Abstract
Rds3p, a component of the U2 snRNP subcomplex SF3b, is essential for pre-mRNA splicing and is extremely well conserved in all eukaryotic species. We report here the solution structure of Rds3p, which reveals an unusual knotted fold unrelated to previously known knotted proteins. Rds3p has a triangular shape with a GATA-like zinc finger at each vertex. Pairs of cysteines contributing to each finger are arranged nonsequentially in a permuted arrangement reminiscent of domain-swapping but which here involves segments of subdomains within a single chain. We suggest that the structure arose through a process of segment swapping after gene duplication. The fingers are connected through β-strands and loops, forming an overall topology strongly resembling a “triquetra knot.” The conservation and surface properties of Rds3p suggest that it functions as a platform for protein assembly within the multiprotein SF3b complex of U2 snRNP. The recombinant protein used for structure determination is biologically active, as it restores splicing activity in a yeast splicing extract depleted of native Rds3p.
Keywords: knot, NMR, splicing
In eukaryotes, the majority of protein coding genes contain multiple noncoding sequences called introns that must be precisely removed from mRNA precursors (pre-mRNAs) to yield translatable mRNAs. This reaction is catalyzed by the spliceosome, which consists of the small nuclear ribonucleoprotein (snRNP) particles U1, U2, U4/U6, and U5 and hundreds of non-snRNP proteins (for reviews, see refs. 1 and 2). U2 snRNP plays an important role at each stage of the splicing cycle. Before the first step of splicing, it binds the pre-mRNA near the branch point adenosine, which is the nucleophile involved in the first trans-esterification reaction in splicing. Yeast U2 snRNP contains U2 snRNA, the seven Sm proteins that form a heptameric ring at the core of all snRNP particles, two U2 snRNP specific proteins (Lea1p and Yip9p), and the protein complexes SF3a and SF3b (ref. 3; for a review, see ref. 4).
Rds3p is a component of the U2 snRNP subcomplex SF3b, which for yeast has been reported to be an assembly of either six or eight proteins (3, 5, 6). It is extremely well conserved in all eukaryotic species and is essential for pre-mRNA splicing. In yeast with a heat-sensitive mutation in Rds3p, splicing is inhibited at nonpermissive temperatures, and spliceosomal assembly arrests at the U1-dependent commitment complex stage (7). Several proteins in SF3b cross-link to pre-mRNA near the branchpoint adenosine and to U2 snRNA toward the 5′ end, surrounding the branchpoint complementary sequence and thereby positioning SF3b, and hence Rds3p, near the catalytic center of the spliceosome (8–10). For human SF3b, there are some electron cryomicroscopy structures (11, 12) and atomic resolution structures of SF3b14a in complex with a small peptide of SAP155 (13, 14). However, there are as yet no structures reported for the yeast SF3b subcomplex or any of its components.
Here, we present the solution structure of Rds3p. Its primary sequence contains 13 cysteine residues, of which 10 are present within five CXXC motifs. Proteins with this particular motif have been annotated as belonging to the superfamily of “plant homeodomain” (PHD)-finger proteins (Interpro accession no. IPR005345), hence the alternative name for Rds3p in higher eukaryotes “PHD finger-like domain-containing protein 5A” (Phf5a). However, our structure shows that Rds3p has a new fold unrelated to known zinc finger proteins. This study sheds light on the role of this essential protein within the SF3b complex.
Results and Discussion
Overall Structure of Rds3p.
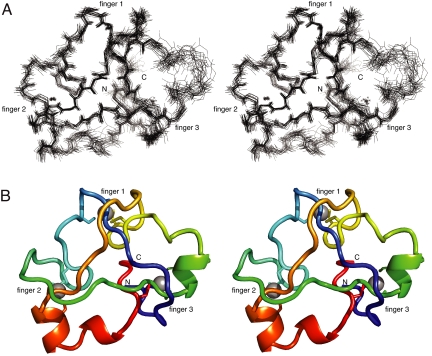
We have determined the solution structure of Rds3p by NMR spectroscopy [Table 1 and supporting information (SI) Text]. Residues Met-10 to Leu-90 form a compact globular domain (Fig. 1 and Fig. S1) whereas the remaining residues are unstructured as evidenced by the near-random coil chemical shift values of their resonances and the lack of any long-range NOE connectivities. The protein behaves as a monomer in solution (analytical gel filtration and native mass spectrometry); some new signals attributable to a dimeric species of Rds3p slowly appeared as samples aged, but this did not interfere with the structure calculation.
Table 1.
Structural statistics for the Rds3p ensemble
| Structural restraints | ||
| NOE-derived distance restraints | ||
| Intraresidue | 814* | 1,616† |
| Sequential | 477* | 814† |
| Medium (2 ≤ |i − j| ≤ 4) | 80* | 243† |
| Long (|i − j| > 4) | 269* | 668† |
| Ambiguous | 3,780* | 1,540† |
| Total | 5,420* | 4,881† |
| Dihedral constraints | ||
| Chi1 | 32 | |
| Statistics for accepted structures | ||
| Accepted structures | 20 | |
| Mean CNS energy terms (kcal·mol−1 ± SD) | ||
| E total | −3,250.3 ± 99.2 | |
| E van der Waals | −290.1 ± 20.3 | |
| E distance restraints | 83.5 ± 30.5 | |
| Restraint violations >0.3 Å (average number per structure) | 1.2 ± 0.9‡ | 1.1 ± 0.9§ |
| RMS deviations from the ideal geometry used within CNS | ||
| Bond lengths, Å | 0.0041 | |
| Bond angles, ° | 0.63 | |
| Improper angles, ° | 1.70 | |
| Ramachandran statistics | ||
| Most favored, % | 77.60‡ | 79.50§ |
| Additionally allowed, % | 18.90‡ | 17.80§ |
| Generously allowed, % | 2.70‡ | 2.40§ |
| Disallowed, % | 0.80‡ | 0.30§ |
| Average atomic RMS deviations from the average structure (± SD) | ||
| N, Cα, and C, atoms, Å | 0.67 ± 0.19‡ | 0.53 ± 0.12§ |
| All heavy atoms, Å | 1.21 ± 0.27‡ | 1.06 ± 0.15§ |
*Input.
†Aria cycle 8.
‡Residues 10–90.
§Residues 10–50, 56–90.
Fig. 1.
NMR ensemble and backbone representations of Rds3p. (A) Stereoview of overlay of deposited ensemble of 20 lowest-energy models of Rds3p. The N and C termini, which are at the rear of the structure as viewed, are marked, as are clusters 1, 2 and 3 (the zinc atom of cluster 1 is obscured in this view). Significant disorder is limited to the region adjacent to cluster 3 (residues 51–55; see Overall Structure of Rds3p) on the far right of the structure as shown. The structure is oriented with the β-triangle facing upward. (B) Corresponding stereoview of the lowest energy structure from the NMR ensemble shown in chainbow coloring as a backbone ribbon with cysteine sidechains.
Mass spectrometry under native and denaturing conditions shows that Rds3p binds three zinc ions (data not shown). Twelve of its 13 cysteines are almost completely conserved in eukaryotes (Fig. 2A and Fig. S2). In contrast, Cys-77 is not conserved and was found not to be involved in any of the zinc binding clusters in our calculated structures (including calculations carried out with no zinc-binding or interligand constraints and no prior NOE cross peak assignments; see Fig. S3 and Table S1).
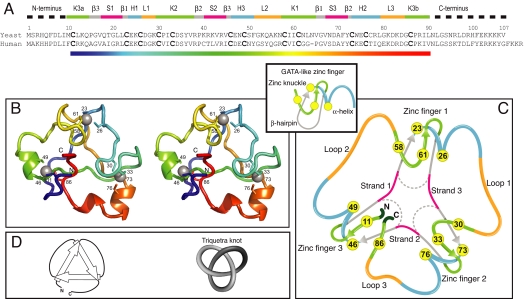
Fig. 2.
Sequence alignment and overall structure of Rds3p. (A) Sequence alignment of human and yeast Rds3p. Components of the structure are shown above the sequence, using the same color code as in 2C (K, zinc knuckle; β, β-hairpin; S, strand; H, helix; L, loop; numbers denote fingers 1, 2, and 3). (B) Ribbon stereoview of the Rds3p structure in chainbow coloring (corresponding to the rainbow color gradient bar in 2A) with the β-triangle facing downwards (180° rotation from Fig. 1). Sidechains of the metal-binding cysteine residues are shown numbered. (C) Schematic showing how Rds3p is built from three GATA-like zinc fingers (shown in same orientation as B). In Rds3p, each β-hairpin is broken and replaced by a strand (magenta) running from one finger to the next, and a loop (orange) is added from the α-helix of one finger to the zinc-knuckle of the next. The zinc knuckle of finger 3 is “interrupted.” (D) Simplest representation of the origin of the knot in the Rds3p structure (same orientation as in B and C).
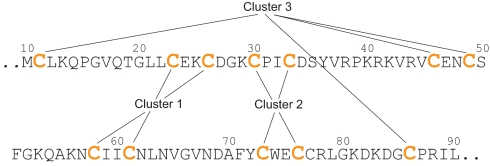
The overall structure of Rds3p has an approximately triangular shape with a 4-cysteine cluster near each vertex (Figs. 1 and 2). These clusters each form part of a structure very similar to that of a GATA-like zinc finger, particularly those in LIM domains (15). Such fingers typically comprise a zinc knuckle (also sometimes called a half-knuckle) followed by a loop, a β-hairpin, and an α-helix. The zinc knuckle and the first turn of the helix usually donate two zinc ligands each (16). In Rds3p the three zinc fingers are connected through loops and β-strands. Interestingly, the pairs of cysteines participating in each finger are not arranged sequentially. Instead, they are permuted between the fingers in an arrangement reminiscent of domain-swapping (17), although here it occurs between subdomains within one chain (Fig. 3). The first (Cys-23 to Cys-26) and the fourth (Cys-58 to Cys-61) CXXC motifs contribute the helical portion (although restricted here to a single turn) and the zinc knuckle respectively of finger 1. Finger 2 is formed from the second (Cys-30 to Cys-33) and fifth (Cys-73 to Cys-76) CXXC motifs, which contribute the zinc knuckle and the helical portion, respectively. This is the finger that most strongly resembles a canonical GATA finger, in that it has the longest and best-defined helix and the best-defined region corresponding to a β-hairpin. The helical portion of finger 3 is formed by the third CXXC motif (Cys-46 to Cys-49). Finger 3 formally lacks a canonical zinc knuckle, but the first and last cysteines (Cys-11 and Cys-86) of the sequence form an “interrupted knuckle.”
Fig. 3.
Sequence of the ordered part of Rds3p showing the nonsequential arrangement of the three zinc clusters.
Near-symmetric structures arising from such “segment swapping” between subdomains within a single chain are not uncommon for cases involving two subdomains (18), but cases involving three subdomains, as for Rds3p, are rare; the only other case we are aware of is that of S-adenosylmethionine synthetase (19) (PDB entry 1XRA). The evolutionary origin of such an arrangement in Rds3p is not obvious, but could perhaps involve multiple gene duplication events to generate four or more repeated fingers, followed by deletions that leave two intact fingers flanked by two fragments of fingers. The process of segment-swapping might then be a mechanism that allows the N- and C-terminal finger fragments to come together and form part of a common structure while reorganizing the CXXC motifs such that each finger in the new arrangement is built from a nonsequential set of motifs. Alternatively, the structure might arise through segment-swapping among three fingers in conjunction with deletions and point mutations.
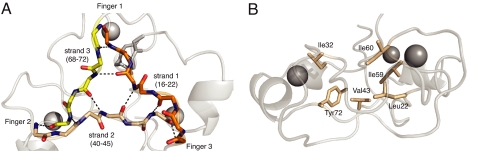
Near the center of the structure, the β-strands that link the fingers come together to form a distinctive, approximately triangular arrangement stabilised by either two or three interstrand hydrogen bonds at each “vertex.” This pattern leaves none of the donors or acceptors on the inwardly interacting edges of these strands unsatisfied (Fig. 4A). The structure is further stabilized by several hydrophobic sidechains (Leu-22, Ile-32, Val-43, Ile-59, Ile-60, and Tyr-72) that project toward the interior from the strands of the β-triangle. These sidechains form a small core that occupies the central space between the fingers (Fig. 4B). The fold is completed by three loops on the outside of the structure that each connect the C terminus of one finger to the N terminus of the next (Fig. 2C).
Fig. 4.
Hydrophobic core of Rds3p. (A) The strands connecting the three zinc fingers form a β-triangle and are highly stabilized by hydrogen bonds. (B) Conserved hydrophobic residues between the face of the zinc fingers and the β-triangle stabilize the core of Rds3p.
Finger 3 was found to have a faster rate of metal exchange than the other two fingers in experiments intended to substitute 113Cd for Zn (see SI Text), suggesting that the chain break may cause additional dynamics in this part of the structure. 15N NMR relaxation experiments did not show any significant differences anywhere in the domain (data not shown), but this is not inconsistent, because metal exchange processes take place on timescales much too slow to influence 15N relaxation rates [time scales of minutes to hours have been reported for Zn-Cd exchange in other zinc fingers (20)]. The poor local order for residues 51–55 (Fig. 1A) is also consistent with this interpretation, because it reflects a low constraint density in this part of the molecule that in turn is probably due to accelerated solvent exchange of nearby amide NH signals (Gln-54 NH is weak and Lys-53 NH is missing altogether from the spectra).
Rds3p Contains a Deep Trefoil Knot.
The polypeptide chain of Rds3p forms a deep topological knot. Resolution of this knot would require scission of the chain at around residue Leu-22 if starting from the N terminus or at around residue Tyr-72 if starting from the C terminus; these are approximately the points at which the chain passes through a loop to form the knot. Thus, starting from the N terminus, there are ≈10 native residues of the tail and 11 residues from the folded domain (including one zinc-binding cysteine, Cys-11) in the portion of the chain that would need to be removed to resolve the knot, whereas, starting from the C terminus, there are ≈17 residues of the tail and 18 residues from the folded domain (including three zinc-binding cysteines, Cys-73, Cys-76, and Cys-86) in the portion that would need to be removed.
Another way to visualize the origin of the knot is to consider the β-triangle. If one views the triangle from the same face as that on which the three zinc-fingers lie, the C-terminal end of each strand of the triangle passes over the N-terminal end of the next strand, forming a cyclic array (Fig. 2D). To create a knot from this arrangement, it is only necessary to link the ends of the strands together with loops. If all of the strands are thus linked to create a cyclic chain, then the arrangement forms a perfect trefoil knot.
As a further confirmation that the fold of the protein was correct, we used three independent methods of calculating the structure (see Materials and Methods). In addition to the main calculations, which used the ARIA protocol (21), calculations were carried out either by using manual assignment of NOE cross peaks or by using the semiautomatic program ATNOS/CANDID (22, 23). Although these calculations were not all refined to the same final precision as the ARIA calculations, in each case they showed the same knotted fold (data not shown). Furthermore, the data converged well on the knotted fold even in early cycles of refinement, showing that the fold was well determined by the data (see also SI Text).
Although not very common, knotted proteins have been found before. The deep trefoil knot is the most abundant type and was discovered in four distinct families: the RNA methyltransferases, carbonic anhydrases, transcarbamylases, and the previously mentioned S-adenosyl methionine synthetases (reviewed in refs. 24–26). Their knotted domain is classed as a β/α-knot fold in the SCOP database (27). Although Rds3p contains a deep trefoil knot, it does not resemble any of these proteins and has a previously unknown fold. Most trefoil knots are right-handed; this preference was associated to the mainly right-handed connection of the βαβ-units of the β/α-class proteins (24). Intriguingly, the chirality of the trefoil knot in Rds3p is left-handed (because, when passing from the N to the C terminus, the direction of the over-crossing strand to the under-crossing strand is left to right). The only other left-handed trefoil found to date is a substructure in ubiquitin hydrolase (24).
The arrangement of the loops and strands of Rds3p bears a striking resemblance to the “triquetra” symbol from Celtic and Nordic art (Fig. 2D); hence, we propose to name this knotted fold as the triquetra motif. We are unaware of any other reported structures of knotted zinc-finger proteins.
Heating of Rds3p Does Not Result in Permanent Denaturation.
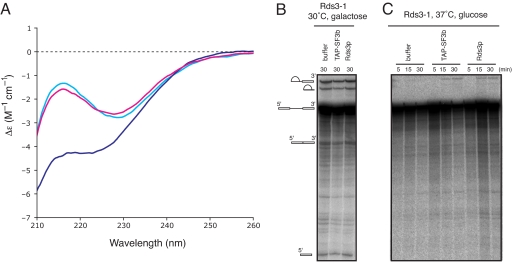
The far UV circular dichroism (CD) spectrum of Rds3p shows that it is folded and that, consistent with the NMR solution structure, it does not show many signatures of secondary structure. Thermal denaturation of Rds3p from 20°C to 95°C was followed by using CD by measuring the change in ellipticity at 215, 222, and 229 nm (data not shown). Analysis of these curves showed the midpoint of the thermal transition (Tm) to be 66 ± 0.3°C and the enthalpy of transition at the midpoint (ΔHm) to be 47 ± 1.8 kcal·mol−1. At all three wavelengths the values for Tm and ΔHm were similar within the error range, which indicates that there is a one step transition between two states and that the protein unfolds as a single cooperative unit. The rather low ΔHm value may reflect the limited secondary structure of Rds3p. Interestingly, at 95°C the far UV-CD spectrum is not typical of a fully unfolded protein (Fig. 5A), because it still shows some helical features at ≈222 nm. After cooling, an almost identical CD spectrum is obtained to the one obtained before heating (Fig. 5A), indicating that the protein folds back into its original state. In contrast, removal of zinc with EDTA results in a completely different CD spectrum, which did not change back into the native spectrum upon removal of the EDTA followed by the addition of zinc (see Fig. S4). This suggests that at higher temperature the knot may persist in a partially unfolded state with the zinc still coordinated to the protein, whereas disruption of zinc binding by EDTA gives rise to irreversible denaturation. Presence of fusions at the N or C termini are apparently not incompatible with folding, because mass spectrometry shows that the N-terminal GST fusion of Rds3p, which we used for bacterial expression, also binds three zinc ions. This strongly suggests that the Rds3p part is folded in the fusion protein. Interestingly, others have shown that Rds3p bearing a C-terminal TAP tag is fully functional and supports splicing (5, 7).
Fig. 5.
CD spectra of Rds3p at different temperatures and in vitro splicing assays. (A) Overlay of far UV CD spectra of Rds3p at 20°C (light blue) after heating to 95°C (dark blue) and after recooling the sample to 20°C (pink). The spectra recorded at 20°C almost perfectly overlap, indicating that Rds3p recovers its original structure after this heating cycle. (B) In vitro splicing assay with TAP-SF3b and recombinant Rds3p. GAL1::rds3-1 strain grown under splicing supporting conditions shows that activity is not affected by addition of TAP-purified SF3b complex or recombinant Rds3p. (C) Time course of splicing activity in an extract of GAL1::rds3-1 strain grown in the presence of glucose at 37°C. No splicing activity can be observed in this extract unless it is supplemented with TAP-purified SF3b complex (as a control) or recombinant Rds3p.
In Vitro Splicing Assay.
To show that our recombinant Rds3p is functional, we tested its ability to restore splicing in an inactivated extract from a yeast strain with a heat-sensitive mutation in the Rds3 gene under the control of a GAL1 promoter (GAL1::rds3-1) (28). In this strain, splicing is inhibited at the nonpermissive temperature and spliceosomal assembly arrests at the U1-dependent commitment complex stage (7). A splicing extract prepared from this strain grown at 30°C with galactose supported splicing in vitro as expected (Fig. 5B). When the strain was grown at elevated temperature and Rds3–1p expression was repressed by glucose, splicing was abolished but could be rescued to a significant level by the addition of our recombinant Rds3p (Fig. 5C).
Surface Properties.
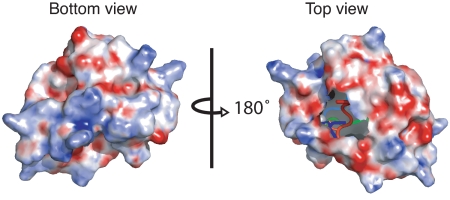
The sequence alignment (Fig. 2A and Fig. S2) shows that there are many highly conserved hydrophobic residues in the ordered part of the Rds3p molecule. As mentioned in Overall Structure of Rds3p, some of these hydrophobic residues form the hydrophobic core of Rds3p. However, these represent well under half the hydrophobic residues present in the ordered region. The remainder are exposed to varying degrees on the surface. Consistent with this, the electrostatic surface potential map is relatively featureless (Fig. 6). Overall, the surface is slightly acidic, and the only pronounced feature is a basic patch on the base of the β-triangle contributed by the lysine and arginine residues on strand 2. In addition, the C-terminal disordered tail (residues 91–107) is highly basic.
Fig. 6.
Electrostatic surface of Rds3p. Regions of positive charge are shown in blue, and regions of negative charge are shown in red. (Left) The bottom view (same orientation as in Fig. 1) shows a basic patch formed by lysine and arginine residues on strand 2 at the base of the triangle. (Right) The top view (same orientation as in Fig. 2 B and C) shows a slightly acidic patch, but otherwise the surface is relatively featureless. The missing area in the top view results from truncation of the flexible tails.
It is known that the SF3b complex cross-links to U2 snRNA; however, a direct cross-link between RNA and Rds3p has never been observed. Therefore, we attempted to establish whether Rds3p can bind nucleic acids. Electrophoretic mobility shift assays (EMSA) of Rds3p with U2 snRNA and single or double stranded unrelated RNAs did not reveal any specific or general RNA affinity for this protein (data not shown and SI Text). It has been suggested that the homolog of Rds3p in rat and S. pombe (Ini1) acts as a transcriptional activator (29, 30) or as a chromatin associated protein in mouse (Phf5a) (31) that binds to DNA. In contrast, we did not observe any general DNA affinity in EMSA, using five different constructs of DNA (data not shown).
Based on its structure, surface properties and lack of general nucleic acid binding, we propose that Rds3p mediates protein–protein interactions within the SF3b complex. The existence of the disordered tails of isolated Rds3p (residues 1–9 and 91–107) is also consistent with this likely function. These tails represent >20% of the total sequence of Rds3p, and it is notable that there is a high degree of sequence conservation within them. This suggests that when Rds3p interacts with its binding partner(s) in the SF3b complex, the N- and C-terminal tails may well fold against the surfaces of these partners.
Conclusions
Rds3p is an essential component of the SF3b complex. We have determined its structure and found that it comprises a previously uncharacterized knotted fold; we know of no other examples of knotted zinc finger proteins. We have shown that, despite the lack of secondary structure, this protein is very stable and that recombinantly expressed protein is functional in an in vitro splicing assay. Based on our structure, we believe that Rds3p functions as a protein platform within the SF3b complex.
Materials and Methods
Recombinant Rds3p.
DNA coding for the His-tagged GST fusion protein of yeast Rds3 was cloned into a pRK172 vector. The protein was expressed in BL21 (DE3) RIL CodonPlus cells (Stratagene) either in rich medium for biochemical studies or in M9 minimal medium to obtain isotope labeled protein for NMR experiments (SI Text). The His-tagged GST fusion protein was cleaved with tobacco etch virus (TEV) protease.
NMR.
Data were acquired at 27°C from samples containing 0.3–0.8 mM Rds3p in NMR buffer [20 mM Tris·HCl (pH 7.0), 1 mM [2H6] DTT, 200 mM NaCl]. A nearly complete set of resonance assignments was made by using a standard suite of triple resonance NMR experiments, and 1H, 15N and 13C chemical shifts were calibrated by using sodium 3,3,3-trimethylsilylpropionate (TSP) as an external 1H reference.
Structures were calculated by using ARIA 1.1.2 (21), which makes use of CNS 1.1 (32) for simulated annealing calculations. Initial calculations were run with no NOE cross peak assignments included in the input and without defining zinc atoms or using interligand constraints; these calculations established the binding connectivity for the zinc ions unambiguously (Fig. S3 and Table S1), and gave the same knotted fold as later calculations that included zinc. Calculations including explicit zinc used a slightly modified version of the ARIA protocol (SI Text).
Mass Spectrometry.
Data were acquired by using either modified LCT or QTOF2 instruments (Waters) configured for nanoflow ESI in positive ion mode. Nanoflow capillaries (drawn and coated with gold in-house) were loaded with 2-μl sample and a low pressure of nitrogen gas was used to initiate flow through the capillary. Other experimental details have been described in ref. 33. Spectra under native conditions were acquired from Rds3p in 400 mM ammonium acetate, 1 mM DTT solution after dialysis. Where necessary, an additional gel filtration step, using MicroBiospin-6 tubes (Bio-Rad) with the same buffer was used. Rds3p was denatured by either the addition of cation exchange beads (AG 50W-W* resin, 100–200 mesh, hydrogen form, Bio-Rad) or an equal volume of 25% formic acid in acetonitrile.
Far-UV CD.
Spectra of 0.2 mg/ml Rds3p in CD buffer [10 mM Na-phosphate (pH 7.2), 100 mM NaCl, 1 mM DTT] were recorded by using a Jasco J-810 spectropolarimeter (Jasco) at 20°C and 95°C in the spectral range from 190 to 260 nm with 0.2-nm resolution. Spectra were recorded as averages of six scans at a scanning speed of 100 nm/min. The response time was 2.0 s with a bandwidth of 1.0 nm. Quartz cuvettes with an optical path of 1 mm were used. The spectra were corrected for the buffer background. Thermal denaturation data were collected at a scan rate of 1°C per minute in the range of 20 to 95°C. Changes in ellipticity were measured at 215, 222, and 229 nm. The data were fit to a standard equation for a two-state transition (34). For the denaturation of Rds3p by removal of zinc, EDTA was added to 0.2 mg/ml Rds3p in Tris CD buffer [5 mM Tris·HCl (pH 7.4), 100 mM NaCl, 1 mM DTT] to a final concentration of 10 mM and incubated overnight at 4°C. The sample was then extensively dialyzed to Tris CD buffer, and 5 molar equivalents of ZnSO4 were added to the sample. CD spectra were recorded of the sample after addition of EDTA followed by dialysis and again after addition of ZnSO4.
Splicing Assays.
Splicing extracts were prepared as described in ref. 35, and splicing assays were carried out as described in ref. 7 with minor adjustments (SI Text).
Supplementary Material
Acknowledgments.
We thank A. Murzin and A. Andreeva for helpful discussions and for suggesting the name “Triquetra motif.” We thank B. Rymond for providing the GAL1::rds3-1 strain; A. Dziembowski and B. Seraphin for providing TAP-tagged SF3b yeast extract; C. Johnson, D. Veprintsev, and M. Nilges for helpful advice; and M. Wiezlak for technical assistance. This work was supported by the Medical Research Council, a Human Frontier Science Program grant, an EMBO fellowship, and a Marie-Curie Fellowship (to A.M.M.v.R.), a Camille and Henry Dreyfus Foundation Faculty Start-up Award (to N.M.L.), and a Wellcome Trust Traveling fellowship and a Medical Research Council Career Development fellowship (to E.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates for the 20 lowest energy models of Rds3p from the final run have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 2K0A). Chemical shift assignments have been deposited to the BioMagResBank (accession no. 15644).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802494105/DCSupplemental.
References
- 1.Jurica MS, Moore MJ. Pre-mRNA splicing: Awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, editor. The RNA World. New York: Cold Spring Harbor Laboratory; 2006. pp. 369–400. [Google Scholar]
- 3.Brosi R, Hauri HP, Krämer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 4.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 5.Dziembowski A, et al. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 2004;23:4847–4856. doi: 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, He J, Lynn B, Rymond BC. Interactions of the yeast SF3b splicing factor. Mol Cell Biol. 2005;25:10745–10754. doi: 10.1128/MCB.25.24.10745-10754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Rymond BC. Rds3p is required for stable U2 snRNP recruitment to the splicing apparatus. Mol Cell Biol. 2003;23:7339–7349. doi: 10.1128/MCB.23.20.7339-7349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dybkov O, et al. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol. 2006;26:2803–2816. doi: 10.1128/MCB.26.7.2803-2816.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golas MM, Sander B, Will CL, Lührmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 12.Golas MM, Sander B, Will CL, Lührmann R, Stark H. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol Cell. 2005;17:869–883. doi: 10.1016/j.molcel.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Schellenberg MJ, et al. Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci USA. 2006;103:1266–1271. doi: 10.1073/pnas.0508048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spadaccini R, et al. Biochemical and NMR analyses of an SF3b155–p14-U2AF-RNA interaction network involved in branch point definition during pre-mRNA splicing. RNA. 2006;12:410–425. doi: 10.1261/rna.2271406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deane JE, et al. Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex. EMBO J. 2004;23:3589–3598. doi: 10.1038/sj.emboj.7600376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grishin NV. Treble clef finger–a functionally diverse zinc-binding structural motif. Nucleic Acids Res. 2001;29:1703–1714. doi: 10.1093/nar/29.8.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreeva A, Murzin AG. Evolution of protein fold in the presence of functional constraints. Curr Opin Struct Biol. 2006;16:399–408. doi: 10.1016/j.sbi.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Takusagawa F, Kamitori S, Misaki S, Markham GD. Crystal structure of S-adenosylmethionine synthetase. J Biol Chem. 1996;271:136–147. [PubMed] [Google Scholar]
- 20.Houben K, et al. Dynamics and metal exchange properties of C4C4 RING domains from CNOT4 and the p44 subunit of TFIIH. J Mol Biol. 2005;349:621–637. doi: 10.1016/j.jmb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Linge JP, O'Donoghue SI, Nilges M. Automated assignment of ambiguous nuclear overhauser effects with ARIA. Methods Enzymol. 2001;339:71–90. doi: 10.1016/s0076-6879(01)39310-2. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 23.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 24.Taylor WR. Protein knots and fold complexity: Some new twists. Comput Biol Chem. 2007;31:151–162. doi: 10.1016/j.compbiolchem.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Virnau P, Mirny LA, Kardar M. Intricate knots in proteins: Function and evolution. PLoS Comput Biol. 2006;2:e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeates TO, Norcross TS, King NP. Knotted and topologically complex proteins as models for studying folding and stability. Curr Opin Chem Biol. 2007;11:595–603. doi: 10.1016/j.cbpa.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 28.Vincent K, Wang Q, Jay S, Hobbs K, Rymond BC. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics. 2003;164:895–907. doi: 10.1093/genetics/164.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oltra E, Pfeifer I, Werner R. Ini, a small nuclear protein that enhances the response of the connexin43 gene to estrogen. Endocrinology. 2003;144:3148–3158. doi: 10.1210/en.2002-0176. [DOI] [PubMed] [Google Scholar]
- 30.Oltra E, Verde F, Werner R, D'Urso G. A novel RING-finger-like protein Ini1 is essential for cell cycle progression in fission yeast. J Cell Sci. 2004;117:967–974. doi: 10.1242/jcs.00946. [DOI] [PubMed] [Google Scholar]
- 31.Trappe R, et al. Identification and characterization of a novel murine multigene family containing a PHD-finger-like motif. Biochem Biophys Res Commun. 2002;293:816–826. doi: 10.1016/S0006-291X(02)00277-2. [DOI] [PubMed] [Google Scholar]
- 32.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 33.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 34.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 35.Lin RJ, Newman AJ, Cheng SC, Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985;260:14780–14792. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.