Abstract
The plasmid origin of replication, oriP, of Epstein–Barr Virus (EBV) was identified in an assay to detect autonomously replicating sequences (ARSs) in human cells. Raji ori, a second origin in EBV, functions in vivo but fails in long-term ARS assays. We examined the initiating element, DS, within oriP and Raji ori to resolve this paradox. DS, but not Raji ori, binds EBNA1; whereas both act as ARSs in short-term assays, with DS being more efficient, only DS can act as an ARS in long-term assays. Surprisingly, we found that DS supported the establishment of a plasmid with Raji ori in cis and that after deletion of DS, Raji ori could now act as an ARS in the long term. This finding explains the frequent failure of ARS assays in mammalian cells. More origins can initially act as ARSs than can be established. We identified one requirement for ARSs to be established: They must function efficiently enough initially to generate a wide distribution of numbers of plasmids per cell. Only the cells that have more than a threshold number of plasmids can survive selections imposed on the cells to retain these replicons.
Keywords: autonomously replicating sequence, DNA replication, Epstein–Barr virus
The study of DNA replication in the budding yeast Saccharomyces cerevisiae has provided much of the foundation of our understanding of DNA replication in eukaryotic cells. In S. cerevisiae, assays for autonomously replicating sequences (ARS) were used to identify the origins of DNA synthesis after their introduction into plasmids having a functional centromere (CEN) (1–5). Unlike ARSs in yeast, origins of DNA synthesis in mammalian cells are poorly understood. Parallel experiments in mammalian cells have not been the standard initially used to identify chromosomal origins of DNA synthesis. One difficulty in developing a mammalian ARS assay, for example, is imposed by the size of mammalian centromeres making their use difficult.
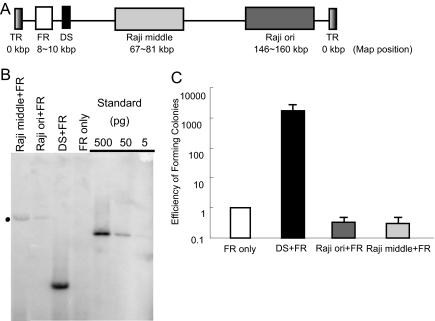
One mammalian origin of DNA synthesis that was identified in an ARS assay in human cells is the Dyad symmetry (DS) of Epstein–Barr virus (EBV) (6, 7). DS is a discrete, licensed origin that functions in a variety of mammalian hosts in the presence of the viral trans-acting element EBNA1 (Epstein–Barr nuclear antigen 1) (8). It requires the family of repeats (FR) from EBV in cis and EBNA1 in trans to provide its segregation mechanism to the replicon (9–11). FR and EBNA1 thus functionally replace the requirement in yeast for a CEN element in ARS assays. A second origin of DNA synthesis has been identified in the EBV genome, which also functions extrachromosomally and is not discrete but does support licensed DNA synthesis (Fig. 1A) (12–15). These latter properties make this origin, which we term “Raji ori,” similar to well studied mammalian chromosomal origins such as the DHFR (16) and β-globin (17, 18). Raji ori was identified by two-dimensional gel electrophoresis and in an assay termed “single molecule analysis of replicated DNA” (SMARD) and was found both to be the predominant origin of DNA synthesis used in the EBV endogenous to the Raji cell line and to function in the EBV DNAs in multiple, additional cell lines (12–14).
Fig. 1.
Raji ori and Raji middle can support extrachromosomal replication transiently but cannot be established as extrachromosomal replicons. (A) Shown are relative positions of Raji ori and Raji middle on a linear representation of the EBV genome (165 kbp). (B) Southern blot analyses detect plasmids replicated transiently. Equal masses of plasmids containing Raji ori and Raji middle fragments or DS with a FR backbone were transfected into Raji cells. Extrachromosomal DNAs were harvested 4 days after transfection, digested with a single-cut enzyme to linearize the DNA and DpnI to remove unreplicated DNA, and detected by Southern blot analysis. There were 100 replicated DS plasmids per transfected cell on average, whereas there were ≈10 Raji ori and Raji middle plasmids (denoted with a dot) per transfected cell 4 days after transfection (average of three measurements). (C) Raji ori and Raji middle plasmids fail in a long-term assay. Raji ori or Raji middle plasmids were transfected into Raji cells (n = 5). The transfected cells were selected with puromycin for 3 weeks. The activity of the plasmid origins to support long-term replication was determined by the ability of the transfected cells to give rise to drug-resistant colonies. The efficiency of cells transfected with FR plasmids giving rise to drug-resistant colonies that result from integration was set to be 1. Cells transfected with either Raji ori or Raji middle plasmids did not give rise to colonies more efficiently than cells transfected with the FR plasmids.
We have analyzed Raji ori in ARS assays comparing it with DS to understand the requirement for a dispersed, licensed origin to score in those assays. In all of our studies, the FR element was provided in cis to support the segregation to daughter cells of all origin-containing replicons analyzed. The studies showed that Raji ori initially functions as an ARS in two cell types expressing EBNA1 but failed to support extrachromosomal replication for extended times under selection. This observation appeared inconsistent with Raji ori's serving as the predominant origin of DNA synthesis within the EBV genome in Raji cells. We resolved this paradox by using DS in cis to establish plasmids containing Raji ori as stable, extrachromosomal replicons and tested whether, after deleting DS, Raji ori could act stably as an ARS. It could, indicating that Raji ori fails to be established but, once established, can act as an ARS to support an extrachromosomal replicon stably.
The process of establishment has been characterized as the early, rapid loss from proliferating cells of newly introduced, synthesized DNAs, followed after 10–20 generations by their relatively stable maintenance (19). Maintenance is determined by the plasmids replicating stably and providing their host cells resistance to a selective agent. This process occurs successfully in ≈1–10% of transfected cell recipients and is mediated epigenetically (19). We have identified one function of DS that is necessary for it to be established that Raji ori lacks. The initial efficiency of DNA synthesis of DS allows the evolution over 10–20 generations of a population of daughter cells with a wide distribution in numbers of plasmids per cell. Such a wide distribution is required to sustain cells' resistance to selection (11). ARS assays thus require peculiarly efficient origins of DNA synthesis in mammalian cells.
Results
Raji Ori Acts as an ARS in Short-Term Assays.
Raji ori has been shown to support extrachromosomal DNA replication in several different strains of EBV (12–14). We tested a plasmid containing Raji ori with FR and a puromycin-resistance gene in short-term assays in Raji cells to determine whether it functions as an ARS element in mammalian cells. Raji ori supported extrachromosomal DNA synthesis 4 days after transfection, albeit inefficiently compared with DS (Fig. 1B). Both of the two known origins of DNA synthesis of latent EBV other than Raji ori, DS, and Rep*, require EBNA1's binding to properly spaced EBNA1-binding sites to initiate licensed, DNA synthesis (20–22). However, we found there are no detectable EBNA1-binding sites within Raji ori that bind sufficiently to support EBNA1-dependent initiation of DNA synthesis [supporting information (SI) Fig. S1]. DNA synthesis initiating within Raji ori is therefore likely to function in an EBNA1-independent manner. We wanted to test whether Raji ori is unique or whether any other 14-kbp DNA fragment from the EBV genome can act as an ARS as does Raji ori. A 14-kbp DNA sequence from EBV that spans BamHI M, S, and L fragments (Fig. 1A), which we term “Raji middle” has been shown to support the initiation of EBV DNA synthesis inefficiently by SMARD (13). Raji middle in a plasmid with a FR-puromycin-backbone, however, did act as an ARS similarly to Raji ori in this short-term assay (Fig. 1B).
Raji Ori and Raji Middle Do Not Act as ARSs in Long-Term Assays.
When Raji cells transfected with Raji ori and Raji middle plasmids were cultured under selection for 3 weeks, neither plasmid yielded drug-resistant colonies above the background characteristic of colonies arising with integrated DNAs (Fig. 1C). Similar, negative results were observed in 293 cells expressing EBNA1 (Fig. S2). These findings indicate that although Raji ori and Raji middle can support extrachromosomal DNA synthesis transiently, they cannot act as ARSs for extended times under selection.
DS in cis Supports the Establishment of Raji Ori and Raji Middle; Once Established, Raji Ori and Raji Middle Act as ARSs.
Raji ori in the context of the EBV genome in Raji cells and DS in multiple plasmid replicons can each support extrachromosomal replication for extended times under selection. Our results show, however, only DS allows plasmids to be established. How then could the EBV DNA endogenous to Raji cells that largely uses Raji ori to initiate its DNA synthesis have become established?
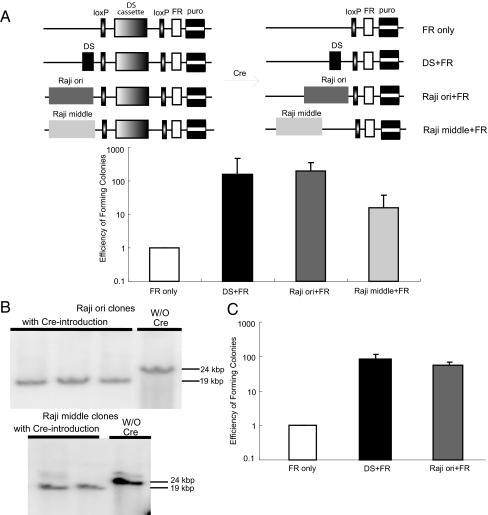
We constructed a FR puro-based vector, which has a DS cassette flanked by loxP sites to test whether, once established by DS, either Raji ori or Raji middle could act as ARSs stably (Fig. 2A). In the presence of the DS cassette, plasmids can be established in EBNA1-expressing cells. Upon introduction of Cre recombinase into the cells, the DS cassette can be deleted. If another DNA fragment added in cis could support continued extrachromosomal DNA synthesis once established, then after deletion of DS, the cells would proliferate and continue to be resistant to puromycin. After the deletion of the DS cassette, Raji cells harboring Raji ori plasmids gave rise to colonies approximately as efficiently as the positive control, a DS plasmid. On the other hand, cells harboring Raji middle plasmids gave rise to drug-resistant colonies only 8.3% as efficiently as the positive control plasmids (Fig. 2A). DNAs from the puromycin-resistant colonies were isolated and subjected to Southern blot analysis to confirm the extrachromosomal status and the successful excision of the DS cassette in these plasmids (Fig. 2B). The harvested DNAs were also transformed into Escherichia coli to confirm their extrachromosomal status (Fig. S3). Similar results were obtained when the experiment was carried out in BJAB cells expressing EBNA1 (Fig. 2C). These results indicate that Raji ori and Raji middle, although able to support transient extrachromosomal DNA synthesis, require their establishment by another ARS to maintain efficient long-term replication.
Fig. 2.
Establishment by virtue of DS in cis is required for Raji ori and Raji middle to act as ARSs for long-term extrachromosomal replication. (A) Shown are the structures of the plasmids (not to scale) confirmed in clones of Raji cells before and after Cre-mediated deletion of the DS cassette used to establish the plasmids initially. Clones of Raji cells harboring the plasmids with the DS cassette and transfected with a vector expressing Cre were tested for their ability to form colonies resistant to puromycin after 3 weeks. They were replated in gancyclovir to ensure the loss of the DS cassette that confers sensitivity to this drug. The efficiency of forming colonies of the cells harboring the plasmids with only a DS cassette but not an additional origin (FR only) was set to be one. Cells harboring Raji ori plasmids gave rise to colonies approximately as efficiently as the positive control, DS+FR plasmid, after the removal of the DS cassette. Cells harboring Raji middle plasmids gave rise to drug-resistant colonies only 8.3% as efficiently as the positive control plasmids after the removal of the DS cassette (n = 5). (B) Confirming the structure of plasmid DNAs. Cell clones harboring either Raji ori or Raji middle plasmids were picked 3 week after deletion of the DS cassette. Extrachromosomal DNAs were harvested by the alkaline lysis method and detected by Southern blot analysis. The decrease in size of the plasmids from 24 kbp to 19 kbp indicates the successful removal of the DS cassette. The slowly migrating signal in Raji middle without Cre is from incompletely digested DNA. (C) Similar experiments to those described in A were performed in BJAB cells expressing EBNA1. The cells harboring Raji ori plasmids gave rise to drug-resistant clones approximately as efficiently as the DS plasmid after the removal of the DS cassette (n = 3).
Raji Ori's and Raji Middle's Functioning as ARSs After Establishment Are Similar to Their Efficiencies Supporting Initiation of DNA Synthesis in Endogenous EBV.
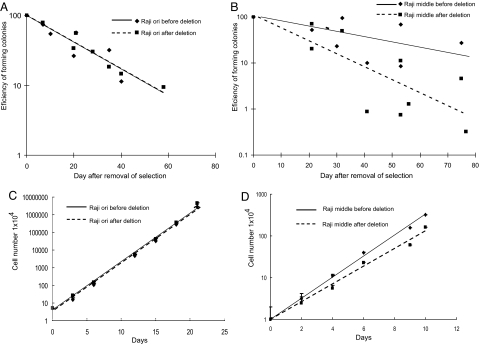
To test whether the different abilities of Raji ori and Raji middle plasmids to serve as ARSs stably after establishment reflect their different efficiencies in supporting DNA synthesis, clones of cells with Raji ori and Raji middle plasmids with the DS cassette deleted were isolated and expanded. The efficiencies of Raji ori and Raji middle to act as stable ARSs were then measured in two ways: (i) Cells were cultured in the absence of selection for increasing times and subsequently plated into selective media in 96-well plates to measure their colony-forming ability. In this experiment, only the efficiency with which the various origins support the initiation of DNA synthesis determines the rate of loss of plasmids from the cells (11). The more efficient the origin is, the more slowly the cells lose the drug-resistant plasmid and the more efficiently colonies arise after selection is reapplied. (ii) The rate at which cells with the different plasmids proliferate was also measured with and without selection. Cells harboring plasmids that have a higher efficiency of initiating DNA synthesis would have an increased ability to maintain drug-resistance and double in a shorter time in the selective media.
In the first assay, Raji ori plasmids with and without the DS cassette were lost at the same apparent rate of 4.4% per generation (Fig. 3A). Raji middle plasmids were lost from the cells at an apparent rate of 6.5 ± 3.1% per generation after the DS cassette was removed, which was substantially more rapid than the rate of 2.4 ± 2.2% per generation when the DS cassette was present (P value 0.04) (Fig. 3B). This result was confirmed in the second assay that found that cells carrying Raji ori plasmids with and without the DS cassette doubled at the same rates (20 h). In the presence of selection, they both doubled more slowly but still at the same rates (26 h) (Fig. 3C). In the absence of selection, cells harboring Raji middle plasmids either with or without the DS cassette had the same doubling time, 26.7 h. In the presence of selection, cells harboring Raji middle plasmids with DS cassette doubled at a rate of once per 28.8 ± 2.2 h, which was faster than the cells harboring Raji middle plasmids after removal of the DS cassette, which doubled every 32.8 ± 2.5 h (P value 0.04) (Fig. 3D). The efficiency of Raji ori and Raji middle to function after establishment reflects their relative efficiency of functioning as initiation sites of DNA synthesis in vivo (13); i.e., Raji middle supports DNA synthesis less efficiently than does Raji ori.
Fig. 3.
Raji ori's and Raji middle's support of DNA synthesis after establishment is similar to their support of DNA synthesis in endogenous EBV. (A) To measure colony formation, the cells harboring Raji ori plasmids with the DS cassette and Raji ori plasmids after deletion of the DS cassette were cultured in the absence of selection. At different times, the cells were plated into selective media, and their efficiency of colony formation was measured. Before the deletion of the DS cassette, Raji ori plasmids were lost at a rate of 4.4 ± 1.8% per cell generation (indicated by the solid regression line). After the deletion of the DS cassette, Raji ori plasmids were lost at the same rate of 4.4 ± 0.1% per cell generation (indicated by the dashed regression line). (B) Similar experiments to those described in A were performed with cells harboring the Raji middle plasmids before and after deletion of the DS cassette. Before deletion of the DS cassette, Raji middle plasmids were lost at a rate of 2.4 ± 2.2% per cell generation (indicated by the solid regression line). After the deletion of the DS cassette, Raji middle plasmids were lost at a higher rate of 6.5 ± 3.1% per cell generation (indicated by the dashed regression line). (C) To measure rates of proliferation, the cells harboring the Raji ori plasmids with the DS cassette and the Raji ori plasmids after deletion of the DS cassette were cultured in the presence of puromycin for selection. The cells were initially plated at 5 × 104 per milliliter. The cells harboring Raji ori plasmids in the presence of the DS cassette doubled in 26.5 ± 0.4 h and doubled in 25.4 ± 0.5 h after deletion of the DS cassette. (D) In experiments parallel to those in C, cells harboring the Raji middle plasmids with the DS cassette and Raji middle plasmids after deletion of DS cassette were cultured in the presence of puromycin for selection. The cells were initially plated at 1 × 104 per milliliter and measured over time. The cells harboring Raji middle plasmids in the presence of DS cassette doubled in 28.8 ± 2.2 h, whereas those lacking DS doubled more slowly, in 32.8 ± 2.5 h. The data were analyzed with the Wilcoxon rank-sum test; P value (two-sided) = 0.04.
Newly Introduced Plasmids Localize to the Nuclear Matrix.
Our ARS assays indicated that DS, which supports the initiation of extrachromosomal DNA synthesis efficiently before establishment, permits establishment of Raji ori in cis, whereas Raji ori alone does not. We have analyzed DS and Raji ori during the phase when DS is established to identify properties it has that allow its establishment.
The origins of DNA synthesis of adenovirus and oriP have been shown to localize to the nuclear matrix (23–25), an operationally defined compartment involved in many nuclear functions (25–31). We tested the hypothesis that localization to the nuclear matrix is required for establishment. DNAs that copurified with the nuclear matrix were separated into two portions, treated with or without DpnI and detected by Southern blot analysis. Two different probes were used in this assay; one probe detected the oriP sequence, which could hybridize both to the transfected oriP plasmids and that of the endogenous Raji oriP sequence as well. The other probe detected EBNA1's ORF. The oriP region in Raji EBV is localized to the nuclear matrix, whereas EBNA1's ORF is not (25). In each sample (days 4, 8, and 11), 80–90% of the endogenous oriP sequence copurified with the nuclear matrix, but <12% of the EBNA1 sequence was detected in the matrix fraction (Fig. S4). Approximately 90% of the newly introduced replicated (DpnI-resistant) DNA copurified with the nuclear matrix 4 days after transfection and throughout the time course of the experiment (Fig. S4). Approximately 80% of the newly introduced pPUR- and FR-only plasmids, which are not capable of supporting DNA synthesis, also localized to the nuclear matrix 4 days after introduction (Fig. S5). These results indicate that plasmids associate with the nuclear matrix upon introduction into the cells independent of their ability to support DNA synthesis; however; the association is maintained for established plasmids. Thus, oriP molecules that will or will not be established home to the nuclear matrix, and this homing property is not peculiar to replicons destined to be established.
DS, but Not Raji Ori, Can Generate a Broad Distribution of Numbers of Plasmids per Cell Shortly After Its Introduction.
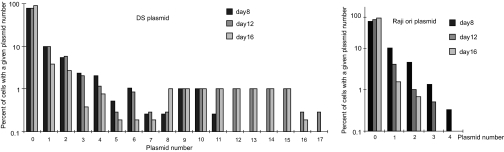
Our recent analysis of EBV plasmids in live cells found that ≈16% of these DNAs fail to be synthesized each S phase (11). EBV compensates for this failure by providing its host cell a selective advantage and by errors in its partitioning that generate cells with a wide distribution of numbers of plasmids per cell. Only those cells with numbers of plasmids higher than some threshold can lose plasmids without losing the selective advantage the plasmids afford them (11). We hypothesized that DS would generate this wide distribution, and Raji ori would not. To test this hypothesis, plasmids with Raji ori or DS, each with FR in cis were transfected into BJAB cells expressing EBNA1. Transfected cells were isolated 2 days after transfection and analyzed 8, 12, and 16 days after transfection by fluorescence in situ hybridization (FISH) to enumerate the plasmids in the transfected cells. Both Raji ori and DS plasmids were lost, as expected after transfection, and increasing numbers of cells without plasmids were detected in the populations throughout the time points. No cells harboring more than three Raji ori-plasmids were found 12 days after transfection. On the other hand, cells harboring more than five copies of DS plasmids were detected on day 8, and cells harboring >10 copies of DS plasmids were detected on days 12 and 16 (Fig. 4). These measurements demonstrate that the DS plasmids, which can be established, can generate a broad distribution of numbers of plasmids per cell in daughter cells, whereas Raji ori cannot.
Fig. 4.
Only plasmids competent to be established can generate a broad distribution of numbers of plasmids per cell. Shown are the distribution of DS and Raji ori plasmid numbers in transfected cells measured 8–16 days after their introduction. The DS and Raji ori plasmids were transfected into BJAB cells expressing EBNA1. Approximately 500 transfected cells were analyzed by fluorescence in situ hybridization to detect the introduced plasmids 8, 12, and 16 days after transfection. Both the DS and Raji ori plasmids were lost throughout the time points, so more cells harboring zero plasmids were detected over time. By day 12, no cells transfected with Raji ori had more than three plasmids, whereas DS plasmids generated a broader distribution, with 10% of the cells harboring more than three plasmids by day 12.
Discussion
The study of mammalian origins of DNA synthesis has been difficult because these DNA elements are difficult to identify operationally and difficult to dissect genetically. ARS assays might provide alternative, powerful tools to analyze the origins of DNA synthesis; however, they have not been usually successful in the study of origins in mammalian cells. We have identified one reason underlying their failure and developed a means to overcome it, thus allowing the facile genetic dissection of additional origins of DNA synthesis.
EBV genomes that are licensed, plasmid replicons in human cells have two characterized efficient origins of DNA synthesis. DS binds EBNA1 to recruit ORC and supports the initiation of DNA synthesis locally. Raji ori spans >10 kbp in length, cannot bind EBNA1 (Fig. S1 and Table S1), and supports the initiation of DNA synthesis at multiple sites (13). These characteristics make Raji ori akin to the zones of initiation of DNA synthesis in mammalian chromosomes such as DHFR and β-globin (16–18, 32).
Raji ori functions as the zone of initiation for DNA synthesis in several strains of EBV, whereas Raji middle rarely serves as the initiation site of DNA synthesis in vivo (13). We have found that both Raji ori and Raji middle act as ARSs transiently (Fig. 1). More surprisingly, plasmids with Raji ori do not survive in an ARS assay to maintain DNA extrachromosomally (Fig. 1). The failure of Raji ori to support long-term extrachromosomal DNA synthesis is striking. This same DNA element serves as the predominant zone of initiation of DNA synthesis within EBV DNA in Raji cells (13). Why does it (and Raji middle) eventually fail in an ARS assay?
One obstacle that a DNA element must surmount to succeed in a long-term mammalian ARS assay is to become established. Even highly-efficient origins of DNA synthesis such as DS usually fail individually to be established, that is to survive in the long-term to yield progeny that continue successfully as extrachromosomal replicons under selection. Between 90% and 99% of the newly introduced plasmids containing oriP support DNA synthesis initially but fail to be established (19). We have found that between 99.99% and 99.999% of newly introduced plasmids containing Raji ori fail to become established, which is the background signal of our assay (Fig. 1). However, when DS is provided in cis to plasmids with Raji ori to support their establishment, and DS is subsequently removed by Cre-mediated recombination, Raji ori does support long-term extrachromosomal replication efficiently under selection (Fig. 2).
These findings indicate that successful establishment is essential for a DNA element to score in a long-term, mammalian ARS assay. They also demonstrate that some DNA elements can support long-term extrachromosomal replication but not establishment, whereas others can do both. What, then, occurs during establishment?
Much data indicate that homing of DNA templates to the operationally defined nuclear matrix correlates with their supporting DNA synthesis (33–36). Consistent with this data, newly introduced plasmids containing oriP homed rapidly to the nuclear matrix (Fig. S4). In these same cells, an endogenous oriP element purified with the nuclear matrix, whereas the EBNA1 ORF, an element not associated with the initiation of DNA synthesis, did not. These data document the robustness of our successful fractionation of the nuclear matrix. Surprisingly, fractions that contained the nuclear matrix also contained all newly introduced DNAs whether or not they had supported DNA synthesis and whether or not they contained oriP. Thus, newly introduced DNAs may need to home to the nuclear matrix to support DNA synthesis but homing to this operationally defined compartment does not ensure an origin's continued function.
A newly appreciated property of established oriP plasmids in mammalian cells is that each analyzed to date in clonal cell populations is present in a wide distribution of numbers per cell (11). This wide distribution reflects three features of these replicons: (i) None supports DNA synthesis in 100% of all S phases; (ii) all depend on the FR element and EBNA1 to mediate their partitioning that is imperfect; and (iii) all can be maintained only as extrachromosomal replicons by selection. For these replicons, failures in DNA synthesis lead to loss of plasmids, but for those individual cells in a population with more than some threshold number of plasmids, loss of one plasmid does not lead to the cells being lost by selection (11). We determined that DNAs with oriP newly introduced into cells are synthesized sufficiently for their progeny to yield a wide distribution in numbers per cell within 16 days of their introduction. Plasmids with Raji ori lack this property crucial for establishment.
This property of oriP depends on the efficiency of its rate of initiation of DNA synthesis and not on the fidelity of its partitioning. We simulated the distribution of plasmids in cells that would arise when the rates of DNA synthesis are varied between the values for oriP measured to be 84% per plasmid per S phase (11) down to 54% or when their faithful partitioning is varied between the values for oriP measured to be 88% for its duplicated DNAs (11) down to 50%. Only reducing the rate of DNA synthesis markedly reduced the range of numbers of plasmids per cell found by day 16, and this reduced rate appears to occur primarily for oriP by day 8 (Fig. S6). One reason that plasmids with Raji ori fail to be established is that their initial rates of DNA synthesis are too low to allow them to generate a sufficiently wide distribution in their numbers per cell to survive selection.
This insight explains the failure of Raji ori to be established and raises the question: “Given that Raji ori supports the initiation of DNA synthesis in recently transfected cells too inefficiently to be established, how can it act as an ARS to maintain extrachromosomal synthesis once established?” One observation that addresses this question comes from measuring the rate of accumulation of plasmids in cells that both have established and newly introduced plasmids. Established oriP plasmids are lost from cells at rates of ≈8% per generation, whereas the newly introduced plasmids are lost at twice that rate (Fig. S7). Failures in partitioning do not affect the accumulation of plasmids in the total population of cells but only their distribution within its members (11). Thus, newly introduced oriP plasmids have lower rates of DNA synthesis relative to that of their established kin. These findings lead us to hypothesize that Raji ori, once it is established, also supports DNA synthesis at an increased rate relative to its rate upon first being introduced into cells. This hypothesis is supported by our finding that plasmids with Raji ori plus or minus DS, once established, are lost from cells in the absence of selection at similar rates (Fig. 3).
Our finding that DS allows the establishment of origins that alone cannot be established means that the latter origins can now be dissected genetically as extrachromosomal replicons. Two additional implications of these findings have broad significance. First, other viruses that are maintained extrachromosomally as licensed replicons such as Kaposi's sarcoma-associated herpes virus must encode the means to become established as does EBV. Second, DS and probably Raji ori increase the efficiency with which they function as origins upon becoming established. This increase must be epigenetic and indicates also that the efficiency of chromosomal origins is likely to be quantitatively regulated epigenetically.
Materials and Methods
Plasmids and cell lines are described in SI Text. The replication of newly introduced DNAs was assayed after their extraction 4 days after their introduction by isolating them by using alkaline lysis and measuring those that had undergone at least two rounds of DNA synthesis by extensive digestion with DpnI endonuclease and subsequent detection by Southern blot analysis. These procedures are detailed in SI Text.
The efficiency of long-term replication of plasmids was measured by their ability to support colonies of cells resistant to the markers encoded by the plasmids after 3 weeks of selection as described in Wang et al. (22) and described in SI Text. The replication of plasmids after deletion of DS cassette flanked by lox sites by expression of Cre was assayed by measuring the ability of cells to form colonies resistant to puromycin as described in the SI Text.
The preparation of nuclear matrices was modified following published methods by using lithium 3′, 5′-diiodosalycilate (26, 28) and is detailed in SI Text.
The distribution of plasmids containing DS or Raji ori newly introduced into Raji cells was measured over time with FISH and is detailed in the SI Text.
Supplementary Material
Acknowledgments.
We thank Arthur Sugden for performing the computer simulations and Norman Drinkwater and Paul Lambert for their insights and suggestions. This work was supported by National Institutes of Health Grant CA022443. B.S. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801378105/DCSupplemental.
References
- 1.Murray A, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 2.Murray AW, Szostak JW. Construction of artificial chromosomes in yeast. Nature. 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- 3.Stinchcomb DT, Struhl K, Davis RW. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 4.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 5.Stinchcomb DT, Mann C, Davis RW. Centromeric DNA from Saccharomyces cerevisiae. J Mol Biol. 1982;158:157–190. doi: 10.1016/0022-2836(82)90427-2. [DOI] [PubMed] [Google Scholar]
- 6.Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein–Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lupton S, Levine AJ. Mapping genetic elements of Epstein–Barr virus that facilitate extrachromosomal persistence of Epstein–Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 9.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein–Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirchmaier AL, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein–Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–4262. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Little RD, Schildkraut CL. Initiation of latent DNA replication in the Epstein–Barr virus genome can occur at sites other than the genetically defined origin. Mol Cell Biol. 1995;15:2893–2903. doi: 10.1128/mcb.15.5.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norio P, Schildkraut CL. Plasticity of DNA replication initiation in Epstein–Barr virus episomes. PLoS Biol. 2004;2:e152. doi: 10.1371/journal.pbio.0020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norio P, Schildkraut CL, Yates JL. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein–Barr virus chromosome after infection of established cell lines. J Virol. 2000;74:8563–8574. doi: 10.1128/jvi.74.18.8563-8574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams A. Replication of latent Epstein–Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dijkwel PA, Hamlin JL. The Chinese hamster dihydrofolate reductase origin consists of multiple potential nascent-strand start sites. Mol Cell Biol. 1995;15:3023–3031. doi: 10.1128/mcb.15.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 18.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human beta-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 19.Leight ER, Sugden B. Establishment of an oriP replicon depends upon an infrequent, epigenetic event. Mol Cell Biol. 2001;21:4149–4161. doi: 10.1128/MCB.21.13.4149-4161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bashaw JM, Yates JL. Replication from oriP of Epstein–Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J Virol. 2001;75:10603–10611. doi: 10.1128/JVI.75.22.10603-10611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein–Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Lindner SE, Leight ER, Sugden B. Essential elements of a licensed, mammalian plasmid origin of DNA synthesis. Mol Cell Biol. 2006;26:1124–1134. doi: 10.1128/MCB.26.3.1124-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith HC, Berezney R, Brewster JM, Rekosh D. Properties of adenoviral DNA bound to the nuclear matrix. Biochemistry. 1985;24:1197–1202. doi: 10.1021/bi00326a022. [DOI] [PubMed] [Google Scholar]
- 24.Angeletti PC, Engler JA. Adenovirus preterminal protein binds to the CAD enzyme at active sites of viral DNA replication on the nuclear matrix. J Virol. 1998;72:2896–2904. doi: 10.1128/jvi.72.4.2896-2904.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jankelevich S, Kolman JL, Bodnar JW, Miller G. A nuclear matrix attachment region organizes the Epstein–Barr viral plasmid in Raji cells into a single DNA domain. EMBO J. 1992;11:1165–1176. doi: 10.1002/j.1460-2075.1992.tb05157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 27.Leonhardt H, et al. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 29.Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng C, et al. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X, et al. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- 32.Aladjem AF, Kowalski D. Eukaryotic DNA replication origins. In: Depamphilis ML, editor. DNA Replication and Human Disease. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006. pp. 31–62. [Google Scholar]
- 33.Amati B, Gasser SM. Drosophila scaffold-attached regions bind nuclear scaffolds and can function as ARS elements in both budding and fission yeasts. Mol Cell Biol. 1990;10:5442–5454. doi: 10.1128/mcb.10.10.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dijkwel PA, Hamlin JL. Matrix attachment regions are positioned near replication initiation sites, genes, and an interamplicon junction in the amplified dihydrofolate reductase domain of Chinese hamster ovary cells. Mol Cell Biol. 1988;8:5398–5409. doi: 10.1128/mcb.8.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amati B, Pick L, Laroche T, Gasser SM. Nuclear scaffold attachment stimulates, but is not essential for ARS activity in Saccharomyces cerevisiae: Analysis of the Drosophila ftz SAR. EMBO J. 1990;9:4007–4016. doi: 10.1002/j.1460-2075.1990.tb07622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.