Abstract
Saccharomyces cerevisiae strains harboring a nonreciprocal, bridge-induced translocation (BIT) between chromosomes VIII and XV exhibited an abnormal phenotype comprising elongated buds and multibudded, unevenly nucleated pseudohyphae. In these cells, we found evidence of molecular effects elicited by the translocation event and specific for its particular genomic location. Expression of genes flanking both translocation breakpoints increased up to five times, correlating with an increased RNA polymerase II binding to their promoters and with their histone acetylation pattern. Microarray data, CHEF, and quantitative PCR confirmed the data on the dosage of genes present on the chromosomal regions involved in the translocation, indicating that telomeric fragments were either duplicated or integrated mostly on chromosome XI. FACS analysis revealed that the majority of translocant cells were blocked in G1 phase and a few of them in G2. Some cells showed a posttranslational decrease of cyclin B1, in agreement with elongated buds diagnostic of a G2/M phase arrest. The actin1 protein was in some cases modified, possibly explaining the abnormal morphology of the cells. Together with the decrease in Rad53p and the lack of its phosphorylation, these results indicate that these cells have undergone adaptation after checkpoint-mediated G2/M arrest after chromosome translocation. These BIT translocants could serve as model systems to understand further the cellular and molecular effects of chromosome translocation and provide fundamental information on its etiology of neoplastic transformation in mammals.
Keywords: DNA integration, yeast, genome expression, genome dynamics, transcription
The genomic rearrangements after a chromosomal translocation often lead to severe cellular defects that can give rise to neoplastic transformation in mammals. In humans, translocation between chromosomes 9 and 22, t(9;22), producing the Philadelphia chromosome, causes myelogenous leukemia (1) because of the increased expression of the protooncogenic c-Abl tyrosine kinase gene, located at the translocation breakpoint (2). In Burkitt's lymphoma, a multiple translocation involving chromosomes 2, 8, 14, and 22 leads to high expression of c-myc protooncogene (3). More translocations have been documented in hematological, mesenchymal, and epithelial cancer (4), and their detection is used in clinical practice for diagnosis and choice of the treatment protocol (5). Translocations and other gross chromosomal rearrangements (GCRs), i.e., deletions and inversions, are often observed in cancer cells (6). Recently, seven pathways that suppress (7–9) and four pathways that are required for the formation of GCRs (10–12) have been identified in yeast. From the great number and redundancy of factors involved in GCR suppression, it can be concluded that any genomic rearrangement is highly deleterious to cells, and, once escaped from control, it can have a massive impact on cellular morphology and physiology.
The availability of a simple method to create ad hoc specific translocation between any two desired genomic loci in a eukaryotic model system allows investigation of in vivo analogous molecular features observed in mammalian cell transformation after chromosome translocation (13).
In this work, we have performed cellular and molecular analysis of Saccharomyces cerevisiae translocants isolated by our bridge-induced translocation (BIT) technique (14) as well as de novo translocants generated in the same way. Some translocants exhibited an abnormal cell phenotype with elongated buds as well as multibudded and unevenly nucleated cells. Several others revealed further genetic rearrangements of the acentric chromosome fragments, whereas expression of the genes located at the translocation breakpoints was increased up to five times, coinciding, in general, with an increased level of the RNA polymerase II binding to their promoters and to the pattern of histone acetylation. In addition to these local, translocation breakpoint-related cis effects, we also observed trans effects attributable to the deregulation of genes not physically involved in the chromosome translocation. One of these is the DNA damage repair gene RAD53, the phosphorylation of which is one of the first events of checkpoint activation. Rad53p was found strongly decreased and not phosphorylated in the most abnormal translocants, whereas by FACS analysis, the majority of cells was found arrested in G1 with fewer at the G2/M transition phase. In some translocants, decreased cyclin B1 protein (but not its mRNA transcript) level could explain the appearance of cells with elongated buds, supporting the presence of a subpopulation of cells arrested at the G2/M transition phase. The study of gene dosage by contour-clamped homogeneous electric field (CHEF) Southern hybridization, quantitative PCR, and microarray expression analysis led to the discovery that, in some cases, the acentric chromosome fragments either duplicated or, in some cases, integrated at other locations on the genome, with preference for chromosome XI. These results suggest that the translocant cells have adapted to the checkpoint response after the initial DNA damage generated by chromosome translocation. Moreover, in most strains, the actin1 protein appeared modified into a slightly larger adduct, in agreement with the abnormal cell morphology.
Results
Configuration of Chromosomal Arms in D10 Small and Big Strains After Translocation.
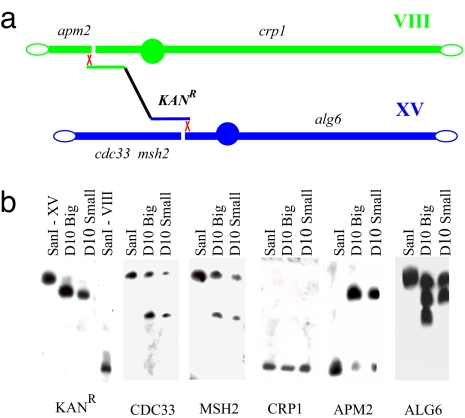
A scheme of BIT translocation between chromosomes VIII and XV is shown in Fig. 1a. After transformation with the linear DNA cassette carrying the G418 resistance marker, which bridges the right, centromeric portion of chromosome XV with a fragment of the left arm of chromosome VIII, site-specific translocants are isolated from KanR transformant cells. Two types of translocants obtained were chosen for in-depth analysis: one, D10 Small, is capable of producing only small colonies on agar plates; the second, D10 Big, is a revertant strain of D10 Small, capable of producing normal-size colonies (14). The configuration of the chromosomal arms after translocation was verified by Southern hybridization probing for genes located on different sides of the translocation breakpoints (Fig. 1b). For wild-type and translocant strains, the probes were specific for KANR, CDC33, MSH2, CRP1, APM2, and ALG6 genes.
Fig. 1.
Structural analysis of BIT translocation between chromosome VIII and XV. (a) Schematic representation of the translocation between chromosomes VIII and XV with the location of the strategic loci tested. The interruption of the chromosome indicates the putative location of the DSB. (b) Southern hybridization of SanI, D10 Big, and D10 Small translocants with a DNA probe for the genes indicated below each panel. In the KANR panel, lanes 1 and 4, the positions of the bands corresponding to chromosome XV and VIII are shown, respectively, whereas lanes 2 and 3 show the bands corresponding to the translocated XV/VIII chromosome in the two strains D10 Big and Small. The positions of these four bands serve as a reference for all panels.
According to Southern blotting and PCR analysis, a new translocant chromosome, consisting of the left portion of chromosome VIII and the right portion of chromosome XV, was generated (Fig. 1b, APM2 and ALG6 panels). Besides the translocated chromosome, the parental copies of chromosomes VIII and XV are still present because the original strain is diploid. In the Southern hybridization reported in Fig. 1b, these wild-type chromosomes are identified by the bands on the D10 translocant lanes corresponding to the band on the parental SanI lanes. The fate of the acentric left fragment of chromosome XV, which is not involved in the translocation, is shown in the CDC33 and MSH2 panels. Two different probes within this fragment indicate that it is recovered by the cell after rearrangement with another chromosome, calculated as chromosome XI on the basis of the best-fitting curve obtained from the migration of chromosomal bands from Fig. 1a and supporting information (SI) Fig. S1 (see Methods). In the CRP1 panel, the hybridization bands demonstrate that the centric fragment of chromosome VIII that did not take part in the translocation is either lost (D10 Big) or it is restored to full size (D10 Small). Therefore, the right portion of chromosome VIII and the left portion of chromosome XV (Fig. 1b, CRP1 and CDC33 panels) do not recombine with each other, making the translocation nonreciprocal. It seems that no genetic information is lost after the translocation event, but it is rearranged in complex ways. It is noteworthy that cytogenetic differences were found not only between parental and translocant strains but also between the same translocants (Fig. 1b, D10 Big and Small lanes in the ALG6 panel). Apparently, once happened, the translocation leads to further genetic rearrangements because it can be inferred from differences between D10 Big and Small strains complex karyotypes. Because of this variability, translocant and parental strains were compared in detail for their cellular and molecular features.
Microscopy Analysis of Parental and Translocant Strains.
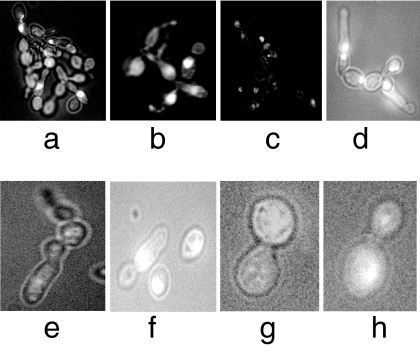
The morphology of parental SanI and D10 translocant strains was compared by light and fluorescence microscopy. It was found that between 10 and 20% of the cell population in the translocant strains had an abnormal phenotype. The most interesting examples of this finding are represented in Fig. 2 a–c in which the loss of the single-cell morphology and the acquisition of a pseudohyphal growth with defective karyocinesis are evident. Another common type of abnormality is the presence of multibudded, multinucleated cells (Fig. 2 d and e) and cell with elongated buds (Fig. 2 f–h). The most severe defects, globally ascribable to cell cycle and cytokinesis deregulation, were found in D10 Small translocant strain, whereas in the D10 Big revertant these were less frequent and evident (data not shown). This implies that there may be differences between strains that might not be linked to the translocation event itself.
Fig. 2.
Microphotography of DAPI-stained D10 Small translocant cells. Different abnormal phenotypes are shown. (Magnification: a–d, ×400; e–h, ×1,000.)
One of the most important cellular structural proteins is actin1. We have checked its distribution in the parental SanI strain and the D10 translocants (Fig. S2) by FITC–phalloidin staining (15). The majority of the translocant cells have normal actin1 distribution typical of that particular phase of the cell cycle, regardless of some morphological defects in M phase, as elongated buds and presence of two buds in one mother cell (Fig. S2, D10 Big and D10 Small rows, M column). However, some D10 translocant cells had actin1 distribution not coinciding with that of the cell cycle phase in which they were observed. Thus, in a portion of the translocant cell population, translocation led to abnormal actin1 distribution patterns.
Expression Profiling of Genes Located at the Translocation Breakpoints.
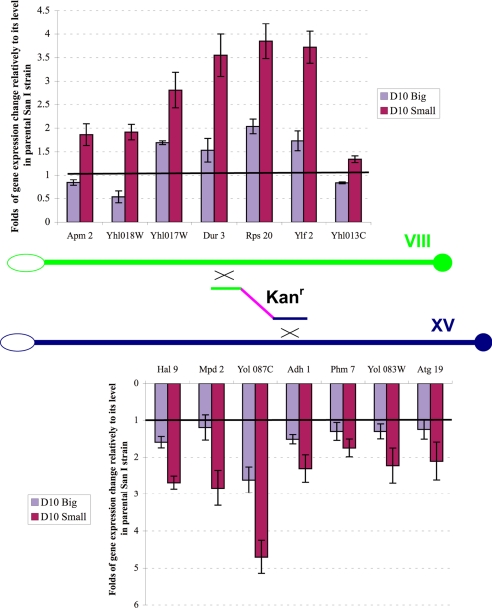
To analyze the effect of translocation on the expression of the genes adjacent to the chromosome breakpoints, we used quantitative RT-PCR profiling of their mRNAs. Three genes on each side of the breakpoints were considered, plus the SPO11 gene on the left side of the breakpoint on chromosome VIII. As a control of constitutive gene expression, the HSC82 gene was chosen as in previous work (16). The names and descriptions of all genes analyzed in the experiment are reported in Table S1. The quantitative RT-PCR amplification of these genes is shown in Fig. S3. After laser-scanning densitometry of the amplified bands and consequent computational analysis of the data, their transcriptional profiling is shown graphically in Fig. 3, with the ratio between the expression level of each of the genes and the level of HSC82 in parental SanI taken as 1. In general, the expression of the genes at translocation breakpoints in D10 translocants was increased. The highest mRNA level changes (4–5 times more than parental SanI) were observed in D10 Small strain for SPO11, YHL017W, DUR3, RPS20, YLF2, and YOL087C genes. In the D10 Big strain, their expression level was lower than in the D10 Small strain, but it was still higher than in the parental SanI strain by a factor up to 2, with the exception of YHL018W, the expression of which was lower than in the wild type. To ascertain whether this change in gene expression correlates with any variation in copy number of the genes involved, we performed a quantitative PCR comparison of their DNA amounts with that of the genes ARK1, PRK1, and HSC82, located on chromosome XIV, IX, and XIII, respectively, in the parental SanI and two D10 translocant strains (Fig. S4). Because it appears clearly, no difference exists between the genes surrounding the translocation breakpoints and that of genes on other chromosomes both in parental and translocant strains.
Fig. 3.
Graphic representation of the quantitative RT-PCR gene expression patterns from Fig. S3 of the various genes on both sides of the translocations breakpoints on chromosome VIII (Upper) and XV (Lower) compared with the level of the reporter gene HSC82 in the wild-type strain taken as 1 (horizontal black line).
To define how far from the translocation breakpoints the effect of increased expression was spread, we checked in the same way the expression level of several genes (CBP2, PPE1, RRP40, TOP1, and CYT1) located farther (50–300 kb) from the translocation breakpoint (Fig. S5 a and b). Detailed information about their roles in the cell and their genomic location is given in Table S1. From this table, expression of the PPE1, RRP40, TOP1, and CYT1 genes located more than 100 kb from the translocation breakpoint appears unchanged, whereas the CBP2 gene, located 50 kb from it at a subtelomeric region of chromosome VIII, had an mRNA level slightly increased 1.4 times. Thus, apparently, this distance could be not enough to quench the effect of the translocation on the expression of the adjacent genes.
Microarray Analysis of Chromosome Transcription Profiling.
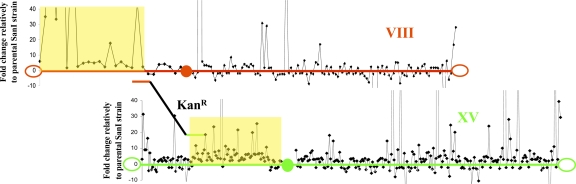
Transcription profiling data from all of the genes located on the two chromosomes VIII and XV were obtained by DNA microarray technology (Agilent; see Methods to access the expression data). According to the results, gene expression enhancement spreads from the breakpoint to the left telomere along chromosome VIII and from the breakpoint to the centromere on chromosome XV (Fig. 4). These regions correspond to the whole left arm of the newly generated translocant chromosome. On the other fragment of the two chromosomes, the leftmost part of chromosome XV and the rightmost part of chromosome VIII, the gene expression pattern appears unchanged, as well as on the right arm of the translocant chromosome. Thus, in the D10 Small strain, BIT resulted in severalfold enhancement of the expression of many genes located up to ≈160 kb from the translocation breakpoints. This increase can be considered as a locus-specific effect of translocation, most likely because of changes in chromatin structure close to the translocation breakpoints, making gene promoters in this area more accessible to the transcriptional machinery. This hypothesis is analyzed in the next subsection.
Fig. 4.
Microarray transcription profiling of genes on chromosomes VIII and XV involved in BIT translocation in D10 Small strain. The genes are indicated by black square dots according to their position along the two chromosomes. The two DNA regions of homology for integration are indicated by the short red and green ends of the KanR cassette. The open oval and filled circles represent the telomeres and centromere, respectively. The yellow areas indicate the regions of gene overexpression.
Chromatin Remodeling at the Translocation Breakpoints.
To check whether accessibility of gene promoters located at the translocation breakpoints in D10 strains to RNA polymerase II binding was increased, we used the chromatin immunoprecipitation (ChIP) technique with antibodies to this enzyme and quantitative PCR with primers complementary to the promoter regions of the genes (Table S2). The results are presented in Fig. S6. In a, the amplified bands without (−IP, Input) and after (+RPII IP) immunoprecipitation are showing a different pattern among the strains analyzed. After laser densitometry and quantitative data analysis (Fig. S6b), we concluded that RNA polymerase II binding to the promoter of these genes in D10 translocants reflects their mRNA level with the exception of the HAL9 and YHL013W (OTU2) genes adjacent to the left and right end of the DNA bridge, respectively. Interestingly, in D10 Small strain, SPO11 mRNA level was drastically increased compared with the parental SanI strain, in agreement with a 11.5-fold increase in RNA polymerase binding level to its promoter, shown by the ChIP experiment (Fig. S6 a and b).
In D10 Big strain, the RNA polymerase II binding to the promoters of all genes analyzed was lower than in D10 Small strain but still higher than in parental SanI strain, coinciding with the RT-PCR data (Fig. 3). Only for YHL013 and HAL9 the relationship was complex: YHL013 mRNA level was almost unchanged, but its RNA polymerase II-binding level decreased; HAL9 mRNA level increased 1.5 times in D10 Big and 2.8 times in D10 Small cells, but its RNA polymerase II binding decreased 5 and 2.3 times, respectively, suggesting that, unlike the other genes analyzed, their regulation may be controlled individually by a posttranscriptional pathway.
Because chromatin structure plays an important regulatory role in gene expression and multiple signaling pathways converge on histones (17), we checked the histone acetylation pattern as one of the most important landmarks of transcriptionally active chromatin. For this analysis, antibodies to acetylated lysine-14 of histone H3 were used, and ChIP was performed. Histone acetylation pattern of gene promoters at the translocation breakpoints in D10 translocants increased for SPO11, YHL017W, DUR3, MPD2, YHL083, and ATG19 (SI Fig. S6 Top, +H3Ac IP bands, and Fig. S6c), coinciding with their increased mRNA level (Fig. 3). On the promoters of APM2, YOL087, ADH1, and PHM7 the acetylation level in D10 translocants decreased ≈2 times compared with the parental SanI strain, even though their mRNA level quantified by RT-PCR increased (Fig. 3).
Cellular Adaptation After DNA Damage Response.
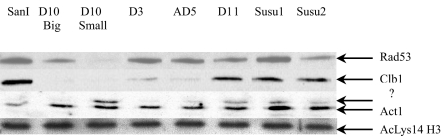
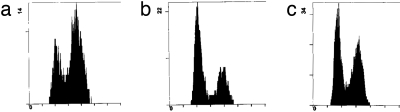
During the process of BIT chromosome translocation, besides the presence of two DSB-mimicking free DNA ends on the transforming linear DNA cassette, at least two DSBs are generated (Fig. 1a). Therefore, we analyzed the expression of one of the key proteins involved in DSB repair, Rad53p (18), by Western blot hybridization. The data are presented in Fig. 5, Top row. In D10 Big, and more evidently in D10 Small translocants, its expression is decreased, whereas it is nearly normal in D3, AD5, D11, Susu1 and Susu2 strains, which do not exhibit noticeable cell cycle or morphological defects (data not shown). Moreover, no phosphorylated forms of Rad53 were detected with specific antibodies (data not shown). Thus, the down-regulation of Rad53 level without its phosphorylation and the G1 arrest of the majority of the cells with a 1C DNA content demonstrated by FACS analysis (Fig. 6), both characteristic hallmarks of DNA damage adaptation (19), support the hypothesis that the D10 translocants could have undergone DNA damage adaptation after the transactivation of the DNA repair pathway caused by the translocation.
Fig. 5.
Western blot hybridization of Rad53, cyclin B1, and actin1 expression levels in SanI, the two D10 translocants, and other translocant strains D3, AD5, D1, Susu1, and Susu2. The Lys-14Ac histone H3 bands were used as protein loading control with anti-acetylated lysine-14 histone H3 antibodies.
Fig. 6.
Histogram representation of FACS analysis data for SanI strain (a), D10 Big (b), and D10 Small (c) translocants.
CyclinB1 Protein Level Was Decreased in Translocants.
As was shown in previous works (20, 21), successful transition from G2 into mitosis depends on Cdc28p (Cdk1p) activity, which in turn is activated by B-type cyclins Clb1p and Clb2p. A decreased level of these cyclins arrests cells in G2/M phase with an elongated-bud phenotype. To check for CLB1 expression level in translocants we used Western blotting with anti-Clb1p antibodies (Fig. 5, Clb1 row). In both translocants analyzed, the cyclin B1 level was almost absent compared with the parental SanI strain. These results could explain the appearance of elongated buds observed in the translocants (Fig. 2 f–h).
Actin1 Protein Was Modified in Several Translocants.
Actin1 is one of the most important structural proteins involved in such critical processes as endocytosis, cytokinesis, cell polarity, and morphogenesis (22). Because cell morphology of several translocant cells was different from the parental SanI cells (see Fig. 2), we analyzed actin1 protein by Western hybridization with anti-actin1p antibodies (Fig. 5, Act1 row). In D10 Small translocants a new band appeared, which migrates 2–3 kDa above the regular Act1p band, corresponding, probably, to a modified form of actin1. This new band was almost invisible in the parental SanI strain and could be partially responsible for the unusual cellular morphology observed in some of the translocant strains (Fig. 2 a–e).
Discussion
In this article, we have described local (cis) effects, ascribable to the genomic location of a specific BIT translocation, and other (trans) effects present in some of the strains studied. The latter are probably caused by cellular adaptation after DNA damage response as well as by overcoming translocation inhibition. Therefore, they should be found in other unrelated translocants. Indeed, when we analyzed Rad53 expression in other BIT translocant strains, D3, D11, and Susu2, these strains showed a comparable down-regulation. Translocants D3, AD5, D11, Susu1, and Susu2 showed a decreased Clb1 expression, whereas all five of them showed the extra Act1 band (Fig. 5).
An important local effect in D10 Big and Small strains was the increased expression level of genes located near the translocation breakpoints, confirmed by quantitative RT-PCR (Fig. 3) and by microarray data. This effect seems to initiate from the translocation breakpoints and to spread over several tens of kilobases in both directions, regardless of the orientation of the genes. However, it appears as if the enhancement of the transcription level cannot cross the centromere region (Fig. 4). This local effect can have cumulative consequences in translocant cells because it may lead to a deregulation cascade of many cellular functions. For example, in D10 translocants, the meiosis-specific SPO11 gene expression has raised 7-fold from its near absence typical of mitotic cells. The SPO11 gene encodes an endonuclease, which initiates meiotic recombination by catalyzing the formation of double-stranded breaks (DSBs) in DNA. Its improper activation could lead to the production of DSBs during mitotic growth, interfering with DNA replication, and, consequently, generating genomic instability in mammalian cells. The amplification of a genomic region containing the SPO11 ortholog gene was indeed documented in some breast and ovarian tumors (23). The genes, the expression of which was increased in D10 translocants, are involved in different cellular processes, possibly explaining why many cellular functions can be affected after even one single translocation event.
The increased expression level of genes located at chromosomal translocation breakpoints was known before. For example, activation via translocation of Abl, Myc, and Mll protein expression leads to myelogenous leukemia (2), Burkitt's lymphoma (3), and mixed-lineage leukemia (13), respectively. However, the molecular mechanism underlying this phenomenon is yet unknown. We found that, besides two exceptions (HAL9 and YHL013), an increased binding of RNA polymerase II to the promoters of the genes at translocation breakpoints could be correlated to their activation, whereas the activation of HAL9 and YHL013 probably depends on different factors. The increased accessibility of DNA to RNA polymerase II at translocation breakpoints may reflect a more open state of their chromatin structure, or its remodeling, which allows the DNA to be more accessible to the transcription machinery. In Fig. S6 a and c, histone H3 lysine-14 acetylation pattern, one landmark of transcriptionally active chromatin (24) was changed in a corresponding manner in the D10 translocants. In this case, juxtaposition of originally distant genomic sites with different chromatin conformations may have led to the remodeling of the newly generated chromatin structure. These conclusions are corroborated by results from mammalian cell cultures, in which the effects of translocation are observed well after its occurrence. For instance, an X chromosome–autosome translocation led to spreading of the X chromosomal gene silencing effect on autosomal genes (25). Moreover, in some Burkitt's lymphoma patients, c-Myc expression was activated by juxtaposition to the Ig heavy-chain gene HS3 enhancer, which actually had increased histone-3 and histone-4 acetylation on P1 c-Myc promoter via the recruitment of the CBP histone acetylase (26).
By using BIT translocations, similar to those described in this work, it would be possible to obtain more information on chromatin remodeling at translocation breakpoints at any locus of the yeast genome, immediately after the initial translocation event.
According to previous works (20, 27), a decreased Clb1p level suggests that the majority of translocants had impairments in the completion of the S phase of the cell cycle. Besides the DSBs necessary for the homologous DNA integration, translocations produced some DNA damage, most likely DSBs mimicked by the free DNA ends of the telomeric fragments that eventually were recovered by ectopic integration on chromosome XI (Fig. 1, CDC33 and MSH2 panels). The cellular checkpoint could have brought the cells to a G2/M arrest until completion of the repair. Indeed, G2/M-arrested cells with elongated buds can be observed in populations of translocants (Fig. 2 f–h). Also, the translocants manifested a growth delay of several hours, and a G1 phase arrest (Fig. 6), which, together with the decrease in Rad53p and the absence of its phosphorylation, is an indication of cell cycle adaptation to the initial DNA damage generated by the translocation. In some cases, the repair of this DNA damage seems to have led to serious genome rearrangements and to the appearance of severely defective cells, some examples of which are presented in Fig. 2 a–c. The heterogeneity of chromosomal translocation processing has been confirmed by selection of D10 Big strain from D10 Small. Indeed, these two translocants are different at least by the rearrangement of the right arm of chromosome XV, which shows some duplicated region in D10 Big strain (Fig. 1b, D10 Big and Small lanes in the ALG6 panel).
Another interesting feature of the translocants is the increased level of a putative modified actin1 form (Fig. 5, Act1 line) found in D10 Small and D11 translocants, which could explain the distortion of the normal cell bud morphology.
A most important conclusions deriving from our observations is that a single translocation event leads to a successive cascade of molecular events eventually ensuing in genomic instability. From an evolutionary point of view, that is probably why GCR formation is normally suppressed by many factors (10) because it is usually strongly deleterious to the cells. However, in some environmental conditions, chromosomal rearrangements like translocations may potentially provide a selective advantage, having represented quantum adaptive steps in species evolution history (28), albeit being mainly reciprocal. In this context, “a more thorough understanding of the mechanisms that cause chromosome translocations will be aided by developing in vitro and in vivo model systems that can generate this type of translocations” (13). In this case, the translocation breakpoints location is known beforehand, eliminating the work for its mapping. The procedure also establishes an evident temporal order of events, preempting the question of whether the translocation caused the cellular defects or it was the result of them.
These first results on the overall effects of a specific, induced chromosome translocation could be instrumental in elucidating the molecular mechanism underlying genome rearrangements generated by DNA integration, such as retrotransposition, characteristic of many types of cancer in mammalian cells.
Methods
Strains and Media.
The strains used in this work were: SanI (MAT a/α ura3–52/ura3–52, lys2–80/+, ade1/+, ade2–10/ade2, ade8/+, trp1-Δ1/trp1–289, his3-Δ200/+, leu2Δ1/leu2, can1/+, ARG4:FRTG-NLS-tetR-GFP/ARG4::FRTG-DsRed), and its derivatives D10, D3, AD5, and D11 obtained previously (14). Strains Susu1 and Susu2 were obtained in this work by BIT, constructed with primers SSU1 For-65 and Suc2 Rev-65 (SI Table S2).
Light and Fluorescence Microscopy.
Light microscopy of live cells was performed with a Leica DMBL photomicroscope equipped with a CCD computer-driven camera at 60× and 100× magnifications. For nuclei staining, cells were embedded in Vectashield mounting medium (Vector Laboratories) according to the manufacturer's instructions. Actin cytoskeleton was visualized with fluoresceinisothiocyanate–phalloidin (FITC–phalloidin; Sigma), according to the manufacturer's instructions.
Bridge-Induced Translocation.
Translocants were obtained by using the BIT technique developed previously (14), exploiting the EUROFAN protocol (29) for PCR-based gene replacements with the lithium acetate transformation method (30). The list of primers used for BIT is given in Table S2. Correct chromosomal integrations were confirmed by PCR and then by sequencing.
Southern Blot Analysis.
Chromosomal separation was performed in a 1% pulse-field certified agarose (Bio-Rad) gel electrophoresis with the CHEF DR-II apparatus (Bio-Rad) as described in ref. 32. Hybridizations were optimized from standard protocols (29). Probes were labeled by using the PCR digoxygenin probe synthesis kit (PCR DIG; Roche).
Chromosome positions were calculated from bands of the gel in Fig. S1 by trinomial equation: y = −7.87465 x3 + 757.298x2 − 55.019x + 1890.16.
Quantitative RT-PCR.
Expression of the genes of interest was analyzed by quantitative RT-PCR. Total RNAs were isolated by using the RNeasy mini kit (Qiagen), and ≈1 μg was used for total cDNAs synthesis by avian myeloblastosis virus reverse transcriptase (Promega). Then PCR was carried out with Thermus aquaticus DNA polymerase (Promega) by a standard quantitative amplification protocol. Sequences of primers used are given in Table S2. As a control of steadily expressed gene, HSC82 mRNA level was chosen as described in ref. 16. The intensity of the bands was analyzed by laser-scanning densitometry (UltroScan XL; Pharmacia LKB) and normalized to the intensity value of the housekeeping gene HSC82. The final data were reported as normalized expression level of every gene analyzed with respect to its parental copy in the SanI strain considered as the unit. The experiments were repeated at least three times, and standard deviations were calculated.
DNA Copy Number Determination.
Copy number of genes at the translocation breakpoints was determined by a standard quantitative PCR with limiting number of cycles (18–22). Sequences of primers used are given in Table S2. As a control of copy number of genes located on other chromosomes, PRK1 (chromosome IX), ARK1 (chromosome XIII), and HSC82 (chromosome XIV) were used. Intensity of the bands was analyzed by laser densitometer (UltroScan XL) and normalized to intensity values of the genes located on chromosomes not involved in the translocation.
Microarray Analysis.
Microarray transcription profiling was performed four times for redundancy with the S. cerevisiae yeast oligonucleotide microarray kit version, 4 × 44K slide platform (Agilent) following the manufacturer's instructions. Only genes that revealed expression changes with a P value <0.01, were considered. Gene expression data are accessible at URL: ftp://ftp/pub/tmp/, folder: Yeast D10 Small ∼ WT SanI.
ChIP.
ChIP on formaldehyde-fixed yeast cells was performed essentially as described in ref. 31 and micrococcal nuclease treatment as in ref. 32. Immunoprecipitation was performed with monoclonal antibodies to RNA polymerase II (Covance) and polyclonal antibodies to acetylated lysine-14 of H3 histone (Upstate Biotechnology). Primers used for amplification were designed to amplify fragments corresponding to positions −250 to +50 of the genes used for RT-PCR profiling at the translocation breakpoints; their sequences are given in Table S2. The bands were analyzed by laser-scanning densitometry (UltroScan XL). Intensity of all bands was normalized first to input values, then to the intensity value of the HSC82 promoter band. Final data were reported as for quantitative RT-PCR.
FACS Analysis.
Flow cytometry cell analysis has been performed as described (33, 34). The samples were analyzed by using a Beckman Coulter fluorescence-activated cell sorter. The DNA content reflects an average from ≈100,000 cells.
Western Blot Analysis.
Western blot analysis was performed by the ECL advanced Western blotting detection kit (Amersham) according to the manufacturer's instructions. The concentration of soluble proteins was determined by the protein assay reagent from Bio-Rad, based on Bradford's technique (35). For the protein detection were used monoclonal antibodies EL7 to all isoforms of Rad53p and F9 to its phosphorylated form; polyclonal antibodies to cyclin B1 (Santa Cruz Biotechnology), and monoclonal antibodies to actin1 (Abcam). As control, antibodies to acetylated lysine-14 of histone H3 were used.
Supplementary Material
Acknowledgments.
We thank Beatrice Rossi (International Centre for Genetic Engineering and Biotechnology) for providing two Susu yeast translocant strains; Tom Petes, Ian Hickson, Michael Breitenbach, and Marco Foiani (Instituto Firc di Oncologia Moleculare, Milan, Italy) for fruitful scientific discussion of the results, and Achille Pellicioli and Marco Foiani for the Rad53 antibodies. D.N. is a postdoctoral fellow supported by the International Centre for Genetic Engineering and Biotechnology. We acknowledge the Functional Analysis and Biochips Group of the University of Ljubljana for technical assistance and fruitful bioinformatic discussion of the array data.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800464105/DCSupplemental.
References
- 1.Nowell PC, Jackson L, Weiss A, Kurzrock R. Historical communication: Philadelphia-positive chronic myelogenous leukemia followed for 27 years. Cancer Genet Cytogenet. 1988;34:57–61. doi: 10.1016/0165-4608(88)90169-0. [DOI] [PubMed] [Google Scholar]
- 2.de Klein A, et al. A cellular oncogene is translocated to the Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1982;300:765–767. doi: 10.1038/300765a0. [DOI] [PubMed] [Google Scholar]
- 3.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowley JD. Chromosome translocations: Dangerous liaisons revisited. Nat Rev Cancer. 2001;1:245–250. doi: 10.1038/35106108. [DOI] [PubMed] [Google Scholar]
- 5.Staudt LM. Molecular diagnosis of the hematologic cancers. N Engl J Med. 2003;348:1777–1785. doi: 10.1056/NEJMra020067. [DOI] [PubMed] [Google Scholar]
- 6.Grady WM. Genomic instability and colon cancer. Cancer Metastasis Rev. 2004;23:11–27. doi: 10.1023/a:1025861527711. [DOI] [PubMed] [Google Scholar]
- 7.Putnam CD, Pennaneach V, Kolodner RD. Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:13262–13267. doi: 10.1073/pnas.0405443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee S, Myung K. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot Cell. 2004;3:1557–1566. doi: 10.1128/EC.3.6.1557-1566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal S, Tafel AA, Kanaar R. DNA double-strand break repair and chromosome translocations. DNA Repair. 2006;5:1075–1081. doi: 10.1016/j.dnarep.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 10.Huang ME, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell. 2005;17:709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Myung K, Smith S, Kolodner RD. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:15980–15985. doi: 10.1073/pnas.0407010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang JY, Smith S, Myung K. The Rad1–Rad10 complex promotes the production of gross chromosomal rearrangements from spontaneous DNA damage in Saccharomyces cerevisiae. Genetics. 2005;169:1927–1937. doi: 10.1534/genetics.104.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aplan PD. Causes of oncogenic chromosomal translocation. Trends Genet. 2006;22:46–55. doi: 10.1016/j.tig.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosato V, Waghmare SK, Bruschi CV. Non-reciprocal chromosomal bridge-induced translocation (BIT) by targeted DNA integration in yeast. Chromosoma. 2005;114:15–27. doi: 10.1007/s00412-005-0332-x. [DOI] [PubMed] [Google Scholar]
- 15.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 16.Anthony C, Zong Q, De Benedetti A. Overexpression of eIF4E in Saccharomyces cerevisiae causes slow growth and decreased α-factor response through alterations in CLN3 expression. J Biol Chem. 2001;276:39645–39652. doi: 10.1074/jbc.M101564200. [DOI] [PubMed] [Google Scholar]
- 17.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 18.Lowndes NF, Murguia JR. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 19.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 20.Grandin N, Reed SI. Differential function and expression of Saccharomyces cerevisiae B-type cyclins in mitosis and meiosis. Mol Cell Biol. 1993;13:2113–2125. doi: 10.1128/mcb.13.4.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surana U, et al. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 22.Moseley JB, Goode BL. The yeast actin cytoskeleton: From cellular function to biochemical mechanism. Microbiol Mol Biol Rev. 2006;70:605–645. doi: 10.1128/MMBR.00013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romanienko PJ, Camerini-Otero RD. Cloning, characterization, and localization of mouse and human SPO11. Genomics. 1999;61:156–169. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 24.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 25.Popova BC, Tada T, Takagi N, Brockdorff N, Nesterova TB. Attenuated spread of X-inactivation in an X;autosome translocation. Proc Natl Acad Sci USA. 2006;103:7706–7711. doi: 10.1073/pnas.0602021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu HM, Kanda K, Zhang L, Boxer LM. Activation of the c-myc p1 promoter in Burkitt's lymphoma by the hs3 immunoglobulin heavy-chain gene enhancer. Leukemia. 2007;21:747–753. doi: 10.1038/sj.leu.2404583. [DOI] [PubMed] [Google Scholar]
- 27.Lee SE, et al. Saccharomyces Ku70, mre11/rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 28.Colson I, Delneri D, Oliver SG. Effects of reciprocal chromosomal translocations on the fitness of Saccharomyces cerevisiae. EMBO Rep. 2004;5:392–398. doi: 10.1038/sj.embor.7400123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 30.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 32.Keller C, Ladenburger EM, Kremer M, Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J Biol Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 33.Bruschi CV, Chuba PJ. Nonselective enrichment for yeast adenine mutants by flow cytometry. Cytometry. 1988;9:60–67. doi: 10.1002/cyto.990090110. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Flanary PL, Altieri DC, Dohlman HG. Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J Biol Chem. 2000;275:6707–6711. doi: 10.1074/jbc.275.10.6707. [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.