Abstract
The discovery of new small molecules and their testing in rational combination poses an ongoing problem for rare diseases, in particular, for pediatric cancers such as neuroblastoma. Despite maximal cytotoxic therapy with double autologous stem cell transplantation, outcome remains poor for children with high-stage disease. Because differentiation is aberrant in this malignancy, compounds that modulate transcription, such as histone deacetylase (HDAC) inhibitors, are of particular interest. However, as single agents, HDAC inhibitors have had limited efficacy. In the present study, we use an HDAC inhibitor as an enhancer to screen a small-molecule library for compounds inducing neuroblastoma maturation. To quantify differentiation, we use an enabling gene expression-based screening strategy. The top hit identified in the screen was all-trans-retinoic acid. Secondary assays confirmed greater neuroblastoma differentiation with the combination of an HDAC inhibitor and a retinoid versus either alone. Furthermore, effects of combination therapy were synergistic with respect to inhibition of cellular viability and induction of apoptosis. In a xenograft model of neuroblastoma, animals treated with combination therapy had the longest survival. This work suggests that testing of an HDAC inhibitor and retinoid in combination is warranted for children with neuroblastoma and demonstrates the success of a signature-based screening approach to prioritize compound combinations for testing in rare diseases.
Keywords: chemical genomics, small molecule screening
Dose intensification has improved outcome for some malignancies, such as neuroblastoma, the most common pediatric solid tumor. Despite this advance, progression-free survival for children with advanced disease is 45% at best, even with double autologous stem cell transplantation (1). For those children whose disease is cured, morbidity from intensive chemotherapy is significant. New approaches to the treatment of this malignancy are needed.
One targeted class of compound of particular interest is the histone deacetylase (HDAC) inhibitor. Histone acetylation is a critical regulatory mechanism of gene expression. Aberrant deacetylation of critical genes involved in differentiation, apoptosis, or cell cycle arrest has been reported to contribute to the pathogenesis of malignancy. Indeed, the development of neuroblastoma is believed to be related, in part, to defects in neural crest cell differentiation, and hence aberrant transcriptional regulation. Chromatin-modifying compounds, such as HDAC inhibitors, are therefore attractive agents for testing in neuroblastoma. Small molecules targeting the HDACs have been shown to have prodifferentiating and apoptotic effects in numerous cancer subtypes in model systems, including neuroblastoma (2, 3). Recently, there has been a marked increase in the number of HDAC inhibitors in clinical development. However, as single agents, these molecules have had limited activity, with the exception of the treatment of cutaneous T cell lymphoma (CTCL). In 2006, the Food and Drug Administration (FDA) approved the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) for adults with CTLC (4). It is likely that the broad efficacy of HDAC inhibitors will only be realized in combination with other drugs.
Combination therapy has been a hallmark of successful cancer treatment. With few exceptions, curative systemic cancer therapy has required the use of multiple drugs. The vast majority of combination strategies have derived from testing an agent with components of existing standards of care in the clinic, as opposed to rationally identifying compound combinations in the laboratory based on mechanism of action or performance in a small-molecule library screen. In a time where an extraordinary number of small molecules are in development, the rational selection of compounds for clinical testing becomes even more important. This is particularly problematic in the pediatric malignancies where the diseases are rare, precluding testing of all possible new agents, let alone all possible combinations. The development of small-molecule library screening approaches to facilitate the rational selection of compound combinations is critical.
In the current study, rather than focusing on cytotoxicity, we have focused on neuroblast differentiation. Prior clinical data suggest that differentiation therapy may play a complementary role in the treatment of neuroblastoma when used in combination with other therapies (5). However, high-throughput assays to quantitatively measure neuroblastoma differentiation as a phenotypic endpoint have been limited. We have developed a potential solution to this challenge in which a multigene signature is used as a surrogate for highly complex cellular states: gene expression-based high-throughput screening (GE-HTS) (6). First, gene expression signatures are defined for each biological state of interest by using genomewide expression profiling. Then, a high-throughput, low-cost assay is performed to evaluate up to 100 marker genes (7).
This work develops a general approach to the identification of combinations of small molecules for therapeutic consideration. Specifically, we now apply GE-HTS to the identification of small molecules that enhance the effects of HDAC inhibitors on neuroblastoma differentiation.
Results
Gene Expression-Based Small-Molecule Library Screen Identifies the Combination of an HDAC Inhibitor and Retinoid for Neuroblastoma Differentiation.
The goal in this work was to screen for compounds that act in concert with HDAC inhibitors to induce neuroblastoma differentiation. Although HDAC inhibitors are weak inducers of differentiation as single agents, we hypothesized that together with other compounds, we might identify synergistic combinations. First, we needed to identify an appropriate HDAC inhibitor to use as the enhancer. We selected valproic acid (VPA) because it was the only HDAC inhibitor with FDA-approval status at the time that we began the screen. VPA has a long history of clinical use in treating patients with seizures and bipolar disease, even before its characterization as an HDAC inhibitor. However, since the initiation of this screen, SAHA and the depsipeptide FR901228 have completed pediatric phase I testing, opening up possibilities for future clinical trials. We chose to work with the neuroblastoma cell line BE (2)-C. Presumably, with N-myc amplification, these cells most closely resemble high-stage neuroblastoma where N-myc is frequently amplified and a poor prognostic marker. To facilitate clinical translation, we focused our screening efforts on a collection of small molecules highly enriched for FDA-approved drugs. We screened in triplicate the National Institute of Neurological Disorders and Stroke (NINDS) small-molecule collection containing 1,040 compounds, three-quarters of which are FDA-approved.
To perform an expression-based screen for neuroblastoma differentiation, we needed to identify an expression signature of the mature neuroblast. Although there is no perfect approach to inducing neuroblastoma differentiation, we chose to use two compounds, all-trans-retinoic acid (ATRA) and phorbol 12-myristate 13-acetate (PMA), and two neuroblastoma cell lines, BE (2)-C and SHSY5Y, to ensure that we did not overfit the signature to any one compound or cell line. By using Affymetrix U133A DNA microarrays, we profiled BE (2)-C and SHSY5Y differentiated for 5 days versus a vehicle-treated control in triplicate for each condition. Differentiation was confirmed by morphology and/or expression of the mature neuronal marker neurofilament medium (NF-M) (data not shown). After standard preprocessing of the data, we identified genes meeting the following criteria: (i) at least a 2-fold variation between vehicle versus differentiation agent in one of the cell lines with P < 0.1 by t test and (ii) the appropriate direction of change in the other cell line. Nine top genes meeting these criteria were selected for the neuroblastoma differentiation signature and one additional gene, IGFBP7, with decreased expression in BE (2)-C treatment but poor overall expression in SHSY5Y was included (Fig. 1A). Importantly, these 10 genes were chosen as a surrogate for the end state of differentiation, not as bona fide targets of the differentiation process. In addition, four genes with stable expression across the conditions were included in the signature as control genes, glyceraldehyde-3-phosphate dehydrogenase (GAPD), tubulin, gamma 1 (TUBG1), general transcription factor IIIA (GTF3A), and heterogenous ribonuclear protein A/B (HNRPAB). These genes were used to correct for well-to-well variability in the small-molecule library screen [see supporting information (SI) Table S1 and Table S2 for raw data file mapping and marker gene probe sequence].
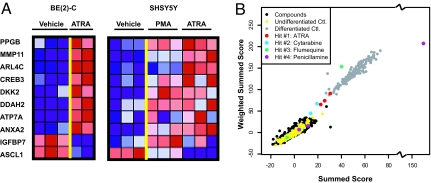
Fig. 1.
GE-HTS identifies the combination of VPA and ATRA for the induction of neuroblastoma differentiation. (A) Ten neuroblastoma differentiation marker genes were chosen to distinguish undifferentiated neuroblastoma from differentiated cells. Blue represents poorly expressed genes and red depicts highly expressed genes. (B) Distribution of the screening data is shown for the summed-score and weighted summed-score metrics. Undifferentiated BE (2)-C controls (DMSO treated) included in the screen are shown in yellow and differentiated controls (5 μM ATRA treated) in gray. The black dots represent the compounds that did not score as hits with either of these two measurements. Hits scoring as differentiated based on the overall scores of all replicates are ATRA (red), cytarabine (blue), flumequine (green), and penicillamine (purple). Each unfiltered replicate is depicted in the figure.
We first tested VPA at the reported HDAC inhibitory concentration of 1 mM in BE (2)-C cells and confirmed an increase in acetylation of histone H3 and H4 in BE (2)-C cells. Next, we confirmed that significant induction of differentiation, as measured by our neuroblastoma 14-gene expression signature, is not induced by VPA alone. BE (2)-C cells were treated with compounds at a final approximate concentration of 20 μM for 3 days. Signature genes were then quantified with a low-cost, high-throughput assay enabling the detection of up to 100 genes. As previously reported, this assay used ligation-mediated amplification (LMA) to amplify marker genes and then a barcode, fluorescent-bead-based system to detect and quantify the amplicons (7). The assay is highly quantitative; it closely recapitulates differential gene expression identified by microarray (7), and the neuroblastoma differentiation signature shows a tight dose–response for control compounds (Fig. S1). Several scoring methods were used to identify hits: a summed-score metric combining expression data for each of the marker genes assuming each gene contributes equal weight, a weighted summed score that weights each gene based on the signal-to-noise ratio determined from the differentiated positive and undifferentiated negative controls, a naïve Bayes classifier, and a KNN classifier. We looked at compounds scoring across multiple scoring metrics in determining hits. Only four compounds scored as differentiated (ATRA, flumequine, cytarabine, and penicillamine) with a probability >0.9 based on the summed score or weighted summed score (Fig. 1B and Table S3). When evaluated more closely, for two of these compounds (flumequine and penicillamine), only one of the three replicates was driving the classification as differentiated, suggesting that these two compounds were false positives. Only one compound, ATRA, scored in all four statistical assessments. We tested each of these compounds with the original assay at doses up to 20 μM in dose–response studies. Only ATRA was confirmed to induce the signature (Fig. S2). ATRA is a reported inducer of differentiation in neuroblastoma, as well as other solid tumors and hematological malignancies in vitro (8–11). As such, perhaps it was not surprising that ATRA scored so highly in the screen. However, as the top hit identified, it raised the important question of whether the HDAC inhibitor enhances the known prodifferentiation effects of ATRA.
The Combination of VPA and ATRA Enhances Neuroblastoma Differentiation.
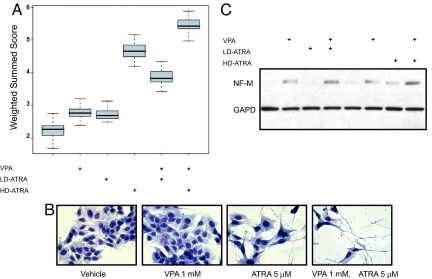
To address whether ATRA potentiates the differentiating effects of VPA, we measured the original differentiation signature with vehicle, single agent VPA or ATRA, or both compounds in combination at 3 days in BE (2)-C cells. With low-dose ATRA (LD-ATRA) (10 nM) and high-dose ATRA (HD-ATRA) (5 μM) there was a greater induction of the original differentiation gene expression signature in combination with VPA than with ATRA alone (Fig. 2A). Next, we determined the extent of differentiation with more standard assays of neurite maturation. ATRA in combination with VPA induced more dramatic morphological evidence of differentiation with neurite extension and extensive branching than either drug alone (Fig. 2B). Furthermore, both compounds when used in combination induced greater expression of neurofilament medium (NF-M), a protein expressed by mature neuronal cells. VPA and HD-ATRA single-agent treatment induced minimal induction of NF-M (Fig. 2C).
Fig. 2.
The combination of VPA and ATRA induces increased differentiation. (A) BE (2)-C cells were treated with combinations of 1 mM VPA, 10 nM LD-ATRA, and 5 μM HD-ATRA and the neuroblastoma differentiation signature evaluated. A box-and-whisker plot demonstrates the distribution of weighted summed scores for each sample type where the heavy lines inside the box show the median, the boxes show the quartiles, and the whiskers show the extremes of the observed distribution of scores. (B) May Grunwald Giemsa staining of BE (2)-C cells treated for 5 days reveals maximal differentiation with both agents in combination. Images were acquired with an Olympus CK40 microscope, 400× magnification, and Qcapture software. (C) Western immunoblot of BE (2)-C cells treated with either vehicle, ATRA (10 nM or 5 μM), VPA (1 mM), or both agents in combination for 5 days and analyzed with antibody to neurofilament medium (NF-M) and GAPD as a control.
The Combination of VPA and ATRA Synergize to Inhibit Neuroblastoma Cell Viability and Induce Apoptosis.
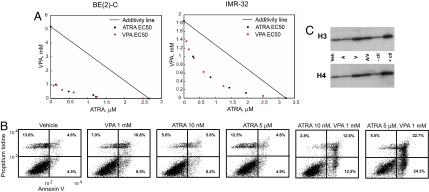
With the induction of terminal differentiation programs, many cells will ultimately undergo apoptosis. In light of their cooperative induction of differentiation, we hypothesized that the combination of an HDAC inhibitor and retinoid would enhance inhibition of neuroblastoma cell viability and the induction of apoptosis. To this end, we determined the effects of ATRA and VPA on cellular viability, measured with an ATP-based assay. In BE (2)-C and in another N-myc amplified cell line, IMR-32, the combination treatment had synergistic effects on viability as determined by isobologram (Fig. 3A). We next addressed whether this effect on cellular viability was, at least in part, related to apoptosis. LD-ATRA and VPA had an additive effect on apoptosis induction, whereas HD-ATRA and VPA had a synergistic effect based on the excess over Bliss independence (Fig. 3B; defined in SI Materials and Methods) (12, 13). The combination of HD-ATRA and VPA exceeded Bliss independence with an excess of 20% induction of apoptosis. These enhanced effects were not related to further increases in hyperacetylation with the addition of retinoids. ATRA alone did not increase histone H3 or H4 acetylation and did not enhance the hyperacetylation induced by the HDAC inhibitor (Fig. 3C).
Fig. 3.
VPA and ATRA show synergistic effects on cell viability and cell death. (A) The combined effect of VPA and ATRA on BE (2)-C (Left) and IMR-32 (Right) cell viability at 5 days, as determined by ATP level, is shown by isobologram. Synergy appears as points below the line of additivity. (B) BE (2)-C cells were treated with compounds for 3 days. Combination treatment induced increased annexin V positive cells consistent with apoptosis with an additive interaction at the low-dose ATRA and synergistic interaction with high-dose ATRA as evaluated by excess over Bliss independence. (C) Western blot analysis of histone acetylation at 6 h in BE (2)-C cells treated with VPA 1 mm (V), ATRA 10 nM (A), and both agents in combination. HeLa cell controls ± butyrate are included.
Differentiation Precedes Apoptosis in HDAC Inhibitor and Retinoid Combination Therapy.
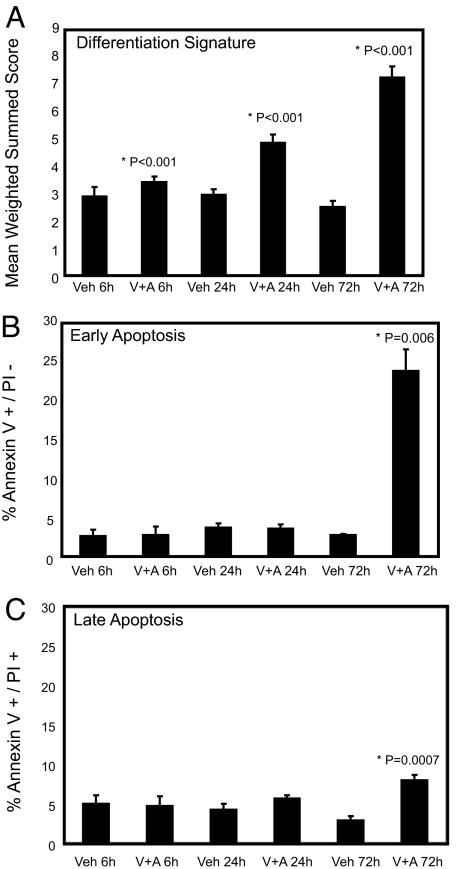
To address whether cells terminally differentiate and then die, we performed parallel evaluation of apoptosis and differentiation in a time course experiment. BE (2)-C cells were treated with vehicle or with the combination of 1 mM VPA and 5 μM ATRA in triplicate for 6, 24, and 72 h. At each time point, cells were evaluated by flow cytometry for Annexin V/FITC and PI staining patterns indicative of early and late apoptosis and for induction of the differentiation signature. As early as 6 h, evidence of differentiation was seen that increased over the 3 days (Fig. 4A). However, apoptosis was not identified until 72 h (Fig. 4 B and C). Although we cannot exclude the possibility that these two are unrelated, the temporal nature of this finding suggests that differentiation occurs and then death by apoptosis follows.
Fig. 4.
Differentiation precedes apoptosis in combination-treated neuroblastoma cells. (A) BE (2)-C cells were treated with either vehicle (veh) or the combination of 5 μM ATRA (A) and 1 mM VPA (V) for 6, 24, or 72 h and the effects on the 14-gene differentiation signature were evaluated. *, statistical significance in a pairwise t test comparing vehicle with drug treatment at each time point. Duplicate biological replicates and 16 technical replicates for each time point and condition were evaluated. BE (2)-C cells were treated in triplicate as above and the effects on early apoptosis (annexin V-FITC positive and PI negative) (B) and late apoptosis (annexin V-FITC positive and PI positive) (C) were evaluated. *, statistical significance in a pairwise t test comparing vehicle with drug treatment at each time point.
The Combination of Other HDAC Inhibitors and Retinoids Enhances Both Differentiation and Cell Death.
VPA is not a potent HDAC inhibitor, with doses of 0.5 to 1 mM necessary to achieve HDAC inhibitory activity. At this high dose, it is possible that the effects on differentiation, in combination with ATRA, are related to a non-HDAC-related mechanism of activity. For example, the antiepileptic effects of VPA are thought to be related to increases in the level of γ-aminobutyric acid (GABA), an inhibitory neurotransmitter (14). To address this issue, we tested two more potent HDAC inhibitors in combination with ATRA: LAQ824 and SAHA. Both of these HDAC inhibitors had synergistic effects on ATRA-related decrease in neuroblastoma cell viability (Fig. S3 A and B) and synergistic effects on ATRA-induced apoptosis (Fig. S3C). Combination treatment also enhanced maturation effects of low-dose and high-dose ATRA in neuroblastoma, measured by the quantitative gene expression signature used in the original screening assay (Fig. S3 D and E). Similarly, 13-cis retinoic acid (13-cis-RA) had synergistic effects with HDAC inhibitors on cellular viability and enhanced differentiation effects (Fig. S4 A and B).
The Combination of SAHA and ATRA Has Enhanced Activity in a Xenograft Model of Neuroblastoma.
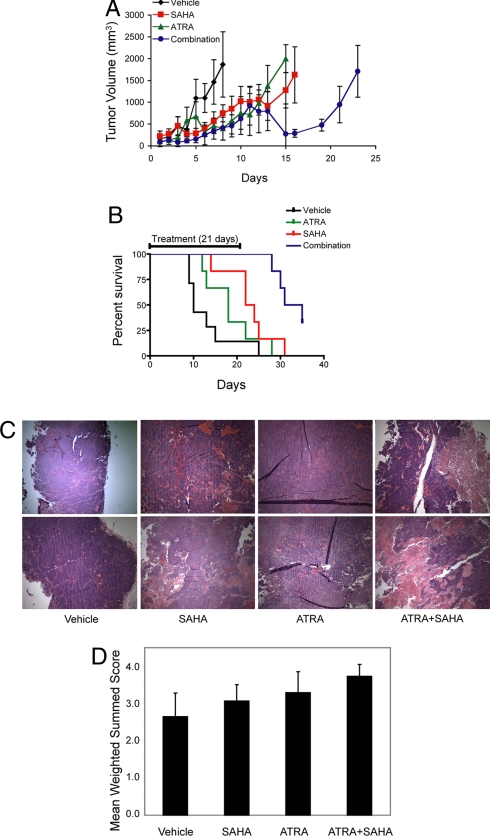
Combination testing was next extended to an in vivo model of neuroblastoma. Neuroblastoma BE-2(C) xenografts were established s.c. in NCr nude mice. For the in vivo work, we chose to use SAHA. Animals were treated with either ATRA at 2.5 mg/kg i.p. (IP) daily, SAHA 25 mg/kg IP daily, both drugs, or vehicle for up to 21 days. Tumor was measured with calipers and volume calculated per standard as V = 0.5 × length × width × width. A statistical difference in survival was demonstrated with SAHA versus vehicle but not with ATRA versus vehicle. The combination of SAHA and ATRA versus any of the other three arms (vehicle, ATRA alone, SAHA alone) showed a significant difference in survival. The longest surviving animals were treated with both drugs (Fig. 5 A and B). We next attempted to address whether the combination treatment was inducing differentiation, cell death, or both in vivo. BE (2)-C xenografts were established in NCr nude mice until tumor volume reached 100 mm3, divided into treatment cohorts, and received either vehicle, ATRA 2.5 mg/kg IP daily, SAHA 25 mg/kg IP daily, or a combination of ATRA and SAHA for 4 days. Tumors were then harvested, formalin fixed, and stained with hematoxylin and eosin (two from each treatment arm). Those treated with combination therapy demonstrated such marked increased cell death by pathological evaluation that it was difficult to determine the exact cause of death in vivo (Fig. 5C). With that said, we attempted to measure gene expression changes in the neuroblastoma maturation signature by using extant RNA. We found the greatest induction of the differentiation signature in the combination-treated tumors, suggesting that differentiation may in part be responsible for the enhanced in vivo effects of combination treatment on survival (Fig. 5D).
Fig. 5.
ATRA and SAHA have enhanced activity in an in vivo model of neuroblastoma. (A) BE (2)-C xenografts were established for 10 days in NCr nude mice. Animals received vehicle, ATRA 2.5 mg/kg IP daily, SAHA 25 mg/kg IP daily, or a combination of ATRA and SAHA for up to 21 days. Error bars show standard error of the mean across six or seven replicates. The x axis represents the days since the initiation of treatment. (B) Percentage of surviving animals is shown. The x axis represents the days since the initiation of treatment. The two-tailed P values of the survival curves were determined by logrank test for pairwise comparisons: vehicle vs. ATRA = NS, vehicle vs. SAHA = 0.05, vehicle vs. combo = 0.003, ATRA vs. SAHA = NS, ATRA vs. combination = 0.001 and SAHA vs. combination = 0.009. (C) BE (2)-C xenografts in NCr nude mice were treated with either vehicle, ATRA 2.5 mg/kg IP daily, SAHA 25 mg/kg IP daily, or a combination of ATRA and SAHA for 4 days. Tumors treated with the combination of drugs demonstrated marked increased areas of cell death by hematoxylin and eosin staining. Images were acquired with an Olympus BX41 microscope, 40× magnification, and Qcapture software. (D) As in C, BE (2)-C xenografts were established and treated. Three to five tumors from each class were harvested, and the neuroblastoma differentiation signature measured for each sample with 16 technical replicates. All drug treatments were statistically elevated compared with vehicle, and combination treatment was statistically elevated compared to single agent treatment (P < 0.001 by t test).
Discussion
Numerous obstacles exist in the development of therapies for pediatric cancer. The rarity of these diseases is a considerable problem. Neuroblastoma is the most common extracranial pediatric solid tumor, yet only 650 children and adolescents will be diagnosed each year in the United States (15). This is in sharp contrast to the most common adult solid tumor, lung cancer, with more than 200,000 new cases diagnosed annually (16). With so few people affected by this disease, there is a reduced market incentive for industry-based drug development. A second challenge, even with drug in hand, is the development of clinical trials adequately powered to address the question of efficacy. When only a limited number of drugs can be tested, rational selection for testing becomes critical. Testing combinations of compounds adds yet another layer of complexity.
Despite both in vitro and clinical trial data suggesting that differentiation is an alternative therapeutic approach for neuroblastoma, limited effort has been placed on the discovery of new differentiation agents. With existing assays it has been difficult to perform quantitative assessments of neuroblastoma differentiation. In fact, we are not aware of any published neuroblastoma differentiation screens, underscoring the limitations of traditional phenotypic and target-based screening. No single marker is yet sufficient to identify neuroblast differentiation in a high-throughput screen. For example, although NF-M is generally considered a marker of neuroblastoma differentiation, in our microarrray profiling, NF-M decreased in SHSY5Y neuroblastoma cells differentiated with PMA rather than increased as would be predicted. Furthermore, with limitations to resources, industry is unlikely to focus on the development of new assays for rare diseases. GE-HTS offered a potential solution to these challenges. Because it is a generic approach, it can be applied to many specific questions without the need to invent an assay each time a small-molecule library screen is performed. Second, the use of a complex signature provides specificity and sensitivity advantages over conventional single-gene reporter assays (17).
The concentrations of drugs used in combination can have significant effects on gene expression as shown in the work by Lamb et al. (18) and Cheok et al. (19) potentially complicating the incorporation of gene expression assays into small-molecule screening. However, the work by Lamb et al. also suggests that with a complex signature, one can see gene expression connections among compounds even without elaborate optimization of compound concentration. This underscores the importance of a sensitive, information-rich primary screening assay (i.e., multigene signature) as opposed to a single gene signature because one cannot evaluate all possible doses in a primary screen. Careful dose–response studies will then be characterized in secondary assays after hits are identified by the primary screen. Here, GE-HTS enabled the quantitative measurement of a complex neuroblastoma differentiation signature.
One of the more striking recent findings in clinical oncology has been the narrow activity of HDAC inhibitors (CTCL) despite their predicted broad effects in cancer cells. An ongoing challenge is to identify combinations of compounds that will broaden the efficacy of this drug class. Because neuroblastoma is associated with defects in neural crest differentiation, aberrant transcription is likely to play a causal role in the pathogenesis of the tumor, implicating this malignancy as a rational testing ground for new HDAC inhibitor combination therapy. Our screen identified the combination of an HDAC inhibitor and a retinoid as prodifferentiating. Earlier studies demonstrated the activity of HDAC inhibitors on neuroblastoma growth in vitro and in vivo xenograft models (20–25) but fewer have evaluated the effects on differentiation (26). More recently, a limited number of reports have evaluated the combination of HDAC inhibitors and retinoids on cell growth in neuroblastoma in vitro (25, 27) and one reported efficacy in combination in a xenograft model of neuroblastoma (28). Our work confirms that the combination indeed has greater effects on cell viability, and ultimately apoptosis, than does either compound alone. Moreover, the presented data reveal an indication for this combination in promoting neuroblastoma differentiation supported by quantitative expression-based assays and traditional assays of neuroblastoma maturation. Multiple HDAC inhibitors (the short-chain fatty acid VPA and the hydroxamic acids SAHA and LAQ824) with either ATRA or 13-cis-RA had differentiating effects when used in combination, suggesting that activity is truly related to these compound classes rather than to an off-target effect. Moreover, the combination of SAHA and ATRA had enhanced activity in a xenograft model of neuroblastoma with prolonged survival of combination-treated animals.
Based on current understanding of retinoic acid receptor regulation, an interaction between HDAC inhibitors and retinoids might be predicted. In the absence of ligand, retinoic acid receptors recruit coregulator complexes with HDAC activity leading to the repression of gene transcription (29). Several preclinical studies have demonstrated an enhanced effect of retinoids with HDAC inhibitors in acute myeloid leukemia. In the acute leukemias, several transcription factor rearrangements result in the abnormal recruitment of HDACs, hypoacetylation, and the repression of transcription of prodifferentiation genes (30, 31). HDAC inhibitors relieve this repression and presumably facilitate the activity of the retinoid (32). For example, in retinoic acid-resistant acute promyelocytic leukemia (APL) with the PLZF-RARα rearrangement, the addition of an HDAC inhibitor has restored ATRA responsiveness in in vitro and in vivo models (33). Furthermore, patient responsiveness to this combination has also been reported (34).
A national Children's Oncology Group phase I study of SAHA for children with recurrent or refractory solid tumors and leukemias followed by a phase I study of SAHA in combination with 13-cis-RA for patients with selected recurrent/refractory solid tumors was recently completed. Our data suggest that a phase II trial testing SAHA and retinoid for patients with high-risk neuroblastoma is warranted. Moreover, this work further develops a generalizable approach to small-molecule library screening, one that can bring compound discovery to many orphan diseases, including the pediatric cancers.
Materials and Methods
Cell Culture.
Neuroblastoma cell lines were purchased from the American Type Culture Collection and grown in Dulbecco's Modified Eagle Medium (DMEM) (Cellgro) supplemented with 1% penicillin-streptomycin-glutamine (Cellgro) and 10% FCS (Sigma-Aldrich).
Marker Gene Selection.
RNA was extracted with TRIzol per the manufacturer's protocol (Invitrogen) from BE (2)-C and SHSY5Y neuroblastoma cells treated in triplicate with vehicle (ethanol) versus 5 μM all-trans-retinoic acid (ATRA) for 5 days and in SHSY5Y treated with16 nM phorbol 12-myristate 13-acetate (PMA) for 5 days. Gene expression was evaluated with Affymetrix U133A DNA microarrays (see SI Materials and Methods for full details). Raw microarray data are available at http://www.broad.mit.edu/cancer/pub/Neuroblastoma_GE-HTS.
Small-Molecule Library Screen Methods.
The GE-HTS assay was carried out as described in ref. 7 and as detailed in the SI Materials and Methods. BE (2)-C cells were plated in 384-well format with 2,000 cells per well and treated with 1 mM VPA. Compounds were added at a final approximate concentration of 20 μM in DMSO and incubated for 3 days. We screened in triplicate the NINDS small-molecule collection (http://www.broad.harvard.edu/chembio/platform/screening/compound_libraries/ninds.htm).
Viability Assay.
Viability experiments were performed in 96-well format in duplicate, and then the experiment repeated two to three times, by using the Promega Cell-Titer Glo ATP-based assay per the manufacturer's instructions. Synergy was assessed by analyzing the IC50 of one drug over a range of concentrations of the other drug and vice versa. The resulting concentration pairs were visualized by isobologram (35).
Morphological Evaluation.
BE (2)-C cells were evaluated by May–Grunwald Giemsa staining with an Olympus CK40 microscope and Q-capture software.
Western Blot Analysis and Histone Extraction.
All proteins were detected by chemiluminescence and antibodies to NF-M (SC-20013, Santa-Cruz) GAPD (Ab 22556, Abcam), anti-acetyl-histone H3 (06-599, Upstate), and anti-acetyl-histone H4 (06-866, Upstate). Control histones were untreated HeLa cell acid extract (13-112, Upstate) and sodium butyrate-treated HeLa cell acid extract (13-113, Upstate). See SI Materials and Methods for histone extraction and Western blot analysis protocols.
Apoptosis Assay.
Annexin V FITC/PI staining was performed with 500,000 cells by using the Annexin V: FITC Apoptosis Detection Kit I (BD PharMingen). Cells were analyzed by flow cytometry with a FACScan flow cytometer (Becton Dickinson) and CELLQuest analytical software.
Chemicals.
Chemicals were obtained from the following sources: VPA, ATRA, and PMA (Sigma), LAQ-824 (Novartis), and SAHA (Broad Chemistry Program).
Xenograft Studies.
Xenograft studies were performed with male NCr nude mice. The four treatment groups were vehicle, ATRA alone (2.5 mg/kg), SAHA alone (25 mg/kg), or a combination of ATRA and SAHA, and animals were treated for either 21 or 4 days. Caliper measurements were used to calculate tumor volume by using the formula: Volume = 0.5 × length × (width)2. All animal experiments were performed after approval from the Institutional Animal Care and Use Committee. See SI Materials and Methods for full details.
Supplementary Material
Acknowledgments.
We thank Paul Clemons, Nicola Tolliday, and Stuart Schreiber for their guidance, David Peck for GE-HTS platform support, Zi Peng Fan for technical support, and Jonathan Jesneck for computational discussions. This work was funded by the Friends for Life Fellowship Program, a Children's Oncology Group Young Investigator Award, and the Claudia Adams Barr Program in Cancer Research (K.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw microarray data have been deposited at http://www.broad.mit.edu/cancer/pub/Neuroblastoma_GE-HTS.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710413105/DCSupplemental.
References
- 1.George RE, et al. High-risk neuroblastoma treated with tandem autologous peripheral-blood stem cell-supported transplantation: Long-term survival update. J Clin Oncol. 2006;24:2891–2896. doi: 10.1200/JCO.2006.05.6986. [DOI] [PubMed] [Google Scholar]
- 2.Furchert SE. Inhibitors of histone deacetylases as potential therapeutic tools for high-risk embryonal tumors of the nervous system of childhood. Int J Cancer. 2007;120:1787–1794. doi: 10.1002/ijc.22401. [DOI] [PubMed] [Google Scholar]
- 3.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nature Rev. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 4.Duvic M, et al. Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL) Blood. 2007;109:31–39. doi: 10.1182/blood-2006-06-025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthay KK, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children's Cancer Group. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 6.Stegmaier K, et al. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 7.Peck D, et al. A method for high-throughput gene expression signature analysis. Genome Biol. 2006;7:R61. doi: 10.1186/gb-2006-7-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pahlman S, Odelstad L, Larsson E, Grotte G, Nilsson K. Neuron specific enolase: a marker for differential diagnosis of neuroblastoma and Wilms' tumor. Int J Cancer. 1981;28:583–589. doi: 10.1016/s0022-3468(82)80494-6. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds CP, et al. Response of neuroblastoma to retinoic acid in vitro and in vivo. Prog Clin Biol Res. 1991;366:203–211. [PubMed] [Google Scholar]
- 10.Sidell N. Retinoic acid-induced growth inhibition and morphologic differentiation of human neuroblastoma cells in vitro. J Natl Cancer Inst. 1982;68:589–596. [PubMed] [Google Scholar]
- 11.Guzhova I, et al. Interferon-gamma cooperates with retinoic acid and phorbol ester to induce differentiation and growth inhibition of human neuroblastoma cells. Int J Cancer. 2001;94:97–108. doi: 10.1002/ijc.1443. [DOI] [PubMed] [Google Scholar]
- 12.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 13.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 14.Johannessen CU. Mechanisms of action of valproate: a commentatory. Neurochem Int. 2000;37:103–110. doi: 10.1016/s0197-0186(00)00013-9. [DOI] [PubMed] [Google Scholar]
- 15.Goodman MT, Gurney JG, Smith MA, Olshan AF. In: Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975-1995. Ries LAG, et al., editors. Bethesda, MD: National Cancer Institute; 1999. pp. 65–72. [Google Scholar]
- 16.Ries LAG, et al., editors. SEER Cancer Statistics Review, 1975-2004. Bethesda, MD: National Cancer Institute; 2006. [Google Scholar]
- 17.Hieronymus H, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Lamb J, et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 19.Cheok MH, et al. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet. 2003;34:85–90. doi: 10.1038/ng1151. [DOI] [PubMed] [Google Scholar]
- 20.Cinatl J, Jr, et al. Sodium valproate inhibits in vivo growth of human neuroblastoma cells. Anticancer Drugs. 1997;8:958–963. doi: 10.1097/00001813-199711000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Jaboin J, et al. MS-27–275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002;62:6108–6115. [PubMed] [Google Scholar]
- 22.Glick RD, et al. Hybrid polar histone deacetylase inhibitor induces apoptosis and CD95/CD95 ligand expression in human neuroblastoma. Cancer Res. 1999;59:4392–4399. [PubMed] [Google Scholar]
- 23.de Ruijter AJ, et al. The novel histone deacetylase inhibitor BL1521 inhibits proliferation and induces apoptosis in neuroblastoma cells. Biochem Pharmacol. 2004;68:1279–1288. doi: 10.1016/j.bcp.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Ouwehand K, de Ruijter AJ, van Bree C, Caron HN, van Kuilenburg AB. Histone deacetylase inhibitor BL1521 induces a G1-phase arrest in neuroblastoma cells through altered expression of cell cycle proteins. FEBS Lett. 2005;579:1523–1528. doi: 10.1016/j.febslet.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 25.De los Santos M, Zambrano A, Aranda A. Combined effects of retinoic acid and histone deacetylase inhibitors on human neuroblastoma SH-SY5Y cells. Mol Cancer Ther. 2007;6:1425–1432. doi: 10.1158/1535-7163.MCT-06-0623. [DOI] [PubMed] [Google Scholar]
- 26.Cinatl J, Jr, et al. Induction of differentiation and suppression of malignant phenotype of human neuroblastoma BE(2)-C cells by valproic acid: enhancement by combination with interferon-alpha. Int J Oncol. 2002;20:97–106. [PubMed] [Google Scholar]
- 27.Coffey DC, et al. Histone deacetylase inhibitors and retinoic acids inhibit growth of human neuroblastoma in vitro. Med Pediat Oncol. 2000;35:577–581. doi: 10.1002/1096-911x(20001201)35:6<577::aid-mpo18>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Coffey DC, et al. The histone deacetylase inhibitor, CBHA, inhibits growth of human neuroblastoma xenografts in vivo, alone and synergistically with all-trans retinoic acid. Cancer Res. 2001;61:3591–3594. [PubMed] [Google Scholar]
- 29.Nagy L, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 30.Lutterbach B, et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 32.Ferrara FF, et al. Histone deacetylase-targeted treatment restores retinoic acid signaling and differentiation in acute myeloid leukemia. Cancer Res. 2001;61:2–7. [PubMed] [Google Scholar]
- 33.He LZ, et al. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J Clin Invest. 2001;108:1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warrell RP, Jr, He LZ, Richon V, Calleja E, Pandolfi PP. Therapeutic targeting of transcription in acute promyelocytic leukemia by use of an inhibitor of histone deacetylase. J Natl Cancer Inst. 1998;90:1621–1625. doi: 10.1093/jnci/90.21.1621. [DOI] [PubMed] [Google Scholar]
- 35.Gessner PK. Isobolographic analysis of interactions: an update on applications and utility. Toxicology. 1995;105:161–179. doi: 10.1016/0300-483x(95)03210-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.