Abstract
The skeletal muscle Ca2+ release channel (RyR1), a homotetramer, regulates the release of Ca2+ from the sarcoplasmic reticulum to initiate muscle contraction. In this work, we have delineated the RyR1 monomer boundaries in a subnanometer-resolution electron cryomicroscopy (cryo-EM) density map. In the cytoplasmic region of each RyR1 monomer, 36 α-helices and 7 β-sheets can be resolved. A β-sheet was also identified close to the membrane-spanning region that resembles the cytoplasmic pore structures of inward rectifier K+ channels. Three structural folds, generated for amino acids 12–565 using comparative modeling and cryo-EM density fitting, localize close to regions implicated in communication with the voltage sensor in the transverse tubules. Eleven of the 15 disease-related residues for these domains are mapped to the surface of these models. Four disease-related residues are found in a basin at the interfaces of these regions, creating a pocket in which the immunophilin FKBP12 can fit. Taken together, these results provide a structural context for both channel gating and the consequences of certain malignant hyperthermia and central core disease-associated mutations in RyR1.
Keywords: Ca2+ release channels, cryo-EM, 3D Structure, modeling
Calcium is a major messenger that mediates a variety of cell functions including muscle contraction, brain functions, chemical senses, light transduction, cell proliferation, fertilization, secretion, and immune response. Ca2+ signaling is based on ability of cells to maintain low levels of Ca2+ (≈10−7 M) under resting conditions and to create a rapid, transient increase of Ca2+ (≈10−6 M) upon the stimulated entry of Ca2+ through the plasma membrane and/or induction of Ca2+ release from intracellular Ca2+ storage organelles such as endoplasmic reticulum/sarcoplasmic reticulum.
Ryanodine receptors (RyR) are members of the Ca2+ release channel superfamily, which includes intracellular tetrameric channels activated by different mechanisms. RyR1 functions primarily as a Ca2+ release channel in the sarcoplasmic reticulum membrane of skeletal muscle. The largest known ion channel (≈2.3 MDa), RyR1 is composed of four identical subunits arranged around a centrally located Ca2+ pathway. Each subunit is organized into two principal regions: a transmembrane (TM) region and a cytoplasmic (CY) region (1, 2). Membrane-spanning portions of each subunit form an ion-conducting pore whereas the CY region regulates gating via interaction with a variety of intracellular messengers, including immunophilin, FKBP12, calmodulin, ATP, Ca2+, and Mg2+ ions. Many mutations found in the CY region of RyR1 are known to affect its gating and underlie diseases such as malignant hyperthermia (MH) and central core disease (CCD) (3). Thus, elucidating the molecular mechanisms by which various stimuli are received in the CY region and regulate the channel gating will be instrumental in discovering means to treat these diseases.
Previously, we were able to determine the structure of RyR1 in a closed conformation to 9.6-Å resolution using single-particle electron cryomicroscopy (cryo-EM) (4). A similar structure of RyR1 was also reported by Samso et al. (5). In this work, we extend our analysis of RyR1 and exploit computational methods to determine plausible mechanisms by which cellular modulators control the channel.
Results
Segmentation of Individual Subunits.
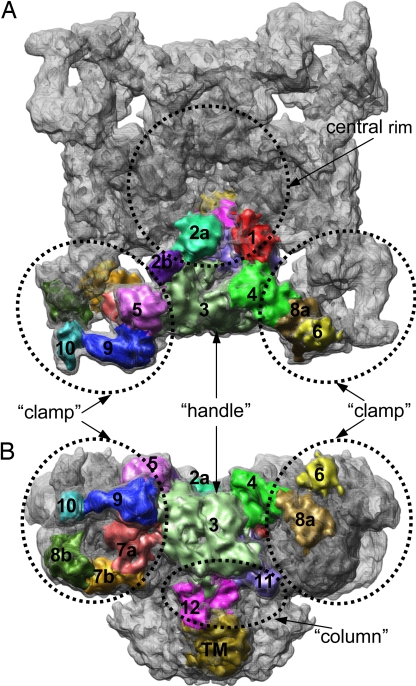
Given the large size of RyR1 and the complex allosteric modulation of its gating by many cellular molecules, the channel is expected to be composed of multiple structural and/or functional domains. Although the quaternary structure of our subnanometer-resolution RyR1 map in the closed state is fully consistent with earlier observations at low resolution (6–8), our higher-resolution density map is sufficiently resolved to identify molecular boundaries between the individual subunits and to divide the CY region into 15 subregions per subunit [Fig. 1 and supporting information (SI) Movie S1]. The subregions are grouped into several morphologically distinct units previously referred to as the “clamps” (6–8), which are connected through the “handle” surrounding the central rim. The “column” regions consist of the bridging densities between the CY and the membrane-spanning regions (Figs. 1B and 2A). Segmentation of the TM region is also in agreement with the pore structure of the RyR1 channel proposed earlier (4).
Fig. 1.
A 9.6-Å resolution cryo-EM density map of RyR1 in closed state. The map is displayed at the threshold level corresponding to channel molecular mass of ∼2.3 MDa and viewed from cytoplasm (A) and in a side view (B). Subregions are shown within one of the RyR1 subunits. The numbering of subregions is adopted according to a previous convention except with finer divisions (43). A subregion is interpreted as compact protein density, and the boundary of subregion tends to be weakly connected with its adjacent densities. A subregion may thus consist of a single/multiple protein fold or of a part of a protein fold. Poorly connected densities between subregions 6 and 8a and between 10 and 8b may be hinges between subregions with high local structural flexibility. The individual subregions are mapped into the distinct morphological units: clamp is formed by subregions 5, 7a, 7b, 8b, 9, and 10 from one monomer and by subregions 6 and 8a from adjacent subunit; handle is formed by subregions 3 and 4; the central rim is formed by subregions 1, 2a, and 2b; and column is formed by subregions 11 and 12.
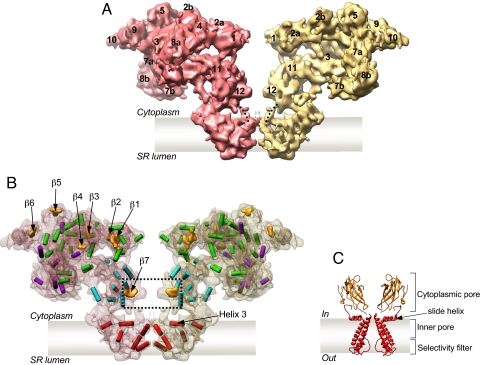
Fig. 2.
Secondary structure elements in the structure of RyR1. (A) Two opposing RyR1 subunits from the tetramer are shown in a side view; thus, two different faces of the RyR1 subunit are seen. Subregions are numbered. Note that densities corresponding to subregion 6 are missing at the chosen high threshold. (B) Two subunits are shown as semitransparent surfaces with identified secondary structure elements. α-Helices are annotated as cylinders colored according to their locations in the map: red, TM region; green, central part of the CY region; purple, clamps; cyan, column region. β-Sheets are shown as orange surfaces. Note that a substantial fraction of the density is not assigned in this interpretation probably because of the structural flexibility or limited resolution. A dashed line indicates β-sheets in the column region that are connected with the TM region through bridging densities marked with dotted lines in A. (C) X-ray structure of KirBac1.1 channel; two subunits are removed to see the structural elements in the conduction pathway in the KirBac1.1 (11) (see also Fig. S1).
Secondary Structure Elements in the CY Region.
At subnanometer resolution, secondary structure elements can be determined by using the feature detection program SSEHunter (9). Our earlier analysis of the TM region revealed the positions of the pore-lining helix and the pore helix (4), which bore a remarkable similarity to those of the MthK channel (10). Extending our computational analysis to the CY region, 36 α-helices with various orientations and seven β-sheets (Fig. 2 A and B and Movie S1) are identified in the CY region of each RyR1 subunit. Three sheets are found in the central part of the CY region: β1 and β2 sheets in subregions 1 and 2a form a central rim, and β3 in domain 4 is bridging the central rim with the clamp. The structure of the clamp is characterized by seven α-helices and three β-sheets (β4, β5, and β6), identified in subregions 8a, 5, and 9, respectively. Eight α-helices are in the column regions and maintain the connection between the TM and the CY regions.
Particularly notable is β-sheet (β7), located in the constricted part of the column region (subregion 12) connecting the TM and CY regions (Fig. 2 A and B). β-Sheets at equivalent locations have been identified in the crystal structures of inward rectifier K+ channels (Kir channels) (11–14) and a cyclic nucleotide-modulated (HCN2) channel (15) (Fig. 2C and Fig. S1). In Kir channels, this β-sheet has been proposed to form a part of the cytoplasmic pore, which is connected to the inner pore. In RyR1, β7 sheet is connected via bridging densities to helix 3, which lies parallel to the membrane/cytoplasmic interface in the TM region (Fig. 2B). Helix 3 has been suggested to play an essential role in gating (4); a helix with a similar orientation close to the membrane surface (designated as the slide helix) was also seen in the KirBac1.1 crystal structure (11) (Fig. 2C). Analogous to Kir channels, subregion 12, containing β7, in RyR1 may play a regulatory role in channel gating by binding to cellular modulators yet to be determined.
Pseudoatomic Model of N-Terminal Domains.
Because of the absence of a high-resolution structure for any domain in RyR1, we used comparative modeling to further interpret the structure. Using sequence and structural analysis (16–18), a structural homolog was identified with high confidence for two segments at the RyR1 N terminus: the ligand-binding suppressor domain [Protein Data Bank (PDB) ID code 1XZZ (19)] for residues Q12–S207 and the IP3-binding core region [PDB ID code 1N4K (20)] of the type 1 inositol 1,4,5-trisphosphate receptor (IP3R1) for residues G216–Y565 (Fig. 3A and Figs. S2 and S3). Initial comparative models for these segments (models 1 and 2, respectively) (Fig. 3A) were constructed with MODELLER 9.0 (21).
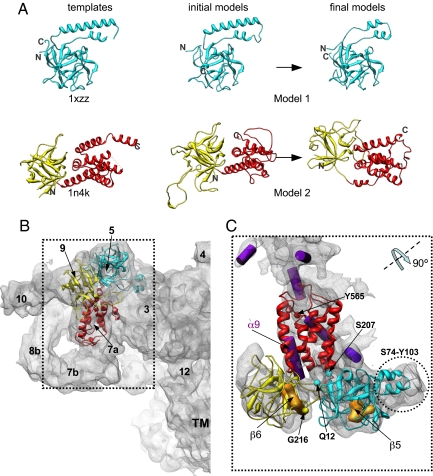
Fig. 3.
Localization of homology models for the N-terminal region of the RyR1 in the 3D map. (A) Comparative models for the N-terminal region of RyR1 and corresponding structural templates. Model 1 (residues Q12–S207) is shown with cyan ribbon; model 2 is composed of two parts, shown with yellow (residues G216–T407) and red (residues A408–Y565) ribbons. Two parts of model 2 were built and refined independently to obtain better fits to the cryo-EM map. Final models were obtained through refinement of initial models using Moulder-EM (22). Dashed lines in template structures indicate regions that were not resolved in original x-ray structures (19). (B) The RyR1 monomer is shown in a side view along with homology models docked to the clamp region. Subregions are numbered. (C) Refined homology models are shown fitted within the density region segmented from the 9.6-Å map. The segmented density region is displayed in the orientation derived from its position in B by rotation to ∼90° around the axis as shown. SSEHunter-identified α-helices and β-sheets are shown with purple cylinders and yellow solid densities, respectively. N- and C-terminal residues in both models are indicated.
The N-terminal sequences of RyR1 have relatively low sequence identity to the template sequences (19% and 16% for models 1 and 2, respectively). Using standard techniques, such low sequence identity would likely result in inaccuracies in the sequence alignment, propagating model errors. Therefore, we applied a constrained homology modeling approach, Moulder-EM, to iteratively optimize target–template alignment, the corresponding model, and its fit into the cryo-EM density map (22) (Fig. S2). The resulting models for the two segments were combined into a single model for the N-terminal region and further refined by energy minimization with the X-PLOR software package (Fig. S4) (23) and real-space cryo-EM fitting with Flex-EM (24). Although the overall fold was preserved in this refinement process, the final models were substantially altered to optimally match the cryo-EM density (Fig. 3A).
Most of model 1 (Fig. 3 B and C and Movie S2) localizes to subregion 5 except for residues S74–Y103, which localize to subregion 3. The N- and C-terminal portions of model 2 (Fig. 3B) are found in subregions 9 and 7a, respectively. Our confidence in placement of the density corresponding to the N terminus of RyR1 within the cryo-EM map is substantiated by previous difference imaging with GST-RyR and GFP-RyR fusion proteins and antibody labeling (Fig. S5) (25–27). Model placement is further validated by the match of the β-sheets β5 and β6 and a long helix (α9) independently derived from the aforementioned SSEHunter results (Fig. 3C). Although there are five other helices in our models, they are two turns or shorter and thus would not normally be detectable at this resolution (Fig. 3C and Fig. S3 A and B). Whereas localization of the N-terminal domain is consistent with our previous work (26), our current models differ from the previously proposed N-terminal model, based on an isocitrate dehydrogenase (4ICD) template. Updated structural databases and advances in fold recognition now predict a more accurate structural template resulting in the current models (Fig. 3).
Mapping of MH- and CCD-Associated Mutations.
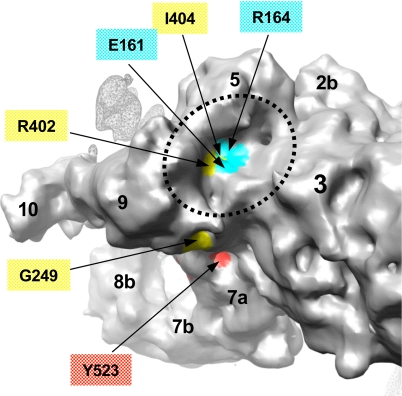
A number of mutations linked to MH and CCD are associated with enhanced sensitivity of the RyR1 channel to activators (3). As mentioned earlier, many of these mutations localize to the CY region and, in particular, to the N-terminal sequence of RyR1 that we have modeled. Using our models, the aforementioned mutations are mapped onto the cryo-EM density map of RyR1 in the closed state (4) (Figs. 4 and 5). Eleven of 15 disease-associated mutations would be surface-exposed when mapped to the density map. Four residues (E161, R164, R402, and I404) would be found in a cluster at the surface of the basin formed by subregions 3, 5, and 9 (Fig. 5).
Fig. 4.
Mapping of MH/CCD-linked residues onto the homology models for the N-terminal region of RyR1. Model 1 (cyan) and the model 2 (red and yellow) are shown docked within the EM density map displayed with a mesh. MH/CCD-associated residues are shown with spheres and are labeled. Labels highlighted with color mark residues exposed on the protein surface.
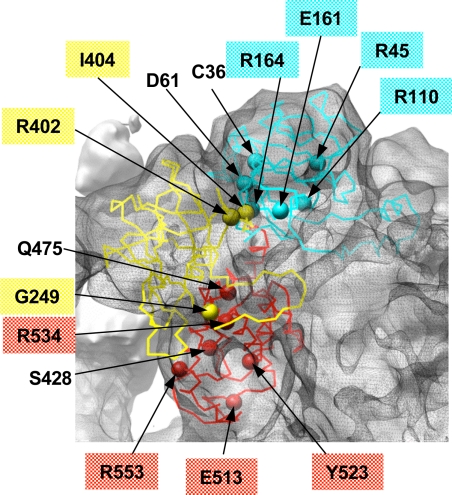
Fig. 5.
Identification of MH/CCD-linked residues in the FKBP12 binding pocket. The surface of the clamp in RyR1 is color-coded based on location of residues associated with MH and CCD mutations in the N-terminal homology models docked to the 3D map of RyR1. Cyan surface corresponds to residues from model 1, and yellow and red correspond to residues from model 2 (see color code used in Figs. 3 and 4). Putative surface-exposed mutation residues are labeled. The basin between subregions 3, 5, and 9 (indicated with dashed line) is proposed to form the FKBP-12 binding site in RyR1 (31, 32) and is identified to include four surface-exposed MH/CCD mutation sites (E161, R164, R402, and I404). The surface of the adjacent RyR1 subunit is shown with a mesh.
Discussion
RyR1 is the largest known ion channel and is among the most challenging proteins to study using single-particle cryo-EM analysis. The use of computational methods for identifying secondary structure elements and comparative modeling combined with cryo-EM density fitting have allowed us to derive a molecular model for the N-terminal domains of RyR1 in the clamp subregions.
The IP3-binding domain and the ligand-binding suppressor domain play an important and well established role in the regulation of the IP3 receptor channel function (28). The significance of the structural similarity within the N terminus of two Ca2+ release channels, RyR1 and IP3R1, is unclear because RyRs lack the ability to bind IP3 (S.L.H., unpublished data). The structural similarity may, however, suggest a structurally conserved mechanism in this channel family for regulating channel activity yet to be determined in this region. Although this regulation mechanism may not yet be known, RyR1 channel opening is associated with changes in the conformations of both the TM and CY regions, and there appears to be a correlation between the conformation of the clamp and channel opening (6, 8). The mapping of MH- and CCD-associated mutations to the clamp subregions suggests that these mutations potentially could destabilize the domain structure and effect conformational motions in the clamp subregions associated with the channel gating.
Furthermore, the subregions of RyR1 in the clamp are implicated in binding various cellular proteins including the immunophilin FKBP12, which is known to stabilize the closed state of the RyR1 channel (29, 30). The binding site for FKBP12 was previously localized to the clamp, proximal to the interface among subregions 3, 5, and 9 (31, 32) (Fig. 1). Given the putative location of the FKBP12-binding site within the 3D map of RyR1, the four MH/CCD-associated mutation sites (E161G, R164C, R402C, and I404M) exposed to the basin formed by subregions 3, 5, and 9 (Fig. 5) may disrupt the RyR1–FKBP12 interactions, thus altering the gating of the RyR1 channel.
In addition, based on freeze fracture studies (33), these and other RyR1 clamp subregions have been implicated to be involved in allosteric modulation of the channel gating through interaction with the voltage-gated L-type Ca2+ channel (also known as dihydropyridine receptor) in the transverse tubule membrane. As such, the RyR1 N-terminal regions may play a role in signal transduction from the dihydropyridine receptor to the TM region.
Using a combination of computational tools, we have been able to provide a structural framework for RyR1, which will help in the design of future experiments to explore the roles of the various domains of RyR1 in channel function and regulation. The structural similarities observed between RyR1 and other ion channels in both the CY and TM domains (4, 5) are striking, and future structures of these channels complexed with cellular regulators should provide more details concerning the mechanisms of cellular signaling in the process of muscle contraction.
Materials and Methods
Cryo-EM and Image Processing.
Specimen preparation, cryo-EM imaging, and 3D reconstruction were described in detail previously (4).
Segmentation of Cryo-EM 3D Map.
Segmentation of individual RyR1 subunits was initially accomplished by using an automated program called segment3d included in the EMAN package (34). The Amira visualization software package (TGS) was used to perform the final, manual segmentation. The segmentation was quite clear over most of the map, particularly in the TM region; however, in the CY region there were several points at which the separation between subunits was ambiguous depending on the parameters used.
Identification of Secondary Structure Elements.
α-Helices and β-sheets in the RyR1 cryo-EM density map were identified by using SSEHunter (9), which identifies α-helices equal to or longer than two turns and β-sheets equal to or larger than two strands. Secondary structure elements were also visually assessed, and only those elements identified by using both quantitative and visual methods are presented in the final interpretation.
Target Identification.
In the sequence homology detection for RyR1 [National Center for Biotechnology Information (NCBI) protein sequence ID gi: 134134] residues M1–L600 was first attempted using a HMM–HMM comparison with HHpred (18). The analysis revealed significant structural similarity to two crystal structures (100% probability, E = 0), both of which contain a β-trefoil fold domain (35). Additional support came from the threading program mGenThreader (16), which identified the same two homologs with very high confidence (P values are 2e-05 and 9e-07). Residues Q12–S207 share a fold with the ligand binding suppressor domain (PDB ID code 1XZZ, sequence identity 19%) (19). Residues G216–Y565 share a fold with the IP3-binding core of the IP3R1 channel (PDB ID code 1N4K, sequence identity 16%) (Figs. S2 and S3) (20).
Model Building and Refinement with Moulder-EM.
Two comparative models of residues Q12–S207 (model 1) and G216–Y565 (model 2) were independently calculated based on sequence structure alignment generated with the program FUGUE, version 2.0 (36) (to 1XZZ and to 1N4K, respectively), using the automodel class in MODELLER 9.0 (Figs. S2 and S3) (21). Fold assessment was performed by using a newly developed protocol optimized specifically for predicting the accuracy of a model in the absence of its native structure (D.E., N. Eswar, M.-Y. Shen, and A.S., unpublished observations). The protocol constructs a model-specific scoring function using a support vector machine, which optimizes the weights of sequence similarity measures and statistical potentials extracted from a tailored training set of models of similar size and the same secondary structure composition as the model being assessed. Models predicted to have native overlap values of ∼0.3 or greater are expected to have the correct fold; native overlap is defined as the fraction of Cα atoms in a model that are within 3.5 Å of the corresponding atoms in the native structure after rigid body superposition of the model to the native structure. The predicted native overlap values for model 1 and model 2 were 0.4 and 0.28, respectively, indicating that both folds are likely correct and that the models are of relatively low accuracy.
These initial models were localized to the cryo-EM density by using the exhaustive fitting program FoldHunter (37), available as a plugin through the AIRS/EMAN package for UCSF Chimera. Individual models were docked to an entire RyR1 monomer without restrictions to any location. The fit of each model was further optimized by using the Monte Carlo optimization protocol of the density-fitting program Mod-EM (38), resulting in a moderately high cross-correlation coefficient between the model and the cryo-EM map of 0.68. The match of the secondary structure elements between the models and the SSEHunter results (9) provided additional support for the localization of the models in this region. Next, we used the density to improve the quality of the models (which were based on low sequence similarity to the template structure) using the Moulder-EM protocol (22). This protocol optimizes a comparative model by iterating over sequence alignment, model building, and model assessment; model assessment is based on a combined score including the cross-correlation score between the fitted model and the density map (38) and an atomic statistical potential score (39).
Residues A311–E343 were excluded from model 2 because of lack of a structural template for this region; no loop modeling for this region was performed because of the large size of the sequence. For each of the loop regions A26–L35, A251–E263, and R493–S514 (which had relative low sequence similarity to the corresponding templates) 500 models were generated by the loopmodel class in MODELLER 9.0 (21, 40). The best loop model was chosen by a combination of the DOPE statistical potential score (41) and the cross-correlation score between the fitted loop and the density map (22).
Energy Minimization.
Two parts of model 2 (residues G216–T407 shown in yellow and residues A408–Y565 shown in red in Fig. S4) were built and refined independently to obtain better fits to the cryo-EM map. When taken together, there were steric clashes as well as a break in the main chain at the interface (residues E396–P430, highlighted with green in Fig. S4) between the N-terminal and the C-terminal portions of model 2. To eliminate these steric clashes and connect the main chain of model 2, the helix encompassing residues G409–D419 (the lower part of the broken helix shown in green in Fig. S4) was manually moved to a nearby position to avoid any clashes with other neighboring residues while maintaining a good fit to the cryo-EM density map. Afterward, residues E396–P430 (highlighted in green in Fig. S4), including helices G409–D419 and S422–S428 and adjacent connecting residues, were energy-minimized with X-PLOR (23) by using default parameters of the CHARMM united-atom force field (version 19) (42). The constrained structures were subjected to 12,000 steps of Powell energy minimization. Finally, the entire model was subjected to a real-space refinement within the density using Flex-EM (24), where the domains, some individual helices, and the atoms connecting them were defined as rigid bodies. The final cross-correlation coefficient between the model and the cryo-EM map improved slightly from 0.68 to 0.70.
Supplementary Material
Acknowledgments.
Movies were produced by Mathew Dougherty (National Center for Macromolecular Imaging, Baylor College of Medicine). This research is supported by National Institutes of Health Grants P41RR02250, PN2EY016525, P01GM99116, P01GM064692, R01AR44864, R01GM072804, and R01AR41802 and National Science Foundation Grants EIA-0325004, CNS 0420984, IIS-0705644, IIS-0705474, and IIS-0705196. M.T. is currently supported by the Medical Research Council Career Development Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The 3D cryo-EM map has been deposited in the European Molecular Biology Laboratory European Bioinformatics Institute database (accession no. 1275).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803189105/DCSupplemental.
References
- 1.Takeshima H, et al. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- 2.Zorzato F, et al. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1990;265:2244–2256. [PubMed] [Google Scholar]
- 3.Durham W, Wehrens X, Sood S, Hamilton SL. In: Calcium Signaling and Disease. Carfoli E, Brini M, editors. New York: Springer; 2007. pp. 273–321. [Google Scholar]
- 4.Ludtke SJ, Serysheva II, Hamilton SL, Chiu W. The pore structure of the closed RyR1 channel. Structure (Cambridge) 2005;13:1203–1211. doi: 10.1016/j.str.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samso M, Wagenknecht T, Allen PD. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat Struct Mol Biol. 2005;12:539–544. doi: 10.1038/nsmb938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlova EV, Serysheva II, van Heel M, Hamilton SL, Chiu W. Two structural configurations of the skeletal muscle calcium release channel. Nat Struct Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- 7.Serysheva II, Hamilton SL, Chiu W, Ludtke SJ. Structure of Ca2+ release channel at 14 Å resolution. J Mol Biol. 2005;345:427–431. doi: 10.1016/j.jmb.2004.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serysheva II, Schatz M, van Heel M, Chiu W, Hamilton SL. Structure of the skeletal muscle calcium release channel activated with Ca2+ and AMP-PCP. Biophys J. 1999;77:1936–1944. doi: 10.1016/S0006-3495(99)77035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–22. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 11.Kuo A, et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science. 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 12.Nishida M, Cadene M, Chait BT, Mackinnon R. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 2007;26:4005–4015. doi: 10.1038/sj.emboj.7601828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishida M, MacKinnon R. Structural basis of inward rectification: Cytoplasmic pore of the G protein-gated inward rectifier GIRK1 at 1.8 Å resolution. Cell. 2002;111:957–965. doi: 10.1016/s0092-8674(02)01227-8. [DOI] [PubMed] [Google Scholar]
- 14.Pegan S, et al. Cytoplasmic domain structures of Kir2.1 and Kir3.1 show sites for modulating gating and rectification. Nat Neurosci. 2005;8:279–287. doi: 10.1038/nn1411. [DOI] [PubMed] [Google Scholar]
- 15.Zagotta WN, et al. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425:200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- 16.McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–881. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 17.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 18.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosanac I, et al. Crystal structure of the ligand binding suppressor domain of type 1 inositol 1,4,5-trisphosphate receptor. Mol Cell. 2005;17:193–203. doi: 10.1016/j.molcel.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 20.Bosanac I, et al. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- 21.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 22.Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J Mol Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- 23.Brunger A. New Haven CT: Howard Hughes Medical Institute and Department of Molecular Biophysics and Biochemistry, Yale Univ; 1992. X-PLOR: A system for X-ray crystallography and NMR. Version 3.1. [Google Scholar]
- 24.Topf M, et al. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, et al. Three-dimensional reconstruction of the recombinant type 3 ryanodine receptor and localization of its amino terminus. Proc Natl Acad Sci USA. 2001;98:6104–6109. doi: 10.1073/pnas.111382798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker ML, et al. The skeletal muscle Ca2+ release channel has an oxidoreductase-like domain. Proc Natl Acad Sci USA. 2002;99:12155–12160. doi: 10.1073/pnas.182058899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R, et al. Localization of an NH2-terminal disease-causing mutation hot spot to the “clamp” region in the three-dimensional structure of the cardiac ryanodine receptor. J Biol Chem. 2007;282:17785–17793. doi: 10.1074/jbc.M700660200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikoshiba K. The IP3 receptor/Ca2+ channel and its cellular function. Biochem Soc Symp. 2007:9–22. doi: 10.1042/BSS0740009. [DOI] [PubMed] [Google Scholar]
- 29.Timerman AP, et al. The calcium release channel of sarcoplasmic reticulum is modulated by FK-506-binding protein. Dissociation and reconstitution of FKBP-12 to the calcium release channel of skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1993;268:22992–22999. [PubMed] [Google Scholar]
- 30.Brillantes AB, et al. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994;77:513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 31.Samso M, Shen X, Allen PD. Structural characterization of the RyR1-FKBP12 interaction. J Mol Biol. 2006;356:917–927. doi: 10.1016/j.jmb.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Wagenknecht T, et al. Locations of calmodulin and FK506-binding protein on the three-dimensional architecture of the skeletal muscle ryanodine receptor. J Biol Chem. 1997;272:32463–32471. doi: 10.1074/jbc.272.51.32463. [DOI] [PubMed] [Google Scholar]
- 33.Paolini C, Protasi F, Franzini-Armstrong C. The relative position of RyR feet and DHPR tetrads in skeletal muscle. J Mol Biol. 2004;342:145–153. doi: 10.1016/j.jmb.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 35.Murzin AG, Lesk AM, Chothia C. β-Trefoil fold: Patterns of structure and sequence in the Kunitz inhibitors interleukins-1β and 1α and fibroblast growth factors. J Mol Biol. 1992;223:531–543. doi: 10.1016/0022-2836(92)90668-a. [DOI] [PubMed] [Google Scholar]
- 36.Shi J, Blundell TL, Mizuguchi K. FUGUE: Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 37.Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: Computational tools for intermediate resolution structure interpretation. J Mol Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- 38.Topf M, Baker ML, John B, Chiu W, Sali A. Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J Struct Biol. 2005;149:191–203. doi: 10.1016/j.jsb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 39.John B, Sali A. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003;31:3982–3992. doi: 10.1093/nar/gkg460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiser A, Do RK, Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brooks BR, et al. CHARMM: A program for macromolecular energy minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 43.Radermacher M, et al. Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.