Abstract
Eukaryotic cells organize their contents through trafficking along cytoskeletal filaments. The leading edge of a typical metazoan cytoskeleton consists of a dense and complex arrangement of cortical actin. A dendritic mesh is found across the broad lamellopodium, with long parallel bundles at microspikes and filopodia. It is currently unclear whether and how myosin motors identify the few actin filaments that lead to the correct destination, when presented with many similar alternatives within the cortex. Here we show that myosin X, an actin-based motor that concentrates at the distal tips of filopodia, selects the fascin-actin bundle at the filopodial core for motility. Myosin X moves individual actin filaments poorly in vitro, often supercoiling actin into plectonemes. However, single myosin X motors move robustly and processively along fascin-actin bundles. This selection requires only parallel, closely spaced filaments, as myosin X is also processive on artificial actin bundles formed by molecular crowding. Myosin X filopodial localization is perturbed in fascin-depleted HeLa cells, demonstrating that fascin bundles also direct motility in vivo. Our results demonstrate that myosin X recognizes the local structural arrangement of filaments in long bundles, providing a mechanism for sorting cargo to distant target sites.
Keywords: fascin, motor navigation, myosin X, filopodia, single-molecule fluorescence
In a dense mesh of cellular actin, if all filaments are functionally equivalent, then myosins will move in a random walk as they translate, detach, and reattach to new filaments. Alternatively, myosins may identify and walk along certain subpopulations of actin that lead to target locations, based on a set of common structural features. One possibility is that different myosin classes may partition among filaments based on actin isoforms. However, no significant difference between βγ-actin versus α-actin has been detected for myosin V, the only motor to be tested on both tracks (1). A second possibility is that actin-associated proteins, in particular tropomyosins, modulate the binding of myosin to actin. Indeed, class I myosins are excluded from tropomyosin-decorated actin in stress-fibers, whereas class II myosins are not (2, 3). In addition to this direct regulatory mechanism, tropomyosins may also direct myosin traffic by stabilizing particular actin tracks (4). However, tropomyosins are not typically found in regions of actively polymerizing, dynamic actin, such as the leading edge of the cell. Therefore, additional mechanisms likely exist for directing myosin traffic.
To identify factors that could direct myosins to specific locations, we searched for myosin classes that are localized to limited populations of actin even in the presence of nearby dynamic actin. The class X myosin meets these criteria for a selective motor. Myosin X travels to and is highly concentrated at the distal tips of filopodia: long, slender projections often found at the leading edge of migrating cells (5–7). Filopodia are used for environment-sensing, substrate-anchoring, and phagocytosis (8). The filopodial core is largely composed of parallel, unidirectional actin filaments that are bundled by fascin (9–11). Myosin X also undergoes intrafilopodial motility, with striking back-and-forth oscillations that arise from the retrograde flow of the bundled actin (5). To reach filopodial tips, myosin X requires the motor domain, neck, and coiled-coil domains. Neither the motor domain alone nor the tail fragment (containing regulatory and cargo binding domains) reaches the tips, suggesting that myosin X localizes under its own power as a motor dimer (5). Two recent studies reached different conclusions on the kinetic mechanism of myosin X. One proposed that it is a motor with a high duty ratio (i.e., fraction of the motor ATPase cycle in the strongly bound state), potentially capable of processive runs (12). The other found a lower duty ratio, but with a significant population of myosin X weakly bound to actin (13). All of these in vitro studies used single actin filaments, rather than the native bundled actin filament assembly where myosin X has been shown to associate in vivo. Here, we show that myosin X identifies and walks processively on bundles of actin filaments, which allows it to reach its target site at filopodial tips.
Results and Discussion
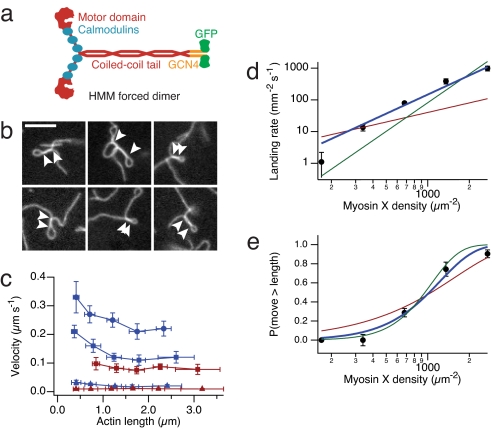
We constructed and purified a forced dimer of myosin X heavy meromyosin (HMM), containing the motor domain, neck, native coiled-coil, an in-register GCN4-p1 leucine zipper, and GFP, but lacking cargo-binding domains (Fig. 1a). Gliding filament in vitro motility assays reveal a number of unusual features of myosin X motility on single actin filaments. At moderate surface densities of myosin X, we routinely observe plectonemes (supercoiled actin filaments) that are several microns long [Fig. 1b and supporting information (SI) Movie S1]. Because filamentous actin has a helical arrangement of actin monomers, supercoiling is a natural consequence of stepping along the helix (14). However, class V and VI myosins avoid spiraling around filaments by matching their step sizes to the actin helical pseudorepeat, which is thought to be a key adaptation for processive motility (i.e., continuous translocation along actin without detachment) (15, 16). At higher densities, myosin X velocities depend strongly on actin filament length, unlike those of other myosins (Fig. 1c). Short actin filaments (<0.5 μm) move up to twice as fast as long filaments (>2 μm), reaching an upper limit of 330 ± 60 nm/s (SD, n = 10). The origin of this velocity dependence on filament length is unclear but may reflect an inhibitory effect of accumulated filament torque on the motor.
Fig. 1.
Motility of myosin X on single actin filaments. (a) Cartoon of the myosin X construct used in this study. The forced dimer was used for all in vitro assays. (b) Images of actin filament plectonemes in the gliding filament assay. Arrowheads indicate the extent of supercoiled segments. (Scale bar, 5 μm.) (c) Myosin X velocity depends on actin filament length. Measured actin filaments were binned by length, and velocities of myosin X (blue) and myosin VI (red) are shown (n = 10 filaments per point, ±SD in velocity and length). ATP concentration; 2 mM (circles), 1 mM (squares), and 1 μM (triangles). (d and e) Actin filament landing rate (d) and fraction of filaments that move greater than their length (e), both as a function of myosin X density on the surface. Shown are fits to models where one (red), two (blue), and three (green) motors are required to propel actin. Best fits are obtained for the two motor model: reduced χ2 = 3.7 (d), reduced χ2 = 1.4 (e). Error bars are standard errors obtained from counting statistics.
To determine whether myosin X is processive on single filaments, we studied the motility over a range of motor densities on the coverslip surface (17). Actin filament landing rates (Fig. 1d) vary as the second power of myosin density, suggesting that continuous motility requires two motors (18). Likewise, the fraction of filaments that travel greater than their own length (which requires a second encounter with a motor) is best fitted by a model that requires two motors for continuous motility (Fig. 1e) (16, 18). Both of these results are signatures of nonprocessive motion, but a typical nonprocessive motor requires at least four motors to sustain motility (19). Apparently, myosin X operates at the boundary between processive and nonprocessive motion. Because the diffusing filament ends are difficult to track with precision, these assays miss occasional short runs (20). However, short runs are likely on single filaments, because supercoiling requires a change in the linking number while maintaining continuous attachments to the surface at either end of the filament. Thus, our landing assay results allow for short processive runs on single filaments, where multiple motors are required to lift the transport distance above the detection threshold.
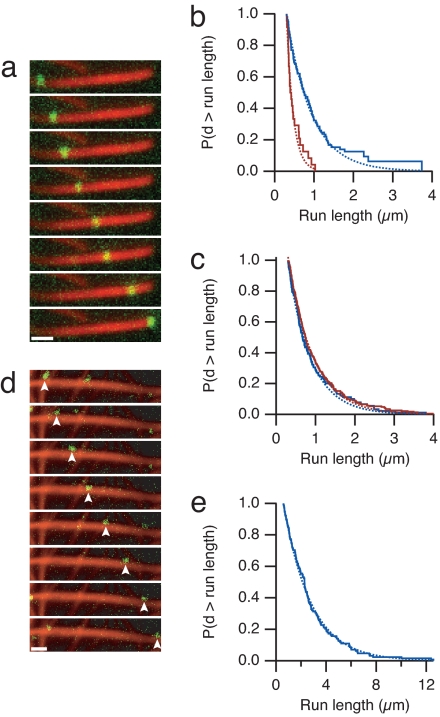
Given the defective motility of myosin X on single actin filaments, we reasoned that other components of filopodia might assist myosin X's motility. Fascin-bundled actin is highly enriched in filopodia (10). To study myosin X on an actin architecture similar to that in native filopodia, we assembled fascin-actin bundles and tracked the motility of single myosin X molecules by using total internal reflection fluorescence microscopy. Myosin X moves processively, with some motors traveling several microns on fascin-actin bundles (Fig. 2a, Movie S2, and Figs. S1 and S2). In contrast, apparent processive runs on single filaments are short and rare. Myosin X moves four times as far (0.63 ± 0.08 μm vs. 0.17 ± 0.05 μm) on bundles than on single filaments (Fig. 2b and Movie S3). Moreover, myosin X initiates processive runs at a 4-fold higher rate on bundles (observed rates of 190 ± 19 mm−2·s−1 on bundles and 50 ± 7 mm−2·s−1 on single filaments). These initiation rates were determined at identical motor concentrations and actin densities, by counting the number of motors that land and move and dividing by the observation time and the viewing area. When we examined myosin X motility on a 10-fold lower density of single actin filaments, we found a 10-fold reduction in the initiation rate (5 ± 1 mm−2·s−1). Thus, initiation on single filaments is first-order with respect to actin, demonstrating that chance colocalization of filaments is not required for the observed runs.
Fig. 2.
Processive motility of myosin X on actin bundles. (a) Time-lapse fluorescence micrographs of a single myosin X motor (green) moving along a fascin-actin bundle (red). (Frame interval, 2.5 s; scale bar, 1 μm.) (b) Run length measurements of myosin X on fascin-actin bundles (blue) and actin alone (red) at 2 mM ATP. The Kaplan–Meier estimate of the run-length survivor function is shown (38). Events are left-truncated at 0.3 μm and are right-censored at bundle ends. Run lengths are estimated from single exponential fits to the empirical survivor function (dotted lines). Run length decay constants are 0.63 ± 0.08 μm (SEM, n = 100) on fascin-actin bundles and 0.17 ± 0.05 μm (SEM, n = 24) on single actin filaments. This difference in run length is significant (P = 0.01, Kolmogorov–Smirnov test). Runs on single filaments were measured on 10-fold less actin that was observed for a 10-fold longer period, allowing us to directly compare the total number of events. Myosin X moved at 340 ± 120 nm/s (SD) on fascin-actin bundles, and 330 ± 120 nm/s (SD) on single filaments. (c) Run length measurements of myosin V on bundles and actin alone, as in b. Run length decay constants are 0.57 ± 0.06 μm (SEM, n = 134) on fascin-actin bundles, and 0.66 ± 0.05 μm (SEM, n = 231) on single actin filaments. Myosin V moved at 270 ± 110 nm/s (SD) on fascin-actin bundles, and 330 ± 100 nm/s (SD) on single filaments. (d) Time-lapse fluorescence micrographs of myosin X (green) moving along methylcellulose-bundled actin (red). Arrowheads indicate the moving spot. (Frame interval, 2.0 s; scale bar, 1 μm.) (e) The run length decay constant on methylcellulose-bundled actin is 2.6 ± 0.2 μm (SEM, n = 128). Myosin X moved at 780 ± 170 nm/s (SD) on methylcellulose-bundled actin. On some methylcellulose bundles, myosin X moved in a back-and-forth manner, suggesting that regions of the branched actin bundle network have mixed polarity. These mixed polarity bundles were excluded from the analysis. In control experiments, methylcellulose did not affect the run length of myosin X on fascin-actin bundles, and reduced the velocity by one-third. Run length standard errors are from fitting 200 bootstrap sampled sets. For single-molecule evidence, see Fig. S1.
To demonstrate that our run length measurements on single filaments are not compromised by attaching filaments to the coverslip surface, we attached filaments to a substrate at widely separated platforms to permit spiral walking paths along actin (21). We failed to observe any processive myosin X runs in this experimental geometry, but this may be because of the extremely low density of filaments on the surface. To force myosin X motor interactions, we used an optical trap to position motor-decorated beads near suspended filaments in a geometry that was described by Ali et al. (22). We identified beads that moved processively on surface-attached fascin-actin bundles, then positioned these beads near suspended single filaments. None of these beads moved processively on the single filaments (zero of five beads tested; see Materials and Methods), but all resumed processive stepping when returned to the fascin-actin bundles. Given that all of these beads had functional motor, we would have expected all to show processive runs if myosin X requires unconstrained access around the single filament.
The high local concentration of actin in the bundle may enhance the processivity of all myosins indiscriminately. However, nonmuscle myosin IIb was nonprocessive on either single filaments or fascin-actin bundles, despite its moderate duty-ratio (no events observed; see Movie S4) (23, 24). Likewise, bundles do not enhance myosin V's processivity. Myosin V moves the same distance (Fig. 2c) and is 4-fold less likely to initiate processive runs on fascin-actin bundles compared to single filaments (rates of 990 ± 85 mm−2·s−1 vs. 4,200 ± 280 mm−2·s−1). Thus, unlike myosin X, myosin V shows a preference to land on unbundled actin. An 8-nm-long myosin head is too large to enter the interior of a fascin bundle where the gap between filaments is approximately 4 nm (Fig. S3a). Therefore, myosins must walk along the outer surface of the bundle. The lower myosin V landing rates on bundles suggest that a large fraction (>75%) of the actin within bundles is sterically occluded and is inaccessible to any myosin.
To facilitate comparisons between myosin V and myosin X, which have different biochemical rates and processive run lengths, we define a dimensionless “motility selection ratio” as s = λb Rb exp((0.3 μm)/λb)/(λs Rs exp((0.3 μm)/λs)). Here, λ is the run length, R is the initiation rate, and subscripts s and b denote single filaments and bundles, respectively. The exponential factors correct for missed events that occur below our 0.3-μm distance cutoff. This motility selection ratio is a measure of a motor's ability to move on bundles in preference to single actin filaments, given equal amounts of actin and motor. From the measured run lengths and initiation rates, we find that s = 4 ± 1 for myosin X, whereas s = 0.2 ± 0.03 for myosin V. Thus, myosin X is 20-fold more selective for fascin-actin bundles than myosin V, when both run lengths and run initiation rates are considered.
Myosin X may identify specific structural features of fascin-decorated actin that are required for its processive motility. Alternatively, myosin X may only require at least two parallel actin filaments in close proximity, with no preference for fascin or any other bundling protein. To distinguish these two possibilities, we nucleated parallel brushes of actin filaments from Limulus acrosomal processes and bundled them by using methylcellulose as a molecular crowding agent in the absence of fascin (Fig. S3b). Remarkably, myosin X moves farther (2.6 ± 0.2 μm) and faster (0.8 ± 0.2 μm/s vs. 0.3 ± 0.1 μm/s) on some of these methylcellulose bundles than on fascin-actin bundles (Fig. 2 d and e and Movie S5). Therefore, myosin X does not strictly require fascin for processive motility but does seem to require multiple filaments.
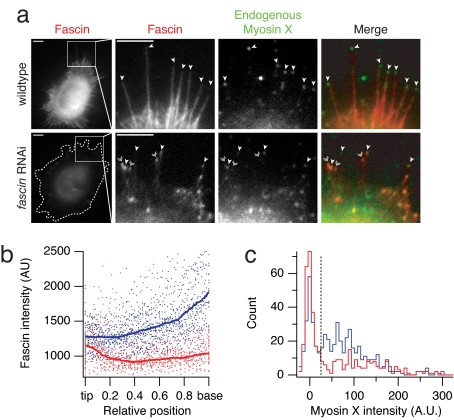
To test whether myosin X relies on fascin bundles to achieve its characteristic localization in vivo, we used RNAi to deplete fascin from HeLa cells (10) and observed either endogenous myosin X (Fig. 3a) or exogenous bovine GFP-myosin X (Fig. S4 and Table S1). Fascin-depletion abolishes half of the filopodia, confirming previous results (Fig. S4e) (10). The remaining filopodia in the depleted cells show an altered distribution of fascin along their length (Fig. 3b), with less fascin at the base of the filopodium where myosin X would enter. This altered fascin distribution is consistent with the more loosely bundled and disorganized appearance of filopodial actin in fascin-depleted cells, observed by Vignjevic et al. (see figure 4 in ref. 10). Likewise, we see a statistically significant alteration in the distribution of myosin X at filopodial tips (Fig. 3c). In wild-type cells, most filopodia are capped with myosin X (Fig. 3a). However, in fascin-depleted cells, significantly fewer of the remaining filopodia are capped with myosin X (Fig. 3a). We propose that many of these remaining filopodia contain bundles that are too disorganized to support wild-type levels of myosin X motility.
Fig. 3.
Fascin is required for in vivo myosin X localization. (a) Epifluorescence images of wild-type and fascin-depleted HeLa cells. We immunostained fascin (red) and myosin X (green), and identified cells transfected with the fascin shRNA from their reduced fascin levels compared to neighboring untransfected cells. Filopodial projections were identified in DIC images (not shown). A dotted outline surrounds the fascin RNAi cell. We rescaled the contrast settings in the close up view of the fascin channel for the fascin RNAi cell, to show the detail in the filopodia. Single arrowheads mark “capped” filopodia with myosin X concentrated at the tip. Double arrowheads mark filopodia that lack myosin X. (Scale bar, 5 μm.) (b) Fascin depletion affects filopodial structure. The plot shows the fascin intensity of 33 individual filopodia from 11 wild-type (blue) and 12 fascin-depleted (red) cells. Position is normalized to the individual filopodial lengths, with the tip at 0.0 and the base at 1.0. Solid lines show the median filtered intensity profiles over a position window of 0.2. In wild-type cells, fascin levels uniformly increase along the filopodium from the tip to the base. In fascin-depleted cells, fascin is found at nearly wild-type levels at filopodial tips but decreases toward the base (compare filopodia shown in a). (c) Fascin depletion affects the filopodial localization of myosin X. The histograms show the (average/sum) intensities of myosin X puncta at the tips of filopodia. For each filopodial tip, we measured the mean pixel value of the fluorescent spot (or a 6-pixel diameter region centered on the tip if no spot was apparent), and subtracted the local background value determined across a 10 × 10 pixel region. Wild-type cells (blue) show a broad distribution of myosin X intensities (n = 526 filopodia from 21 cells). In fascin-depleted cells (red), the distribution of myosin intensity shifts to lower values (n = 400 filopodia from 21 cells). The fraction of filopodial tips falling below the background threshold level of 25 (dotted line) increases from 31% (wild type) to 57% (fascin RNAi). This shift in distributions is statistically significant (P = 3 × 10−7, Wilcoxon two-tailed rank-sum test).
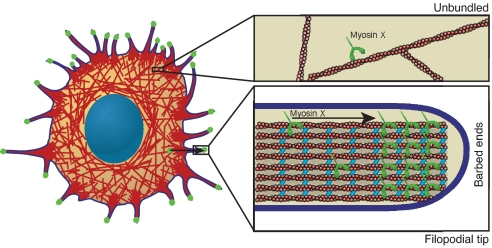
Fig. 4.
A myosin X selection mechanism. A cartoon of a surface attached HeLa cell, with a dense mesh of actin filaments (red). Numerous filopodia project beyond the edge of the cell. Myosin X concentrates at the tips of these filopodia (green). A highly simplified view of a single filopodial tip (lower right) shows the core of actin bundled by fascin (cyan), surrounded by the plasma membrane (blue). Actin barbed ends are oriented toward the filopodial tip. Myosin X (green) is shown traveling through the filopodium toward the barbed ends. Once cytoplasmic myosin X encounters a fascin-actin bundle, it takes a long processive run and enters the filopodium, eventually concentrating at the tip. On unbundled actin (upper right), long runs do not occur. Thus, myosin X identifies and travels on the tracks that lead to the proper destination. One possible identification mechanism is illustrated here. On bundles, the two heads of myosin X may bind to two separate filaments. The neck region of myosin X is too short to allow both heads to bind to one filament, which would preclude processive runs on unbundled single filaments.
Filopodia are formed from lamellopodial actin filaments that are bundled by fascin (10), creating a structurally distinct actin population. Although many other proteins are associated with filopodial actin, including espin, fimbrin (10), Ena/VASP, and formins (8), fascin appears to be a key component that directs myosin X to filopodia. Given the short neck of myosin X, which is half that of myosin V, myosin X likely steps short of the actin pseudohelical repeat and would be forced to spiral around single filaments. We cannot rule out in vivo roles for myosin X on single actin filaments. However, myosin X cannot walk in a spiral path on a filament in a tightly cross-linked bundle for two reasons: (i) the interfilament spacing is smaller than a myosin head and (ii) the tail of myosin X is anchored to the plasma membrane via its PH and FERM domains (or to an optically trapped bead in our in vitro assays). We expect that myosin X is structurally constrained to straddle adjacent filaments along the surface of a bundle, with one head tracking along one filament while the second head tracks a second filament. This straddle mechanism would explain the preference for bundled actin, even with artificial bundling agents such as methylcellulose. The requirement for at least two closely spaced filaments would allow myosin X to identify its target actin bundle in vivo (Fig. 4).
Our results provide insight into how myosin X promotes the formation of filopodia. The Λ-precursor model of Svitkina et al. proposes that filopodia originate from a reorganization of lamellopodial actin. On occasion, two antiparallel fascin bundles come into contact at the plasma membrane, and reorient until they merge and project (25). Myosin X moves laterally along bundles that are parallel to the plasma membrane (Fig. S6), as first observed by Sousa et al. (26) and later by Tokuo et al. (27). At locations where two antiparallel bundles meet, myosin X may form a bridge, which would allow the bundles to pivot at the cross-over point until they merge into a larger, parallel bundle. However, our results argue against the specific model proposed by Tokuo, where myosin X first moves along single filaments toward the plasma membrane before reorganizing and bundling filaments (27).
Parallel, bundled actin filaments are ubiquitous in eukaryotes, appearing in structures that include sensory bristles, microvilli, and stereocilia (28). Because bundle systems often have their own specifically targeted myosins with various numbers of IQ domains in the neck region [including myosins I (29), VII (30), and XV (31)], a similar bundle selection process may occur at these sites as well. Our results emphasize the importance of using the native arrangement of cytoskeletal filaments when studying molecular motors, because key adaptations may be obscured when only individual filaments are used.
Materials and Methods
Proteins.
Bovine GFP-HMM myosin X in pEGFP-C2, containing N-terminal GFP and amino acids 1–943 of myosin X, including the head, neck, and predicted coiled-coil was used as provided (5). The HMM forced dimer was prepared by overlap extension PCR, and contained the myosin X sequence from the N terminus to residue Leu-920, followed by residues 4–32 of GCN4-p1 to ensure dimerization, followed immediately by GFP and a FLAG tag. The forced dimer includes the first 17 native heptad repeats of predicted coiled coil followed by 4 heptads of in-register GCN4 leucine zipper.
The myosin X HMM forced dimer construct was used to create recombinant baculovirus in Sf9 insect cells, expressed in Hi5 insect cells, and purified by FLAG affinity chromatography. Myosin V and myosin VI HMM-GCN4-GFP-Flag constructs were likewise expressed and purified in Sf9 cells by using baculovirus expression system as previously described (16). Chicken nmIIb (no insert) HMM was made by truncation after residue 1233 and adding GFP and FLAG tags in that order by overlap extension PCR. Virus production (including light chains), purification from Sf9 cells, and myosin light-chain kinase reactions (≈95% complete, by glycerol-urea gel electrophoresis) were as described (32). This nmIIb moves in vitro at ≈30 nm/s and decorates both actin and fascin-actin in an ATP-dependent manner. Motor stock concentrations were determined from GFP absorbance. Exchange reactions with Cy5-labeled calmodulin were performed by a calcium pulse as previously described with a 3-fold molar excess of labeled calmodulin over myosin (33). Fascin was prepared as previously described (34). Fascin-actin bundles were created by incubating 8 μM F-actin with 3 μM fascin for ≈2 days (4°C, in F-buffer) to ensure fully formed and ordered bundles. Bundles are stable for 2 weeks at 4°C. Methylcellulose bundles were prepared in situ by polymerizing 0.5 μM G-actin (with 10% G-actin covalently labeled with TMR at Cys-374) in the presence of Limulus sperm acrosomal processes (which nucleated unidirectional filaments) (35) and 2% (wt/vol) 15 cP methylcellulose (which was also used in all subsequent assay buffers) (see Fig. S3b).
Gliding Filament Motility Assays.
Motility assays were performed at 23°C in flow chambers constructed of a glass slide, two strips of double-sided tape and a nitrocellulose coated coverslip. All reagents were prepared in assay buffer (AB) containing 25 mM imidazole, pH 7.5, 25 mM KCl, 1 mM EGTA, 4 mM MgCl2, and 10 mM DTT. Reagents were added to the flow chamber in 10-μl volumes in the following order: 50 ng/μl anti-GFP (Qbiogene), 1 mg/ml BSA, myosin (variable concentration), AB, 100 nM TRITC-phalloidin actin, AB, and motility buffer. Motility buffer contains variable ATP, 0.86 mg/ml glucose oxidase, 0.14 mg/ml catalase, 9 mg/ml glucose in AB. We performed landing assays as described (17), by using 100 nM TRITC-phalloidin actin in the motility buffer. Control landing and continuous movement assays performed with myosin VI showed the expected n = 1 dependence. All solutions were incubated in the flow chamber for 2 min except the assay buffer washes. We assayed myosin X as above, but the motor was diluted in AB with 300 mM KCl and all reagents added to flow chamber after myosin X contained 20 μg/ml calmodulin. Actin filaments were imaged in epifluorescence on a Zeiss Axiovert 200 by an Andor Luca CCD. Images were analyzed in ImageJ. We manually tracked the leading or trailing ends of moving filaments, omitting those filaments that formed plectonemes.
Single-Molecule Imaging.
We imaged myosin motility using a custom-built objective-type total internal reflection microscope. Images were collected with a ×100, 1.65 NA objective (Olympus) and an EMCCD camera (iXon; Andor Technologies). Frames were collected at 2 Hz with a pixel size of 60 nm. We prepared phalloidin-stabilized F-actin that was 10% biotinylated at Cys-374. Flow chambers were coated with neutravidin (0.5 mg/ml), and then blocked with BSA (1 mg/ml). We applied to the flow cell either single filaments or fascin-bundles and immobilized them through the biotin–neutravidin interaction. We used a total actin concentration of 1.6 μM for fascin bundles and 0.16 μM for single filaments. We applied motility buffer (as above) containing 60 nM labeled motor (a concentration estimate that excludes losses from the calmodulin exchange procedure), with 1 mg/ml BSA and 1% Triton X-100 to further block nonspecific adsorption. Actin was imaged for four to six frames, then excitation was switched to the Cy5 channel to image motility. Overlaying the first few frames with the rest of the movie facilitated the identification of bundle ends. We identified moving spots by eye and then tracked them using a spot-tracking algorithm developed for ImageJ (36). Data for spots that moved <300 nm or <1.5 sec were discarded from analysis to eliminate misidentified diffusive events. Reported run lengths are uncorrected for photobleaching. At our measured Cy5 photobleaching rate (0.04 s−1), we estimate a correction of <10%. Single-molecule observations were verified from stepwise photobleaching and initial intensity distributions (see Fig. S1).
Optical Trapping.
We used a custom-built optical trap with bead position detection performed in the condenser back focal-plane (37). The details of the instrument will be provided elsewhere. We decorated 0.5-μm-diameter beads with myosin X via anti-GFP antibodies. The myosin density was chosen such that 30% of the beads bound or moved on surface attached fascin-actin bundles, establishing single-motor conditions. We introduced these beads into a flow chamber that had surface-attached fascin-actin bundles. The chamber also contained surface-attached 1-μm-diameter streptavidin beads. Biotinylated actin filaments (17) were attached to these beads by solution flow, as described in (22). We trapped a motor bead and identified one that moved processively on a fascin actin bundle. This step establishes that the bead under test contains a functional motor. We moved this same bead to a supported single actin filament, and interrogated the filament for 5–10 min. No processive runs were observed. We then returned the same bead to a fascin-actin bundle, to establish that functional motor was still present. Processive runs (traveling ≈100 nm) were observed within 30–60 s. We repeated this test cycle for five separate motor beads, and observed processive stepping only on the fascin-actin bundles, not the single filaments.
Cell Culture and Imaging.
HeLa cells were maintained in DMEM supplemented with 10% FBS and 10 μM l-glutamine at 37°C. We transiently transfected with the fascin RNAi or the control RNAi hairpin plasmids [called Th or Tm, respectively, in Vignjevic et al. (10)] 5 d before plating by using Lipofectamine 2000 (Invitrogen). Coverslips were coated with 20 μg/ml mouse laminin (Sigma) for 2 h and then blocked with DMEM plus 10% FBS for 20 min. We allowed cells to attach to the coverslip surface for 2 h before fixing with −20°C methanol for 4 min, washing three times with 0.5% Tween 20, and blocking with 5% FBS for 30 min at room temperature. Fixed cells were incubated for 1 h at room temperature sequentially with the following antibodies: rabbit anti-bovine myosin X (1:100; gift of R. E. Cheney), Cy5-conjugated donkey anti-rabbit (1:300; ChemiCon), mouse anti-fascin (1:100; DakoCytomation), FITC-conjugated goat anti-mouse (1:300; ChemiCon). Images were collected on a Zeiss Axiovert 200 microscope with an Andor Luca CCD. We counted filopodia [projections extending at least 0.5 μm beyond the cell perimeter in differential interference contrast (DIC) images] in ImageJ. Transfection with fascin RNAi reduced fascin by 75%, as judged from the integrated fascin stain compared to nearby untransfected cells (see Fig. S5).
Supplementary Material
Acknowledgments.
We thank R. E. Cheney (University of North Carolina, Chapel Hill, NC) for providing the myosin X pEGFP-C2 construct and helpful discussions, G. G. Borisy and S. Kojima (Northwestern University, Chicago, IL) for the fascin expression and fascin shRNA plasmids, R. Adelstein and J. Sellers (National Institutes of Health, Bethesda, MD) for the nmIIb construct and light-chain virus, and M. Glotzer and A. Piekny for assistance with cell culture and immunocytochemistry. M. Glotzer and D. Robinson provided comments on the manuscript. R.S.R. is supported by a Burroughs Wellcome Career Award at the Scientific Interface and grants from the American Heart Association and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802592105/DCSupplemental.
References
- 1.De La Cruz EM, Wells AL, Sweeney HL, Ostap EM. Actin and light chain isoform dependence of myosin V kinetics. Biochemistry. 2000;39:14196–14202. doi: 10.1021/bi001701b. [DOI] [PubMed] [Google Scholar]
- 2.Tang N, Ostap EM. Motor domain-dependent localization of myo1b (myr-1) Curr Biol. 2001;11:1131–1135. doi: 10.1016/s0960-9822(01)00320-7. [DOI] [PubMed] [Google Scholar]
- 3.Fanning AS, Wolenski JS, Mooseker MS, Izant JG. Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil Cytoskel. 1994;29:29–45. doi: 10.1002/cm.970290104. [DOI] [PubMed] [Google Scholar]
- 4.Ostap EM. In: Tropomyosins. Gunning P, editor. Austin, TX: Landes Bioscience; 2008. in press. [Google Scholar]
- 5.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 6.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113(19):3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 7.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Vignjevic D, Kojima S, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, Gerisch G. Organization of actin networks in intact filopodia. Curr Biol. 2007;17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Homma K, Ikebe M. Myosin X is a high duty ratio motor. J Biol Chem. 2005;280:29381–29391. doi: 10.1074/jbc.M504779200. [DOI] [PubMed] [Google Scholar]
- 13.Kovacs M, Wang F, Sellers JR. Mechanism of action of myosin X, a membrane-associated molecular motor. J Biol Chem. 2005;280:15071–15083. doi: 10.1074/jbc.M500616200. [DOI] [PubMed] [Google Scholar]
- 14.Nishizaka T, Yagi T, Tanaka Y, Ishiwata S. Right-handed rotation of an actin filament in an in vitro motile system. Nature. 1993;361:269–271. doi: 10.1038/361269a0. [DOI] [PubMed] [Google Scholar]
- 15.Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 16.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rock RS, Rief M, Mehta AD, Spudich JA. In vitro assays of processive myosin motors. Methods. 2000;22:373–381. doi: 10.1006/meth.2000.1089. [DOI] [PubMed] [Google Scholar]
- 18.Howard J, Hudspeth AJ, Vale RD. Movement of microtubules by single kinesin molecules. Nature. 1989;342:154–158. doi: 10.1038/342154a0. [DOI] [PubMed] [Google Scholar]
- 19.Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock RS, Purcell TJ, Spudich JA. In: The Enzymes. Hackney DD, Tamanoi F, editors. Vol 23. New York: Elsevier Academic; 2003. pp. 55–87. [Google Scholar]
- 21.Okten Z, Churchman LS, Rock RS, Spudich JA. Myosin VI walks hand-over-hand along actin. Nat Struct Mol Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 22.Ali MY, Uemura S, Adachi K, Itoh H, Kinosita K, Jr, Ishiwata S. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat Struct Biol. 2002;9:464–7. doi: 10.1038/nsb803. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Kovacs M, Hu AH, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of nonmuscle myosin IIB - Functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 24.Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci USA. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa AD, Berg JS, Robertson BW, Meeker RB, Cheney RE. Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J Cell Sci. 2006;119:184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 27.Tokuo H, Mabuchi K, Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J Cell Biol. 2007;179:229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartles JR. Parallel actin bundles and their multiple actin-bundling proteins. Curr Opin Cell Biol. 2000;12:72–78. doi: 10.1016/s0955-0674(99)00059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coluccio LM. Myosin I. Am J Physiol. 1997;273:C347–C359. doi: 10.1152/ajpcell.1997.273.2.C347. [DOI] [PubMed] [Google Scholar]
- 30.Tuxworth RI, Weber I, Wessels D, Addicks GC, Soll DR, Gerisch G, Titus MA. A role for myosin VII in dynamic cell adhesion. Curr Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 31.Rzadzinska AK, Schneider ME, Davies C, Riordan GP, Kachar B. An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. J Cell Biol. 2004;164:887–897. doi: 10.1083/jcb.200310055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pato MD, Sellers JR, Preston YA, Harvey EV, Adelstein RS. Baculovirus Expression of Chicken Nonmuscle Heavy Meromyosin II-B. J Biol Chem. 1996;271:2689–2695. doi: 10.1074/jbc.271.5.2689. [DOI] [PubMed] [Google Scholar]
- 33.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc Natl Acad Sci USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vignjevic D, Peloquin J, Borisy GG. In vitro assembly of filopodia-like bundles. Methods Enzymol. 2006;406:727–739. doi: 10.1016/S0076-6879(06)06057-5. [DOI] [PubMed] [Google Scholar]
- 35.Sun S, Footer M, Matsudaira P. Modification of Cys-837 identifies an actin-binding site in the beta-propeller protein scruin. Mol Biol Cell. 1997;8:421–430. doi: 10.1091/mbc.8.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sage D, Neumann FR, Hediger F, Gasser SM, Unser MM. Automatic Tracking of Individual Fluorescence Particles: Application to the Study of Chromosome Dynamics. IEEE Trans Image Proc. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
- 37.Visscher K, Gross SP, Block SM. Construction of multiple-beam optical traps with nanometer-level position sensing. IEEE J Sel Top Quant Electr. 1996;2:1066–1076. [Google Scholar]
- 38.Kaplan EL, Meier P. Nonparametric-Estimation from Incomplete Observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.