Abstract
Golgi α-mannosidase II (GMII) is a key glycosyl hydrolase in the N-linked glycosylation pathway. It catalyzes the removal of two different mannosyl linkages of GlcNAcMan5GlcNAc2, which is the committed step in complex N-glycan synthesis. Inhibition of this enzyme has shown promise in certain cancers in both laboratory and clinical settings. Here we present the high-resolution crystal structure of a nucleophile mutant of Drosophila melanogaster GMII (dGMII) bound to its natural oligosaccharide substrate and an oligosaccharide precursor as well as the structure of the unliganded mutant. These structures allow us to identify three sugar-binding subsites within the larger active site cleft. Our results allow for the formulation of the complete catalytic process of dGMII, which involves a specific order of bond cleavage, and a major substrate rearrangement in the active site. This process is likely conserved for all GMII enzymes—but not in the structurally related lysosomal mannosidase—and will form the basis for the design of specific inhibitors against GMII.
Keywords: cancer therapy, enzyme mechanism, N-glycosylation pathway, x-ray crystallography, glycoside hydrolase
The N-linked glycosylation pathway is a complex yet ubiquitous phenomenon in eukaryotic cells (recently reviewed in ref. 1). Eukaryotic N-linked glycosylation begins in the endoplasmic reticulum (ER) with the transfer of a preformed oligosaccharide from dolichol to an asparagine residue on a nascent polypeptide. This oligosaccharide is sequentially modified by enzymes in the ER and Golgi apparatus resulting in a final oligosaccharide structure. The modification steps involve trimming by glycosyl hydrolases and extension by glycosyl transferases.
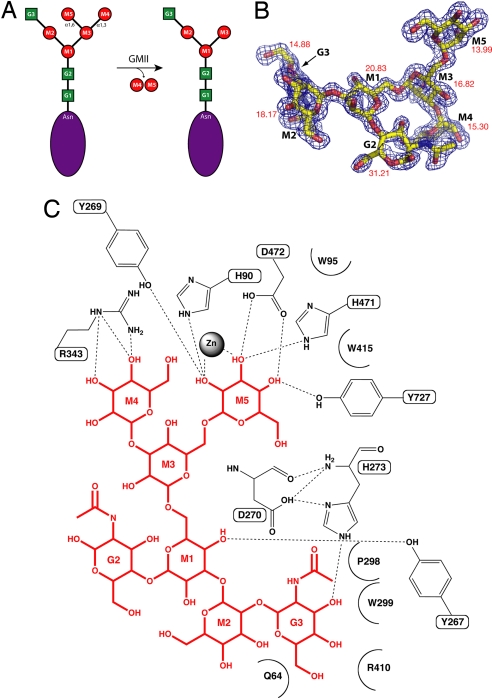
Golgi α-mannosidase II (GMII) is a glycosyl hydrolase that resides in the Golgi apparatus of eukaryotes and plays a key role in the N-linked glycosylation of proteins (2, 3). GMII has a high degree of sequence conservation among many eukaryotes. It has been classified as a family 38 glycosyl hydrolase and catalyzes the removal of both α-1,3-linked and α-1,6-linked mannoses from GnMan5Gn2 to yield GnMan3Gn2 (Gn, N-acetylglucosamine), which is the committed step of complex N-glycan synthesis (Fig. 1A) (4). The reaction requires the presence of the terminal Gn added to the glycan by N-acetylglucosaminyltransferase I (GnT I) and is hypothesized to proceed via a covalent glycosyl–enzyme intermediate, preceded and followed by oxocarbenium ion transition states resulting in retention of the stereochemistry of the substrate. Previous work has determined that whereas GMII is able to cleave two chemically distinct mannosyl linkages, it does so within a single catalytic site.
Fig. 1.
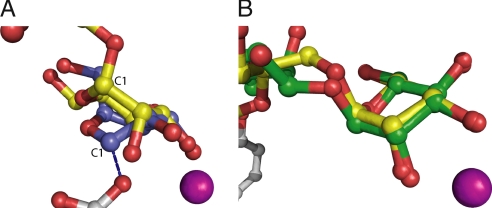
Golgi α-mannosidase II–substrate interactions. (A) Golgi α-mannosidase II catalyzes the cleavage of two mannosyl linkages, an α1,3-linkage between M3 and M4 and an α1,6-linkage between M3 and M5, converting GnMan5Gn2 to GnMan3Gn2. (B) GnMan5Gn fitted to the Fo − Fc electron density in the dGMII active site. The electron density is contoured to 3.0 σ. The crystallographic temperature factors represent the average values for the carbon and oxygen atoms of the saccharide. (C) dGMII–GnMan5Gn interactions as determined by HBPLUS/LIGPLOT.
Swainsonine, a plant-derived indolizidine alkaloid, is a potent inhibitor of GMII, with a Ki of 40 nM against the Drosophila melanogaster enzyme (dGMII) (5–7). Clinical trials suggested that swainsonine has therapeutic value because it reduces metastasis and improves clinical outcome in cancers of the colon, breast, and skin (8, 9). Unfortunately, there are side effects resulting from swainsonine treatment because it also inhibits a structurally related lysosomal mannosidase involved in catabolic processes. A greater understanding of the GMII catalytic process will allow for the development of highly specific inhibitors and thus, more efficacious chemotherapeutics.
Here we present structures of dGMII bound to its natural substrate (GnMan5Gn2) and an oligosaccharide (Man5) lacking the key terminal N-acetylglucosamine residue that is required for the reaction. The substrate was isolated and purified through the use of multiple enzymatic treatments and separation by chromatography. Crystallization and complex formation made use of a catalytically inactive nucleophile mutant. Analysis of the substrate–enzyme interactions provides compelling evidence of an evolutionarily conserved mode of substrate binding and catalysis. These results allow for the formulation of the complete catalytic process, which involves the sequential cleavage of two different linkages in the same catalytic site by substrate rearrangement. They provide a structural explanation for the requirement of the key N-acetylglucosamine for the reaction to occur, despite its distance from the scissile bonds. Furthermore, the results strongly support a model in which the α1,6-linked mannose is cleaved before the α1,3-linked saccharide. Finally, the possibility of exploiting differences between the lysosomal and Golgi variants of the enzyme for the purposes of greater specificity in chemotherapy is explored.
Results and Discussion
Substrate Binding and Catalysis.
The Fo − Fc electron density map shows continuous electron density in the active site at 1.4-Å resolution and allows for the unambiguous placement of all atoms of the substrate oligosaccharide GnMan5Gn [Fig. 1B and supporting information (SI) Table S1]. Hydrogen bonds and hydrophobic interactions are presented in Fig. 1C and Table S2.
The GnMan5Gn oligosaccharide binds in a large groove on the surface of the enzyme. This groove contains the site of the nucleophile (Asp-204), acid/base catalyst (Asp-341), and zinc ion and represents a region of extremely high degree of amino acid conservation (see Fig. 5A and Fig. S1). When this structure is compared with the unliganded D204A dGMII refined to 1.3-Å resolution, it is clear that substrate binding does not result in any noticeable conformational change.
Fig. 5.
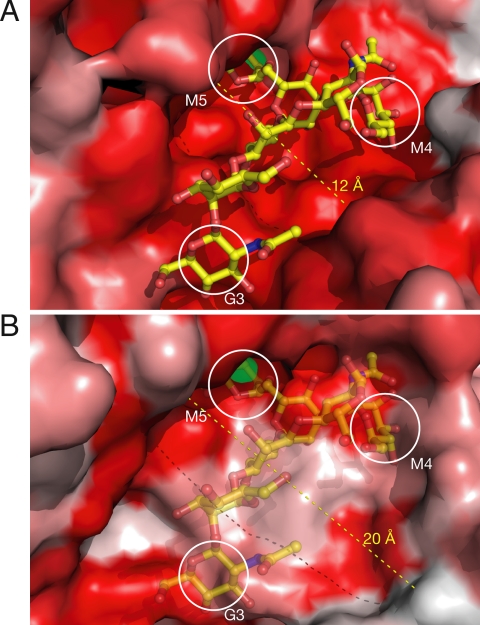
Comparison of the dGMII and bLM active site clefts. (A) A surface model of dGMII depicting the substrate-binding site and colored according to a multiple sequence alignment. More intense red shading indicate increased sequence conservation. Note the high degree of conservation in the cleft where the substrate (yellow) binds. The catalytic (M5), holding (M4), and anchor (G3) sites are indicated. (B) The bLM active site aligned in the same orientation as A above. The dGMII substrate is indicated as partially transparent. There is a lack of sequence conservation at both the holding and the anchor sites and an increased width of the active site cleft between dGMII and bLM.
Interestingly, almost all of the protein–carbohydrate interactions take place at three saccharide positions: M5, M4, and G3 (Fig. 1C and Table S2). Additionally, the ligand B factors as determined by the crystallographic structure solution indicate that M5, M4, and G3 are the most stably bound in the crystal structure (Fig. 1B). These three positions represent the saccharides most distal from the nascent protein. Substrate recognition by sensing the distal positions of the oligosaccharide is similar to what is seen with lectins (10).
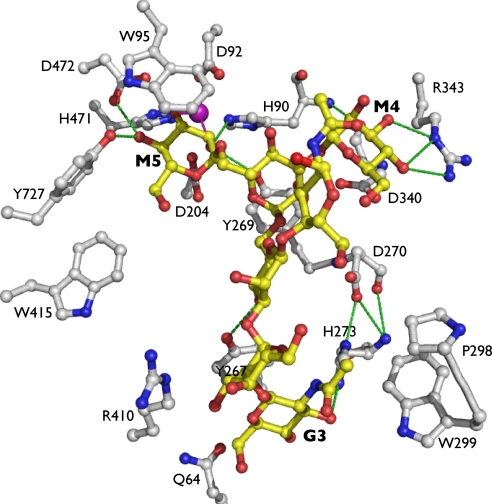
The active site of dGMII is made up of three sugar-binding sites: the catalytic, holding, and anchor sites (Fig. 2).
Fig. 2.
GMII–substrate. GnMan5Gn (yellow) is bound in the dGMII(D204A) active site (white). Hydrogen bonds are depicted as green dashed lines, and a single zinc ion is presented in purple and aids to coordinate M5 in the catalytic site.
Catalytic Site.
The α1,6-linked mannose (M5) is tightly bound in the catalytic site, forming many hydrogen bonds and stacking interactions with highly conserved residues in that region of the active site. Hydroxyls at positions 2, 3, and 4 hydrogen-bond to two conserved tyrosines (Tyr-269 and Tyr-727), a histidine (His-471), and an aspartic acid (Asp-472). In addition, the hydrophobic face of the saccharide ring forms a stacking interaction with Trp-95, and there is some hydrophobic contribution from Tyr-727 as well. The oxygens at positions 2 and 3 coordinate the stably bound zinc ion in the active site, which is consistent with many GMII inhibitors we have studied (7, 11, 12). The site of nucleophilic attack (C1 of M5) is positioned between the nucleophile and acid/base catalyst, indicating that the hydrolysis reaction occurs at this site exclusively.
Holding Site.
The α1,3-linked mannose (M4) occupies a holding site some 9 Å from the Asp-204 nucleophile and is multiply coordinated by a GMII-conserved arginine (Arg-343). The architecture of this holding site is fairly open, with no stacking contribution to binding whatsoever. Previous work with this enzyme has not revealed any inhibitor binding in this site.
Anchor Site.
The required N-acetylglucosamine (G3) is stably bound in a tight pocket 13 and 14 Å from M5 and M4, respectively. It forms strong stacking interactions with a GMII-conserved tyrosine residue (Tyr-267), its acetyl position is buried in a hydrophobic patch formed by a conserved tryptophan (Trp-299) and proline (Pro-298), and it forms a hydrogen bond with a conserved histidine (His-273). The necessity of this nonhydrolyzed anchor is clear when considering the nature of the substrate. Oligosaccharide molecules are highly flexible both about their glycosidic bonds and in the conformation of their saccharide rings. The presence of a stabilizing anchor such as the G3 assists in binding and orienting the substrate for the hydrolysis reaction as well as increasing the local concentration of the linkages to be cleaved by GMII.
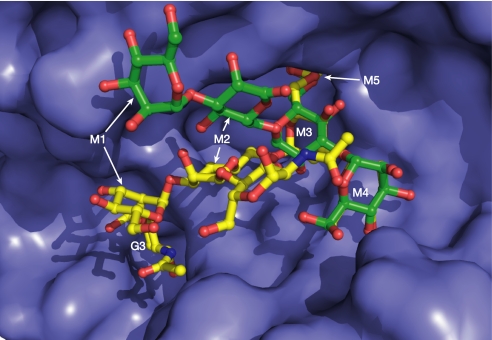
The Fo − Fc electron density map from the dGMII(D204A)-Man5 structure refined to 1.6-Å resolution shows clear electron density for the oligosaccharide (Fig. S2). The G3-lacking Man5 oligosaccharide binding to dGMII also demonstrates the necessity of the anchor site in proper substrate orientation. The α1,6- and α1,3-linked sugars bind to the catalytic and holding sites, respectively, in a manner nearly identical to that seen in GnMan5Gn. However, because of the lack of the anchor saccharide, the other mannose positions are quite different, with the M1- and M2-containing oligosaccharide “tail” extending out of the active site cleft with O4 of the M1 saccharide-binding Asp-873 (Fig. 3). Thus, the enzyme is able to bind the Man5 substrate in the correct orientation even without the G3, and yet Man5 is not cleaved. Recent in vitro work with dGMII indicates that there is an 80-fold greater activity against GnMan5Gn vs. Man5Gn (13). This finding indicates that the G3 residue is critical not only in binding to the M4 and M5 sites but also in the catalytic process itself (see below for further discussion). Recent work studying complexes of dGMII with synthetic S-linked oligosaccharides has contributed preliminary insights into substrate binding (13). The structure of a synthetic mannose tetrasaccharide bound to the active site of dGMII (Protein Data Bank structure ID: 3BVV) suggests that the catalytic site of dGMII can accommodate both α1,6-linked and α1,3-linked mannoses. An α1,6- and α1,3-containing tetrasaccharide binds in the active site with the α1,3-linked mannose in the catalytic site because of the presence of a sulfur linkage to the α1,6-linked sugar. Importantly, the M3 swivel position is shifted significantly from its position in the substrate complex (see discussion below). The α1,3-linked mannose adopts precisely the same conformation and makes identical interactions with catalytic site residues as does the α1,6-linked mannose of the full substrate (Fig. 4B). Therefore, we suggest that the mode of mannose binding in the catalytic site is identical for the α1,6- and α1,3-linked mannoses in the GnMan5Gn2 substrate during catalysis.
Fig. 3.
Comparison of GnMan5Gn and Man5 bound to dGMII. The absence of the G3 anchor position in Man5 (green) results in a very different mode of binding, particularly distal from the catalytic and holding sites. The saccharide shifts are 11 Å for M1 and 5 Å for M2.
Fig. 4.
The M5 saccharide compared with a covalent reaction intermediate and a synthetic tetrasaccharide. (A) Position M5 in the catalytic site (yellow) superimposed on 5-fluorogulosyl fluoride (slate), which is covalently attached (blue dashed line) to OE2 of Asp-204. A zinc ion is in shown in purple. The distance between the C1 positions of each saccharide moiety is ≈2 Å. Such a distance could be traversed by a distortion from a 4C1 to a 1S5—which is what is proposed to occur concurrent with nucleophilic attack by Asp-204. (B) A superposition of the mannosyl moieties in the catalytic site of dGMII from the full substrate (yellow) and a synthetic tetrasaccharide (green).
Initial Substrate Orientation.
Biochemical evidence has previously shown that GMII shows a preference for M5 cleavage over M4 (14, 15). Until now, however, there has been no structural explanation for this specificity. Our results clearly show that the catalytic site exclusively binds the α1,6-linked mannose, whereas the α1,3-linked mannose resides in a holding site. In the context of the full GMII substrate, the holding site cannot accommodate the longer α1,6-linked arm while maintaining G3 in the anchor site because highly conserved residues pack against and make significant contacts with the α1,3-linked mannose in the holding subsite (Fig. S3). It appears that the steric restrictions in the dGMII holding site favor binding of the α1,6-linked sugar in the catalytic site initially. Thus, the chief determinant of bond cleavage order appears to be the holding site. This is consistent with the notion that the catalytic site requires a certain degree of promiscuity to cleave both the α1,6-linked and the α1,3-linked mannose residues, whereas the holding site, in its sole binding of the α-1,3-linked mannose, does not.
First Hydrolysis Reaction.
Previous work with this enzyme containing a wild-type Asp-204 and a trapped covalent intermediate suggests that the catalytic mechanism of GMII proceeds via a B2,5 intermediate because the saccharide ring of 5-fluorogulosyl fluoride is covalently bound to the OE2 in a 1S5 conformation in the catalytic site (Fig. 4A) (16). This observation indicates that when Asp-204 is present in the wild-type enzyme, the M5 saccharide undergoes distortion concurrent with nucleophilic attack by OE2 of Asp-204. This attack forms the glycosyl–enzyme intermediate, which is released when Asp-341 abstracts a proton from a water molecule, leading to nucleophilic attack by a hydroxyl ion at position C1 of the M5 saccharide. This regenerates the enzyme and is followed by release of the M5 mannose.
Intermediate Substrate Rearrangement.
Release of the M5 saccharide is followed by a substrate rearrangement that results in the positioning of the M4 saccharide in the catalytic site for hydrolytic cleavage. Such rearrangements are reminiscent of those reported in the context of processing endoglycanases (17, 18). In previous dGMII structures, we have seen mannose binding at the M3 (+1) position to be quite variant. We have termed the M3 position the swivel position owing to this conformational variability as well as the open character of the active site cleft in this region.
Internal Communication Between Subsites.
An important issue in the catalysis is to what extent the occupancy of each of the sites affects the characteristics of the other sites. In particular, as proposed above, how does the occupancy of the anchor site have an effect on substrate catalysis? There are numerous hydrogen bonds between the conserved residues Asp-270 and His-273 (Figs. 1C and 2). Although it does not make direct contacts with the substrate, Asp-270 acts to coordinate His-273 in the proper orientation to form a hydrogen bond with O3 of the G3 N-acetylglucosamine. In addition, Asp-270 lies in a region of the active site that makes it equidistant from the G3-binding site and the initial M4-holding site. This may indicate that, through proton transfer, the Asp-270 position could facilitate communication between the anchor and holding subsites.
Additionally, the presence of a histidine (His-273) at a key position of the active site suggests that the communication proceeds via a proton shuttle (19–22). Histidine residues are well suited to be both donors and acceptors of protons at physiological pH, and the presence of His-273 in coordinating the active site and making contact with Asp-270 may indicate such a role. We propose that in the absence of a substrate moiety in the anchor site, His-273 is able to use the basic nitrogen of its imidazole ring to abstract a proton from the water molecules in the anchor region, which is followed by a donation of a proton from the acidic nitrogen of the imidazole to the well-positioned side chain of Asp-270. When the anchor site is occupied, Tyr-269 is properly positioned, which, in effect, primes the catalytic site for binding and catalysis. Therefore, the enzyme is able to translate information from two of the key sites in the active site cleft, the anchor site, and catalytic site. This signal transfer must be precisely orchestrated such that the catalytic site remains competent until the second linkage is cleaved, at which time the product is released.
Second Hydrolysis Reaction and Product Release.
The second hydrolysis reaction of the α1,3 linkage follows the same mechanism as the first, with nucleophilic attack being concurrent with distortion of the M4 saccharide ring from a 4C1 to a B2,5 followed by proton abstraction of water by Asp-341 and deglycosylation restoring the catalytic site to its initial state.
Evolutionary Significance of Binding and Catalysis.
It has been known for some time that GMII enzymes from a wide variety of species are conserved, but these results indicate some of the reasons that certain highly conserved amino acids are crucial for substrate binding and catalysis. Nonbinding residues in the catalytic, holding, anchor, and swivel positions all retain a high degree of sequence conservation among GMII enzymes. The residues involved in the putative internal communication are also highly conserved. It follows that the sequence of catalytic events and intermediates is also conserved across species. This allows for greater confidence when using model systems such as D. melanogaster GMII in human drug discovery efforts. This conservation speaks to the high degree of precision required to orchestrate the series of events as described, as well as the physiological importance of this complicated function.
Comparison with the Lysosomal α-Mannosidase.
Knowledge of the structure of a lysosomal α-mannosidase from Bos taurus (bLM) solved by Heikinheimo et al. (23) in conjunction with the data presented here permits detailed structural comparison and will assist in development of specific GMII inhibitors.
The active site cleft of bLM is much wider than that of dGMII (12 Å vs. 20 Å) but still contains a tightly bound zinc in the catalytic position as shown in Fig. 5 and Fig. S4. There is a striking amount of sequence similarity between the two enzymes in some regions and almost none in others. There is a lack of sequence conservation at both the holding and the anchor sites and an increased width of the active site cleft in bLM (Fig. 5B). In examining the surface representation of the two enzymes, it seems that the size and shape of the dGMII active site cleft is well contoured to the size and shape of the GnMan5Gn2 oligosaccharide, whereas the bLM site is much wider.
Almost all of the key residues from dGMII that are involved in M5 binding and Zn coordination are present in the bLM active site, in strikingly similar positions (Fig. S4 and Table S3). The residues involved in M5 hydrogen bonding (Tyr-269/261, His-471/446, Asp-472/447, and Tyr-727/660) are present in both enzymes and adopt similar conformations and would ostensibly coordinate saccharide moieties in a nearly identical manner. The position of the zinc ion is almost invariant, with key residues coordinating the ion (His-90/72, Asp-92/74, His-470/445, and His-471/446) being conserved in both sequence and atomic position. The Trp-95 position in dGMII that is believed to be extremely important for saccharide binding has an equivalent residue in bLM (Trp-77) (24). Last, the nucleophile (Asp-204/196) adopts a similar position in the two enzymes and likely performs a very similar role.
In two active sites this similar to one another, one can surmise that the mechanism of nucleophilic attack, regeneration by the acid/base catalyst, and product release are quite similar. This lends credibility to the notion that one must look beyond the catalytic site as an inhibitor target if one wishes to distinguish between the enzymes by small-molecule inhibition.
Instead of a conserved arginine in the holding site, the bovine enzyme contains two weakly conserved residues, a glutamine (Gln-321) and a leucine (Leu-272). Due to their arrangement, it is difficult to envision a mode by which they would be able to coordinate the hydroxyl groups of the M4 saccharide (Fig. S5A). In addition, the Golgi conserved aspartic acid that provides some saccharide-binding ability by its hydrophobic face in dGMII (Asp-340) is not present in bLM. Instead, there is a serine some distance away. Overall, there do not seem to be residues that would be suited to coordinating saccharide moieties, and there is little sequence conservation in this area.
The anchor site does not seem to exist in bLM. There is instead a pocket of moderately conserved residues, none of which possess the acidic or aromatic character present in the dGMII anchor site (Fig. S5B). The formation of the anchor site in dGMII depends on the extension of two loops from the main body of the protein. The first loop carries Tyr-267, which makes a crucial stacking interaction with the G3 position and a hydrogen bond with O4 of the M1 saccharide. The tyrosine exists on a loop that is stabilized in position by a short α-helical segment immediately preceding it. This tyrosine, along with the loop and helical segment, is completely absent in bLM. A second loop in dGMII carries a proline (Pro-298) and a tryptophan (Trp-299), which are uncharacteristically at the protein surface. These residues form a small hydrophobic patch, which interacts with the N-acetyl group of G3. The loop extension is completely absent in the bLM structure. In fact, there are no bLM residues nearby that could form the same hydrophobic patch seen in dGMII. The fact that bLM lacks these two key loops, and consequently the residues necessary to form the anchor site, indicates that there likely is no element of anchor binding taking place.
The expanded active site cleft and limited sequence conservation is consistent with the substrate specificity of LM, which cleaves all of the α-linked mannose residues from high-mannose oligosaccharides, whereas GMII only cleaves the terminal α1,3- and α1,6-linked residues from GnMan5Gn2 (2, 25).
The two unconserved regions, the holding site and, in particular, the G3 anchor site, are appealing targets for deriving specificity against GMII over the lysosomal counterpart.
Conclusions
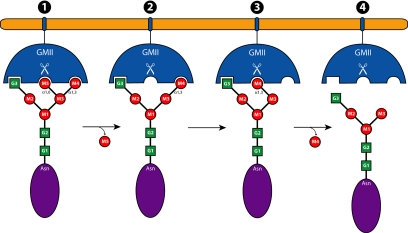
The model for this enzyme of the cleavage of two different linkages in the same site by conformational rearrangement of the substrate is unprecedented (Fig. 6). With a detailed trajectory of the GMII reaction, including putative transition states, we can now envision a variety of approaches to study this fascinating reaction in further detail. Many of the conserved residues mentioned, especially those possibly implicated in internal communication, could be excellent candidates for mutagenesis followed by enzymatic and structural studies.
Fig. 6.
A proposed schematic of the GMII reaction. The reaction begins with the binding of the GnMan5Gn2 oligosaccharide to GMII, with the M5 position binding in the catalytic site, M4 occupying the holding site, and G3 in the anchor site (1). After the hydrolysis of M5 by the enzyme, the catalytic site is free (2). Oligosaccharide rearrangement ensues to allow M4 to enter the catalytic site (3). M4 is subsequently hydrolyzed in the catalytic site by a similar mechanism, and the M4 release is concurrent with product release (4).
In addition, we have been able to clearly define three subsites in the GMII active site, each with unique characteristics, which suggests an approach to glycosidase inhibition. With advanced techniques of chemical synthesis of oligosaccharides and their analogs, designed inhibitors may be of the form that span the unique active site of GMII. One can envision a “three-headed” inhibitor based on a flexible linker with saccharide-type moieties for targeting the catalytic, holding, and anchor sites. Targeting multiple binding sites with a single inhibitor molecule should lead to strong specific GMII inhibition. This avidity-based approach would allow for the required specificity and efficacy for eventual use in the clinic.
Materials and Methods
Protein Expression and Purification.
Wild-type dGMII and the D204A nucleophile mutant protein were expressed and purified in D. melanogaster S2 cells as described previously (13, 15).
Crystallization.
Wild-type dGMII crystals were grown by using the Nextal crystallization tool (Qiagen) by hanging-drop vapor diffusion. Concentrated protein solution (12 mg/ml) was mixed in a 1:1 ratio with reservoir solution containing 0.1 M Tris, pH 7, 8.5% PEG 8000, and 2.5% 2-methyl-2,4-pentanediol and suspended over the reservoir for 18 h. The resulting single crystals were used to prepare wild-type dGMII microseeds by using the SeedBead protocol (Hampton Research). These microseeds were used to seed crystal growth of D204A protein in the above conditions. The resulting D204A crystals were once again subjected to the SeedBead protocol to make mutant crystal seeds and minimize the amount of wild-type enzyme in the crystals. The final D204A microseeds were used to nucleate crystals of the D204A mutant.
Substrate Isolation and Purification.
Bovine pancreatic ribonuclease B (Sigma–Aldrich) was dissolved in a denaturing solution of 0.5% SDS and 40 mM DTT to a concentration of 12 mg/ml and incubated at 95°C for 30 min. The denatured RNase B was treated with sodium citrate to a final concentration of 50 mM, pH 5.5, at which point 10,000 units of endoglycosidase Hf (New England Biolabs) was added, and the mixture was incubated at 37°C for 18 h to release the high-mannose oligosaccharides. Endoglycosidase Hf catalyzes the hydrolysis between the first and second N-acetylglucosamines of high-mannose oligosaccharides. The oligosaccharide release was monitored by SDS/PAGE. The mixture was applied to a 15 × 200-mm BioGel P-10 column (Bio-Rad) and run by gravity. Fractions (1.0 ml) were collected and tested for protein content by the BCA protein assay (Pierce Biotechnology) and for carbohydrate content by the phenol/sulfuric acid assay (26). Carbohydrate-containing fractions were pooled and lyophilized. The resulting residue was dissolved in 1.0 ml of H2O and applied to a PD-10 column equilibrated with H2O for the purposes of buffer exchange. The carbohydrate-containing fractions, as determined by the phenol/sulfuric acid assay, were pooled, lyophilized, and dissolved in 0.6 ml of H2O. To add the GlcNAc required for GMII function (G3 in Fig. 1A), the RNase B oligosaccharides were treated with GnT I in a reaction similar to the one performed by Chen et al. (27). Briefly, the oligosaccharides were dissolved to a concentration of 1 mM in a mixture containing 50 mM Mes, pH 6.5, 60 mM GlcNAc, 3 mM AMP, 15 mM MnCl2, and 2 mM UDP-GlcNAc. The mixture was prewarmed to 37°C, and then Sf9 culture supernatant containing secreted GnT I [a kind gift of Harry Schachter (Hospital for Sick Children, Toronto)] was added to a total volume of 10%. The reaction was incubated at 37°C for 24 h and then was stopped by the addition of an equal volume of 0.2 M glycine, pH 10. The reaction mixture was then applied to a Bio-Gel P-10 column as above, and the carbohydrate-containing fractions were pooled, lyophilized, and redissolved in 0.5 ml of H2O. The carbohydrates were then applied to a Bio-Gel P-4 column as above, and the fractions containing oligosaccharides were pooled, lyophilized, and dissolved in 0.2 ml of H2O. Man5 was obtained from Sigma–Aldrich and was solubilized in water before crystallization experiments.
Crystal Complex Formation.
Cocrystallization of mutant dGMII(D204A) and GnMan5Gn proved to be unsuccessful. Freshly grown D204A crystals were soaked in a solution of GnMan5Gn in reservoir buffer for 18–24 h. The molar ratio of GnMan5Gn to dGMII was calculated to be at least 10:1. The dGMII(D204A)-Man5 crystal complex was obtained by crystal soaking in a similar manner.
Data Collection.
D204A crystals were cryoprotected by soaking in increasing concentrations of 2-methyl-2,4-pentanediol in reservoir buffer (5 → 10 → 15 → 20 → 25%). The cryoprotected crystals were mounted on a 10-μm MicroMesh (MiTeGen) or 0.4-mm CryoLoops (Hampton Research) followed by flash freezing in a nitrogen cold stream set at 100 K (Oxford Cryosystems). The crystals were exposed to X-rays at Station A1 of the Cornell High Energy Synchrotron Source. Data were collected at a wavelength of 0.9777 Å at 100 K, and diffraction patterns were collected on an ADSC Quantum-210 CCD detector. Typically, 400 images were collected per crystal with an oscillation of 0.5° per frame.
Structure Determination.
The data obtained from the diffraction experiment were integrated and scaled by using DENZO and SCALEPACK, respectively, within the HKL2000 graphical interface (28). Model building, refinement, and visualization were accomplished by using REFMAC within the CCP4 suite and Coot (29, 30). In the case of the oligosaccharide complexes, the Fo − Fc σA-weighted electron density map was used to fit GnMan5Gn and Man5 into the model (Fig. 1B and Fig. S2). Molecular topology for the oligosaccharides was generated by using the PRODRG server (31). Images were prepared by using the PyMOL molecular graphics system (32).
Sequence Alignment and Mapping to Structure.
MULTALIN was used to align amino acid sequences for Golgi α-mannosidase II enzymes from several organisms (D. melanogaster, Anopheles gambia, Homo sapiens, Mus musculus, Xenopus laevis, Arabidopsis thaliana, and Caenorhabditis elegans). The multiple sequence alignment was used in combination with ConSurf to produce an atomic coordinate file highlighting conserved residues on the dGMII structure (33, 34).
LIGPLOT.
To determine the protein residues directly involved in substrate binding, an analysis was performed by using HBPLUS/LIGPLOT (35, 36). Hydrogens were added to the substrate and protein by using HBPLUS, and then hydrogen bonds and hydrophobic interactions were visualized by using LIGPLOT.
Supplementary Material
Acknowledgments.
We thank Harry Schachter for providing the GnT I used in the oligosaccharide preparation and for helpful discussions. The mutant enzyme used in this study was produced by T. Signorelli and R.-A. Stewart. This work is based on research conducted at the Cornell High Energy Synchrotron Source, which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under award DMR-0225180. N.S. was funded by a postgraduate scholarship from the Natural Sciences and Engineering Research Council of Canada. This work was supported by grants from the Canadian Institutes for Health Research (MOP42452 and MOP79312) and the Mizutani Foundation (080030).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3CZN, 3CZS, and 3CVS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802206105/DCSupplemental.
References
- 1.Roth J. Protein N-glycosylation along the secretory pathway: Relationship to organelle topography and function, protein quality control, and cell interactions. Chem Rev. 2002;102:285–303. doi: 10.1021/cr000423j. [DOI] [PubMed] [Google Scholar]
- 2.Moremen KW. Golgi α-mannosidase II deficiency in vertebrate systems: Implications for asparagine-linked oligosaccharide processing in mammals. Biochim Biophys Acta. 2002;1573:225–235. doi: 10.1016/s0304-4165(02)00388-4. [DOI] [PubMed] [Google Scholar]
- 3.Chui D, et al. Alpha-mannosidase-II deficiency results in dyserythropoiesis and unveils an alternate pathway in oligosaccharide biosynthesis. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 4.Henrissat B, Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbein AD, Solf R, Dorling PR, Vosbeck K. Swainsonine: An inhibitor of glycoprotein processing. Proc Natl Acad Sci USA. 1981;78:7393–7397. doi: 10.1073/pnas.78.12.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorling PR, Huxtable CR, Colegate SM. Inhibition of lysosomal alpha-mannosidase by swainsonine, an indolizidine alkaloid isolated from Swainsona canescens. Biochem J. 1980;191:649–651. doi: 10.1042/bj1910649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah N, Kuntz DA, Rose DR. Comparison of kifunensine and 1-deoxymannojirimycin binding to class I and II alpha-mannosidases demonstrates different saccharide distortions in inverting and retaining catalytic mechanisms. Biochemistry. 2003;42:13812–13816. doi: 10.1021/bi034742r. [DOI] [PubMed] [Google Scholar]
- 8.Goss PE, Reid CL, Bailey D, Dennis JW. Phase IB clinical trial of the oligosaccharide processing inhibitor swainsonine in patients with advanced malignancies. Clin Cancer Res. 1997;3:1077–1086. [PubMed] [Google Scholar]
- 9.Baptista JA, et al. Measuring swainsonine in serum of cancer patients: Phase I clinical trial. Clin Chem. 1994;40:426–430. [PubMed] [Google Scholar]
- 10.Gabius HJ, Siebert HC, André S, Jiménez-Barbero J, Rüdiger H. Chemical biology of the sugar code. ChemBioChem. 2004;5:740–764. doi: 10.1002/cbic.200300753. [DOI] [PubMed] [Google Scholar]
- 11.van den Elsen JM, Kuntz DA, Rose DR. Structure of Golgi α-mannosidase II: A target for inhibition of growth and metastasis of cancer cells. EMBO J. 2001;20:3008–3017. doi: 10.1093/emboj/20.12.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawatkar SP, Kuntz DA, Woods RJ, Rose DR, Boons GJ. Structural basis of the inhibition of Golgi α-mannosidase II by mannostatin A and the role of the thiomethyl moiety in ligand-protein interactions. J Am Chem Soc. 2006;128:8310–8319. doi: 10.1021/ja061216p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong W, et al. Probing the substrate specificity of Golgi α-mannosidase II using synthetic oligosaccharides and a catalytic nucleophile mutant. J Am Chem Soc. 2008 doi: 10.1021/ja711248y. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren J, Castellino FJ, Bretthauer RK. Purification and properties of α-mannosidase II from Golgi-like membranes of baculovirus-infected Spodoptera frugiperda (IPLB-SF-21AE) cells. Biochem J. 1997;324:951–956. doi: 10.1042/bj3240951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altmann F, März L. Processing of asparagine-linked oligosaccharides in insect cells: Evidence for alpha-mannosidase II. Glycoconj J. 1995;12:150–155. doi: 10.1007/BF00731359. [DOI] [PubMed] [Google Scholar]
- 16.Numao S, Kuntz DA, Withers SG, Rose DR. Insights into the mechanism of Drosophila melanogaster Golgi α-mannosidase II through the structural analysis of covalent reaction intermediates. J Biol Chem. 2003;278:48074–48083. doi: 10.1074/jbc.M309249200. [DOI] [PubMed] [Google Scholar]
- 17.Ludwiczek ML, Heller M, Kantner T, McIntosh LP. A secondary xylan-binding site enhances the catalytic activity of a single-domain family 11 glycoside hydrolase. J Mol Biol. 2007;373(2):337–354. doi: 10.1016/j.jmb.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 18.Li C, et al. Acarbose rearrangement mechanism implied by the kinetic and structural analysis of human pancreatic α-amylase in complex with analogues and their elongated counterparts. Biochemistry. 2005;44:3347–3357. doi: 10.1021/bi048334e. [DOI] [PubMed] [Google Scholar]
- 19.Elder I, et al. Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III. Proteins. 2007;68:337–343. doi: 10.1002/prot.21403. [DOI] [PubMed] [Google Scholar]
- 20.Fisher Z, et al. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry. 2005;44:1097–1105. doi: 10.1021/bi0480279. [DOI] [PubMed] [Google Scholar]
- 21.Huang S, et al. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J Mol Biol. 1998;283:301–310. doi: 10.1006/jmbi.1998.2077. [DOI] [PubMed] [Google Scholar]
- 22.Lindskog S. Structure and mechanism of carbonic anhydrase. Pharmacol Ther. 1997;74:1–20. doi: 10.1016/s0163-7258(96)00198-2. [DOI] [PubMed] [Google Scholar]
- 23.Heikinheimo P, et al. The structure of bovine lysosomal α-mannosidase suggests a novel mechanism for low-pH activation. J Mol Biol. 2003;327:631–644. doi: 10.1016/s0022-2836(03)00172-4. [DOI] [PubMed] [Google Scholar]
- 24.Sundari CS, Balasubramanian D. Hydrophobic surfaces in saccharide chains. Prog Biophys Mol Biol. 1997;67:183–216. doi: 10.1016/s0079-6107(97)00016-3. [DOI] [PubMed] [Google Scholar]
- 25.Park C, et al. Characterization of a human core-specific lysosomal α1,6-mannosidase involved in N-glycan catabolism. J Biol Chem. 2005;280:37204–37216. doi: 10.1074/jbc.M508930200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubois M, Gilles K, Hamilton JK, Rebers PA, Smith F. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Zhou S, Sarkar M, Spence AM, Schachter H. Expression of three Caenorhabditis elegans N-acetylglucosaminyltransferase I genes during development. J Biol Chem. 1999;274:288–297. doi: 10.1074/jbc.274.1.288. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 32.DeLano W. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. version 1.0. [Google Scholar]
- 33.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau M, et al. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 36.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.