute value from the calculations cannot be converted to thermodynamic units of entropy, correlations have been observed between thermal stabilities for a given family of proteins and their overall LSE values (14).
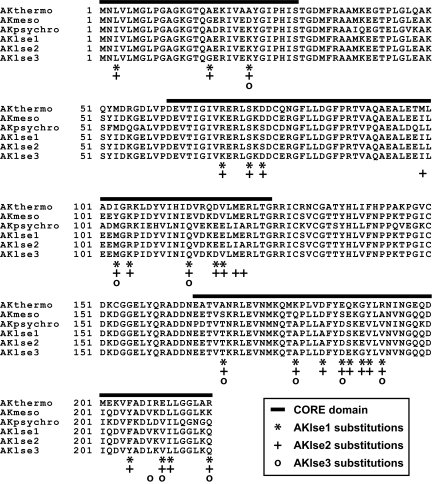
The effectiveness of ICE was tested on a commonly studied enzyme, adenylate kinase (AK). We aligned the sequences of AKs from a thermophile, Bacillus stearothermophilus (AKthermo), a mesophile, Bacillus subtilis (AKmeso), and a psychrophile, Bacillus globisporus (AKpsychro) (Fig. 1). Despite the differences in their stabilities, the amino acid sequences of these proteins are ∼70% identical and their three dimensional structures are very similar (16, 17). Previous studies indicated that amino acid substitutions in one of the enzyme's three domains, the CORE domain, are mainly responsible for the stability differences (18, 19).
Fig. 1.
Sequence alignment of the WT AKs and variants. The variants were designed from AKmeso and AKpsychro by optimizing LSE (see text).
Using ICE, we designed thermally stable AK variants from AKmeso and AKpsychro. We subsequently analyzed the designed variants for temperature dependence of stability and activity to test the validity of our method. The results demonstrate the advantages of ICE and the significance of structural entropy in protein stability.
Results
Computational Design of Thermally Stable AK Variants.
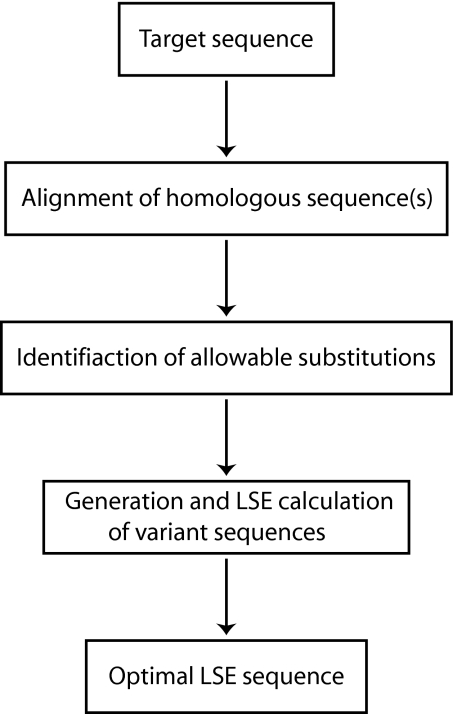
The schematic of ICE is presented in Fig. 2. First, one aligns the homologous sequence(s) to a target protein sequence. The number of homologous sequences to be aligned can be as small as one (in addition to the target sequence). The alignment reveals conserved residues and a set of allowable substitutions in variable residues that do not affect the overall protein fold or function. At each variable position, amino acids are chosen from the set of allowable substitutions to make variant protein sequences. For example, an alignment of two sequences (a target and a homologue) with N variable positions results in 2N possible variant sequences. Among these sequences, one with the optimal (lowest) LSE is selected.
Fig. 2.
Schematics of ICE for designing thermally stable variants.
For an initial test, a more stable protein was designed by substituting residues present in AKmeso with ones in AKpsychro. This may seem backwards because the less thermally stable psychrophilic sequence was used for the substitutions instead of the more stable thermophilic homologue. To evaluate the effectiveness of ICE, we did not directly take nature's adaptations for higher stability found in AKthermo. We also chose mutations only in the CORE domain of the protein (Fig. 1) because the previous biochemical study indicated that mutations in the other regions did not have a considerable effect on overall stability and would not be appropriate for testing the effectiveness of the method (19).
In the CORE domain, AKmeso and AKpsychro have 53 different residues. Among the 253 possible variant sequences, a sequence (AKlse1) having the lowest average LSE was found to have 23 substitutions (Fig. 1). In AKlse1, the substitutions are spread over its sequence and do not show particular patterns in their side chain properties such as polarity and hydrophobicity. It is noteworthy that a significant number of residues in the psychrophilic homologue were predicted to improve the thermal stability of the mesophilic target. We also selected a closely related but slightly less optimized variant having 26 substitutions (AKlse2; Fig. 1) for further in vitro characterization. This variant has the most substitutions in the top 20 most LSE-optimized variant sequences, and therefore shares the highest sequence identity with AKpsychro.
Thermal Stabilities of AK Variants.
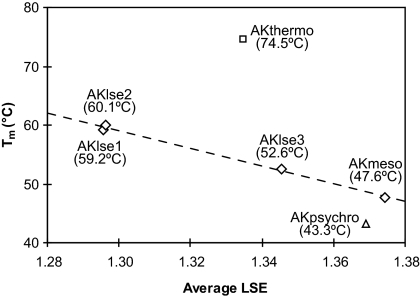
To experimentally validate the result of the LSE optimization, we produced the AKlse1 and AKlse2 proteins and measured their thermal stabilities. Instead of making the variant genes by multiple rounds of site-directed mutagenesis, we used synthetic genes [supporting information (SI) Fig. S1]. Using Escherichia coli, we expressed and purified the proteins and determined their apparent melting temperature (Tm) values using differential scanning calorimetry. As predicted by the computational analysis, the substitutions in the AK variants result in substantial stabilization. The Tm values of AKlse1 and AKlse2 are higher than that of AKmeso by 11.6°C and 12.5°C, respectively (Fig. 3 and Fig. S2).
Fig. 3.
Plot of Tm vs. average LSE for the WT AKs and variants. The average LSE values were calculated for the CORE domain. Tm values are shown in parentheses, and the values for the WT AKs are from previous studies (16, 17). The regression line was drawn for the three AK variants and AKmeso.
AK Variant Lacking “AKthermo Substitutions.”
There are residues conserved only between AKthermo and AKpsychro including five of the substitution sites in AKlse1 and AKlse2 (residues 17, 69, 73, 105, and 205). For example, AKthermo and AKpsychro share Ala-17, but AKmeso has a glycine residue at the same position (Fig. 1). Although the glycine-to-alanine substitution at this position for AKlse1 and AKlse2 was originally selected based on the comparison with AKpsychro, the mutation increases the variant's similarity to AKthermo regardless. One could argue that the increased stability of these variants is not caused by optimizing LSE but instead by substituting amino acids at the five positions with those from AKthermo.
To exclude this possibility and confirm the effectiveness of ICE, another variant was designed. In this experiment, we allowed the same mesophile-to-psychrophile substitutions but only in the positions where amino acids are different in all of the three wild-type (WT) sequences. There are 19 such residues, and ICE produced the most LSE-optimized variant sequence (AKlse3) with 10 substitutions (Fig. 1). Because of the significantly smaller search space for an optimal sequence in this experiment than the previous one (219 vs. 253), AKlse3 has a higher average LSE than AKlse1 and AKlse2, suggesting less thermal stabilization. As expected, the Tm of AKlse3 was lower than those of AKlse1 and AKlse2, but still considerably higher (5.0°C) than that of AKmeso (Fig. 3 and Fig. S2).
Catalytic Activities of AK Variants.
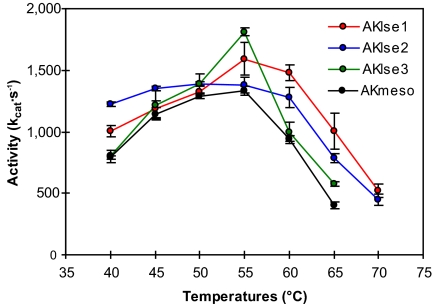
It is possible that the catalytic properties of enzymes are compromised in the process of engineering their thermal stabilities. Because the amino acid substitutions for our AK variants were allowed only at the variable residues in the alignment of homologous sequences, it is likely that the enzyme's intrinsic structure and function would not be adversely affected and the AK variants would retain their catalytic activities. To confirm this, we performed activity assays of our variants at several different temperatures and compared the resulting temperature–activity profiles with that of WT AKmeso. As shown in Fig. 4, all three AK variants maintained their catalytic activity. The measured activities were either higher than or at least equal to that of AKmeso at all of the tested temperatures. The profiles of AKlse1 and AKlse2 clearly indicate that their activities are increased at higher temperatures when compared with that of AKmeso. Although the measured activity of AKlse3 is higher than that of AKmeso at their optimal temperatures (55°C), their profiles at higher temperatures are similar. This could be due to the smaller increase in thermal stability for AKlse3 than for AKlse1 and AKlse2.
Fig. 4.
Activity profiles for the three AK variants and AKmeso. The activity profile of AKmeso is from ref. 19. Means ± SE are shown.
Discussion
As dramatically demonstrated by our experiments, it is not necessary to use a thermophilic homologue, multiple homologous sequences, or the three-dimensional structure to engineer increased thermal stability in a protein. We were able to make more stable proteins by choosing amino acid substitutions from a single less-stable homologue that optimized LSE. Precisely three variants were experimentally tested, and all showed substantial increases in thermal stability when compared with the WT. Introducing a large number of mutations was not practically difficult because the cost of creating synthetic genes has dramatically decreased recently.
The results of our experiments are summarized in Fig. 3. Our AK variants not only have considerably higher Tm values than AKmeso from which the variants were designed, but they also display a correlation in the plot of Tm vs. average LSE. This indicates that increases in their stabilities as their LSE values were optimized. However, AKthermo and AKpsychro deviate from the regression line, suggesting that they have mechanisms other than or in addition to changing their LSE for modifying stability. For AKthermo, electrostatic and hydrophobic interactions bridging distant regions of the protein sequence were suggested as stabilizing factors in the previous comparative studies (17, 18).
Other methods have been used to increase thermal stability of AKmeso. By rationally designing electrostatic and hydrophobic interactions onto AKmeso based on the structural comparison with AKthermo, we were able to increase its Tm by 8.5°C (E.B. and G.N.P., unpublished data). In a directed evolution study using modified B. stearothermophilus in which the original AKthermo was replaced with AKmeso, Counago et al. (20) derived several more stable mutants whose largest Tm increase was 13.8°C. The increases in thermal stability by ICE in this study are comparable with those by the different approaches while ICE was more efficient and required less information about the target.
The tests of ICE on AKs also highlight its differences and advantages vis-à-vis another sequence-based method, the consensus approach. This approach is based on the hypothesis that at a given position in alignment of multiple homologous sequences, the consensus amino acid contributes to stability more than the nonconsensus one. It requires many homologous sequences for alignment and often encounters situations where it is not straightforward to identify the consensus amino acids, thus requiring additional heuristics. On the other hand, ICE optimizes a physical property of a protein, and only uses the sequence alignment to generate diversity in sequence choices. In contrast to the consensus approach, ICE needs a minimum of only two sequences and does not require any arbitrary constraints. In fact, when an additional sequence (AKthermo) is considered, many nonconsensus amino acids were found in our variants (Fig. 1). When designing AKlse3, we allowed substitutions only in the positions where all three WT AKs have different amino acids, which make it impossible to implement the consensus approach.
Substantial differences have been found between our variants and a sequence resulting from a consensus analysis using a larger number of homologous sequences (R.M.B. and G.N.P., unpublished data). For example, a sequence obtained from a consensus analysis with 11 Bacillus AKs has 18 different residues when compared with AKmeso. Among the 18 substitutions, only four of them are the same as those in our AKlse1 variant. The other 14 are either replaced with different amino acids (6 positions) or not substituted (8 positions) in AKlse1. As a retrospect analysis, we also compared our three variant sequences with another thermophilic Bacillus species, Bacillus licheniformis (21). Only four of the substitutions in our variants were also found in the B. licheniformis AK sequence. All of these data suggest strongly that the basis for the thermal stabilization of our three variants is different from that of the consensus approach.
The strength of ICE is that one can learn from nature which sequences are consistent with the required intrinsic structure and function, and choose a sequence from the allowed substitutions by empirically evaluating the degeneracy in structural states of short amino acid segments. We dramatically demonstrated the effectiveness of ICE by making considerably more thermally stable proteins from only the sequence information of one psychrophilic homologue. This indicates a broad applicability of ICE and the importance of entropy optimization in engineering protein thermal stability.
Materials and Methods
LSE Optimization of AK Sequences.
The protocol for calculating the LSE value for a particular amino acid tetramer is described in ref. 14. In that study, the researchers examined how often each possible amino acid tetramer was found in eight different protein secondary structures (“β-bridges,” “extended β-sheets,” “310-helices,” “α-helices,” “π-helices,” “bends,” “turns,” and “others”) in the Protein Data Bank. If a tetramer appeared in many of these secondary structures, it was given a higher LSE value than that value assigned to a tetramer that appeared in only one or two secondary structures.
For a sequence with length n, there are n − 3 segments of length four. We summed the LSE values for these n − 3 segments and divided the total by n − 3 to calculate the average LSE. Using the LSE tetramer values derived from a particular set of nonredundant structures (http://sdse.life.nctu.edu.tw/db-download/scop-35–4-ss.txt), we used a brute-force method to calculate the average LSE for all of the possible AK variants in the CORE domain. We chose to mutate residues only in the CORE domain because our previous study indicated that mutations in the other regions did not cause considerable changes in overall thermal stability (19) and because we wanted to reduce the noise level of our test. The LSE optimization algorithm is designed such that mutations in the other regions would not affect the choice of mutations in the CORE domain and vice versa. Thus, we would have the same mutations in the CORE domain regardless of whether we included the other regions or not.
Generation of AK Variants.
Three AK variant genes were commercially synthesized by Geneart and provided in pET11a expression vectors. The variant proteins were overexpressed in E. coli and purified by a two-step procedure involving affinity chromatography and gel filtration as described in ref. 17.
Tm Measurement.
Tm curves for the three AK variants were obtained by differential scanning calorimetry. The detailed method for collecting the Tm data is described ref. 17. In these experiments, the measured values were obtained from irreversible thermal denaturation. They are more relevant as a practical measure of protein thermal stability than those from simple reversible unfolding often found in model systems (4).
Activity Assay.
Activity assays were performed in the direction of ATP formation as described in ref. 19. Briefly, the enzyme reaction at each temperature was started by adding AK to the reaction buffer (1 mM glucose, 0.4 mM NADP+, 100 mM KCl, 2 mM MgCl2, 50 mM Hepes, pH 7.4) containing ADP and stopped by adding the inhibitor P1,P5-di(adenosine-5′)pentaphosphate. The amount of ATP produced by the reaction was determined by using ATP-dependent reduction of NADP+ to NADPH by the coupling enzymes hexokinase and glucose-6-phosphate dehydrogenase at room temperature. We used this end-point method because the activities of the coupling enzymes are temperature-sensitive.
Supplementary Material
Acknowledgments.
We thank Darrell R. McCaslin for help in differential scanning calorimetry and Craig A. Bingman, Dmitry A. Kondrashov, Ed Bitto, Christopher M. Bianchetti, and Elena J. Levin for comments on the manuscript. This work was supported by the Wisconsin Alumni Research Foundation and BACTER Department of Energy Training Grant DE-FG2-04ER25627.
Footnotes
Conflict of interest statement: The authors would like to disclose that they have a patent pending on this method.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800938105/DCSupplemental.
References
- 1.Schoemaker HE, Mink D, Wubbolts MG. Dispelling the myths—Biocatalysis in industrial synthesis. Science. 2003;299:1694–1697. doi: 10.1126/science.1079237. [DOI] [PubMed] [Google Scholar]
- 2.Unsworth LD, van der Oost J, Koutsopoulos S. Hyperthermophilic enzymes—Stability, activity and implementation strategies for high temperature applications. FEBS J. 2007;274:4044–4056. doi: 10.1111/j.1742-4658.2007.05954.x. [DOI] [PubMed] [Google Scholar]
- 3.Turner P, Mamo G, Karlsson EN. Potential and utilization of thermophiles and thermostable enzymes in biorefining. Microb Cell Factories. 2007;6:9. doi: 10.1186/1475-2859-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eijsink VG, et al. Rational engineering of enzyme stability. J Biotechnol. 2004;113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Bommarius AS, Broering JM, Chaparro-Riggers JF, Polizzi KM. High-throughput screening for enhanced protein stability. Curr Opin Biotechnol. 2006;17:606–610. doi: 10.1016/j.copbio.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Vieille C, Zeikus GJ. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wintrode PL, Arnold FH. Temperature adaptation of enzymes: Lessons from laboratory evolution. Adv Protein Chem. 2000;55:161–225. doi: 10.1016/s0065-3233(01)55004-4. [DOI] [PubMed] [Google Scholar]
- 9.Eijsink VG, Gaseidnes S, Borchert TV, van den Burg B. Directed evolution of enzyme stability. Biomol Eng. 2005;22:21–30. doi: 10.1016/j.bioeng.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Korkegian A, Black ME, Baker D, Stoddard BL. Computational thermostabilization of an enzyme. Science. 2005;308:857–860. doi: 10.1126/science.1107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo P, et al. Development of a cytokine analog with enhanced stability using computational ultrahigh throughput screening. Protein Sci. 2002;11:1218–1226. doi: 10.1110/ps.4580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann M, Wyss M. Engineering proteins for thermostability: The use of sequence alignments versus rational design and directed evolution. Curr Opin Biotechnol. 2001;12:371–375. doi: 10.1016/s0958-1669(00)00229-9. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Buckle AM, Foster NW, Johnson CM, Fersht AR. Design of highly stable functional GroEL minichaperones. Protein Sci. 1999;8:2186–2193. doi: 10.1110/ps.8.10.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan CH, et al. Relationship between local structural entropy and protein thermostability. Proteins. 2004;57:684–691. doi: 10.1002/prot.20263. [DOI] [PubMed] [Google Scholar]
- 15.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser P, et al. Zinc, a novel structural element found in the family of bacterial adenylate kinases. Biochemistry. 1992;31:3038–3043. doi: 10.1021/bi00127a002. [DOI] [PubMed] [Google Scholar]
- 17.Bae E, Phillips GN., Jr Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 18.Bae E, Phillips GN., Jr Identifying and engineering ion pairs in adenylate kinases. Insights from molecular dynamics simulations of thermophilic and mesophilic homologues. J Biol Chem. 2005;280:30943–30948. doi: 10.1074/jbc.M504216200. [DOI] [PubMed] [Google Scholar]
- 19.Bae E, Phillips GN., Jr Roles of static and dynamic domains in stability and catalysis of adenylate kinase. Proc Natl Acad Sci USA. 2006;103:2132–2137. doi: 10.1073/pnas.0507527103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Counago R, Chen S, Shamoo Y. In vivo molecular evolution reveals biophysical origins of organismal fitness. Mol Cell. 2006;22:441–449. doi: 10.1016/j.molcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Claus D, Berkeley RCW. Section 13. Endorespore-forming gram-positive rods and cocci. In: Sneath PHA, editor. Bergey's Manual of Systematic Bacteriology. Vol 2. Baltimore: Williams & Wilkins; 1986. pp. 1105–1139. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.