Abstract
Plant form is shaped by a complex network of intrinsic and extrinsic signals. Light-directed growth of seedlings (photomorphogenesis) depends on the coordination of several hormone signals, including brassinosteroids (BRs) and auxin. Although the close relationship between BRs and auxin has been widely reported, the molecular mechanism for combinatorial control of shared target genes has remained elusive. Here we demonstrate that BRs synergistically increase seedling sensitivity to auxin and show that combined treatment with both hormones can increase the magnitude and duration of gene expression. Moreover, we describe a direct connection between the BR-regulated BIN2 kinase and ARF2, a member of the Auxin Response Factor family of transcriptional regulators. Phosphorylation by BIN2 results in loss of ARF2 DNA binding and repression activities. arf2 mutants are less sensitive to changes in endogenous BR levels, whereas a large proportion of genes affected in an arf2 background are returned to near wild-type levels by altering BR biosynthesis. Together, these data suggest a model where BIN2 increases expression of auxin-induced genes by directly inactivating repressor ARFs, leading to synergistic increases in transcription.
Keywords: cross-regulation, Arobidopsis, growth, phytohormones
In plants, physiology and development are inextricably linked: internal age and metabolic cues are integrated with information about the external environment to direct morphogenesis. During seedling growth, the hormone auxin relays information about endogenous factors, such as where cells are within the plant body (1), as well as environmental stimuli, such as light and gravity (2). Another class of hormones, the brassinosteroids (BRs), act synergistically with auxins to promote cell elongation, and mutants in either pathway show similar phenotypes, including dramatic growth defects (3). Auxin and BRs share a number of early target genes, many of which are involved in growth-related processes (4–7).
Despite the strong evidence for synergism and interdependence between auxin and BR transcriptional responses (3), no direct molecular link between them has yet been reported. The path from auxin perception to transcriptional response is short and acts through alleviation of transcriptional repression (8). Auxin binding to F-box proteins of the AFB family, such as TIR1, triggers the degradation of transcriptional repressors called Aux/IAAs. Aux/IAAs bind to a family of transcription factors, called Auxin Response Factors (ARFs), which show either repressor or activator activity (9). Current models propose that auxin-mediated removal of Aux/IAA proteins increases activity of ARFs and thereby triggers the auxin genomic response (9). A potential molecular link between auxin and BR pathways comes from the discovery that regulatory regions of BR-responsive genes are enriched with predicted ARF-binding sites (6, 10). DR5, a synthetic multimerized version of ARF-binding sites, is induced by both auxin and BRs, and full induction by either pathway depends on both pathways (6, 11).
Genomic BR response requires another family of transcription factors, which includes BES1 and BZR1 (7, 12–14). BES1 can bind and activate transcription on the promoters of genes like SAUR-15, common to both auxin and BR pathways (13). BRs regulate the activity of BES1 and BZR1 by modulating their phosphorylation status, dependent on the coordinate action of the BIN2 GSK3 kinase and the BSU1 phosphatase, and respective family members (7, 15–18). Phosphorylation by BIN2 inhibits homodimerization by BES1, thereby blocking DNA-binding and transcriptional activation (17), and modulates the dynamics of BZR1 in the cell by interaction with a 14-3-3 protein (19, 20). In the presence of BRs, BES1/BZR1 family members are hypophosphorylated and promote BR responses in the nucleus by direct binding to BR-responsive gene promoters (12, 13).
Multiple hormones interact to direct morphogenesis, and no simple hierarchical relationship exists among these factors (21, 22). Auxin and BRs are among the best characterized hormone pathways and serve as a useful test case for elaborating plant signaling networks. Here we quantify and model BR effects on cellular sensitivity to auxin. We show that, for many shared target genes, exposure to both hormones provokes a distinct effect from exposure to either hormone alone. We describe a mechanistic link between the auxin and BR pathways: the BR-regulated BIN2 kinase directly modulates ARF2 transcriptional activity. These data suggest a new model for coordination of auxin and BR transcriptional effects. The activity of BIN2 contributes to a synergistic increase in auxin-induced gene expression by facilitating the removal of a repressor (ARF2), leaving regulatory sites more accessible to activator ARFs that promote transcription.
Results
Brassinosteroids Sensitize Seedlings to Exogenous Auxin.
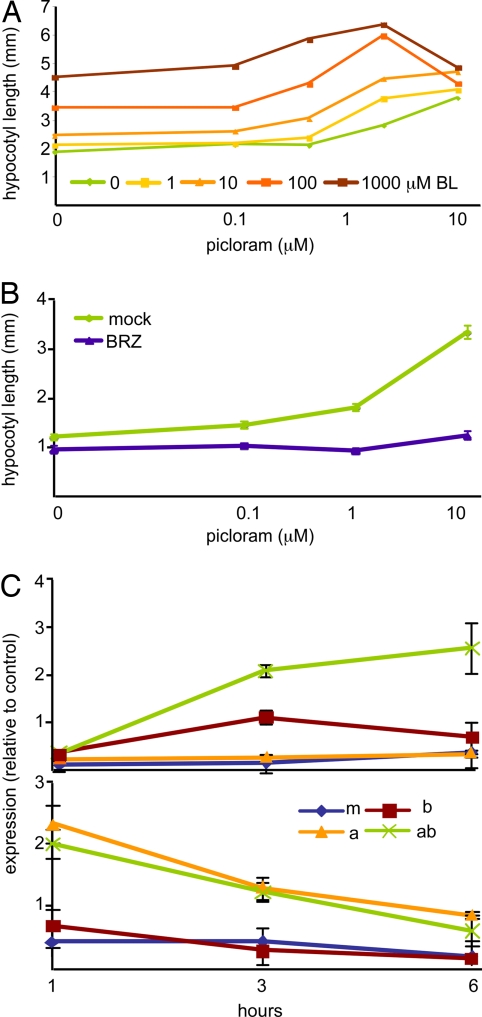
BR treatments have dramatic effects on auxin response (23). One limitation for earlier work is that indole-3-acetic acid (IAA), the naturally occurring form of auxin, does not promote hypocotyl elongation when supplied exogenously, likely as a result of insufficient transport to the shoot. Picloram (pic), a synthetic auxin, phenocopies the effects observed when endogenous auxin levels are increased (24). We have used pic treatments to rigorously test a model of auxin:BR synergism. In seedlings grown on a range of doses of pic, exposure to BRs increases absolute growth in a nonadditive manner and decreases the amount of pic needed to promote hypocotyl elongation (Fig. 1A). In the presence of even higher levels of pic alone, growth is inhibited, an effect observed at the highest levels of added BRs in this experiment. Multiple linear regression indicates that the pic:BR interaction is highly significant (P ≪ 0.001). If BR biosynthesis is inhibited with brassinazole (BRZ), the growth-promoting activity of pic is greatly reduced (Fig. 1B). Thus, normal auxin response depends on an intact BR pathway.
Fig. 1.
Auxin response is greatly enhanced by and dependent on BRs. (A) BR treatment increases sensitivity to exogenous auxin and synergistically enhances auxin-induced hypocotyl elongation. (B) Reducing BR synthesis with BRZ causes a dramatic reduction in auxin response. Average hypocotyl lengths of 10–15 light-grown 4-day-old seedlings are shown in A and B. (C) Quantitative RT-PCR experiments were performed by using RNA extracted from 5-day-old light-grown seedlings treated for 1, 3, or 6 h with mock (m), BR (b), IAA (a), or combined hormone (ab) treatments. Representative examples of observed patterns are shown. Many genes showed an increase in magnitude and duration of expression when both hormones were present (as shown here for At1g62440); other genes were insensitive to BR effects (as shown here for At4g32280). Error bars indicate standard error.
Global transcriptome studies show a large overlap in auxin and BR early response genes (25). Quantitative RT-PCR studies indicated a unique effect on gene expression when both hormones are given together [Fig. 1C and supporting information (SI) Table S1]. Seven-day-old seedlings exposed to mock, IAA only, BR only, or IAA and BR treatments were harvested after 1, 3, or 6 h. Expression of 12 genes previously identified as auxin- and/or BR-responsive were analyzed. In seven of 12 genes, combined BR and IAA treatment increased the magnitude of gene induction (Table 1 and Table S1). In five of these seven cases, the duration of detectable gene response was also increased. For the remaining genes in the study, IAA or BR effects were still measurable after 6 h, so any effect of the combined treatment could not be assessed. These results demonstrate that the combination of auxin and BRs together provoke a distinct cellular signal for a subset of target genes. Promoter analysis revealed significant overrepresentation of the AuxRE-related sites TTGTCT and TGTCT in the upstream region of genes affected by combined hormone treatment (P < 0.001; Promomer) (26) and not in the upstream region of genes affected by IAA only.
Table 1.
Genes analyzed by quantitative PCR
| AGI | BL | IAA | Annotation | Greater | Equal | Longer | TGTCTC | TGTCT | TTGTCT |
|---|---|---|---|---|---|---|---|---|---|
| At4 g08950 | 3.2 | NC | Phi-1-like | + | ND | 1 | 1 | 0 | |
| At3 g45970 | 3.9 | NC | AtEXPL1 | + | ND | 1 | 1 | 1 | |
| At1 g62440 | 2.9 | NC | LRX2 | + | + | 4 | 5 | 2 | |
| At1 g29490 | 4.7 | 24.9 | SAUR-68 | + | + | 0 | 5 | 1 | |
| At4 g30290 | 3.7 | 10.9 | AtXTH19 | + | + | 1 | 3 | 3 | |
| At1 g52830 | 3.6 | 3.4 | IAA6 | + | + | 1 | 3 | 1 | |
| At3 g55840 | 4.1 | 2.6 | Hs1pro-1-like | + | ND | 0 | 1 | 0 | |
| At2 g23170 | NC | 147.8 | GH3.3 | + | ND | 1 | 3 | 0 | |
| At4 g32280 | NC | 27.0 | IAA29 | + | ND | 0 | 0 | 0 | |
| At4 g14560 | NC | 20.6 | IAA1 | + | ND | 0 | 2 | 1 | |
| At5 g02760 | NC | 11.5 | PP2C | + | + | 0 | 2 | 1 | |
| At5 g47370 | NC | 10.0 | Homeobox-leucine zipper | + | ND | 0 | 0 | 0 |
Brassinolide (BL) and IAA columns show level of induction observed in global transcriptome experiments after 2.5- or 3-h hormone treatments, respectively (6). Expression in the presence of both hormones was characterized as either greater than either hormone alone or equal to the level achieved under a single hormone treatment. Increased duration of expression was assessed for genes where single hormone treatments returned to mock levels by the 6-h time point. The number of times each variant of the ARF-binding site appears in the 1,000 bp upstream of the translation start site of each gene is listed. NC, no change; ND, could not be determined.
The BR-Regulated Kinase BIN2 Interacts with an Auxin Response Factor.
BRs, like auxin, are capable of regulating transcription through isolated ARF-binding sites (11). BES1 levels and phosphorylation state are unchanged by auxin treatments (7), suggesting that any effects of the auxin pathway on BR-mediated gene regulation are either downstream of BR regulation of BES1 or parallel to it. BRs have no effect on the stability of Aux/IAAs (6, 27), although they do have modest effects on the transcript accumulation of some ARFs (25). This suggests that BR effects on auxin-mediated gene regulation are downstream of the Aux/IAAs, likely at the level of ARFs themselves.
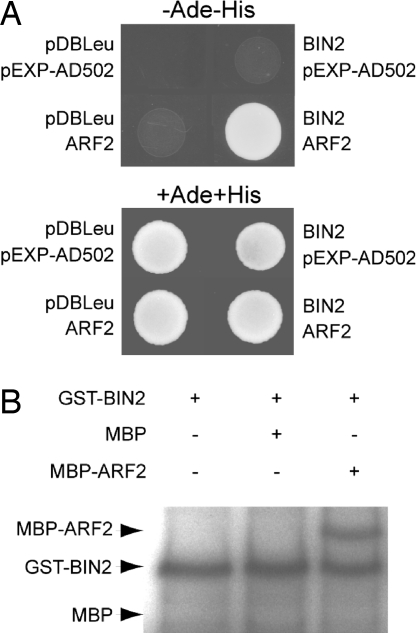
A yeast two-hybrid screen using the GSK3-type kinase BIN2 as bait provided the first potential molecular link between the auxin and BR pathways. A fragment of ARF2 was isolated as a BIN2-interacting protein. The ARF2 region interacting with BIN2 is restricted to the 540 aa in C terminus, encompassing the ARF-Aux/IAA dimerization domain (28, 29). A full-length ARF2 clone was independently verified to interact with BIN2 (Fig. 2A). In silico analysis indicates that ARF2 protein harbors 23 putative GSK3 recognition sites, 14 of them located in the longest fragment isolated in the screen. In vitro kinase assay demonstrated that BIN2 is able to bind to and phosphorylate ARF2 (Fig. 2B), consistent with the presence of numerous GSK3 phosphorylation sites. This suggests a possible regulatory role for the interaction of the downstream auxin and BR signaling components ARF2 and BIN2. No interaction could be detected between BES1 and ARF2 (Fig. S1).
Fig. 2.
BIN2 interacts with and phosphorylates ARF2. (A) BIN2 interacts with ARF2 in yeast. Yeast was transformed with a bait construct and a prey construct. The bait constructs contain GAL4-DNA binding domain fused with BIN2, and the prey constructs contain GAL4-activation domain fused with ARF2. Interactions between each pair of test proteins were determined by selection for growth on −Ade −His medium containing 5-bromo-4-chloro-3-indolyl-α-d-galactoside (X-α-Gal). (B) In vitro kinase assays using GST-BIN2 and MBP or MBP-ARF2. The autophosphorylation of GST-BIN2 serves as a loading control. Arrows show theoretical mobility of MBP, phosphorylated MBP-ARF2, and autophosphorylated BIN2 kinase.
BIN2 Regulates ARF2 Binding to DNA and Repressor Activity.
ARF2 has been shown to act as a repressor in transient expression studies in cell culture (30), suggesting that BRs might increase auxin responsiveness by alleviating repression on auxin-induced promoters. We hypothesized that phosphorylation may modify ARF2 stability, activity, and/or ability to bind DNA. Recently, ARF2 has been shown to be degraded rapidly after ethylene treatment (31). We therefore investigated the effect of BRs on ARF2 stability. Multiple Western blots performed under a variety of hormone conditions and in a number of mutant backgrounds showed no evidence for altered ARF2 stability by BRs (Fig. S2).
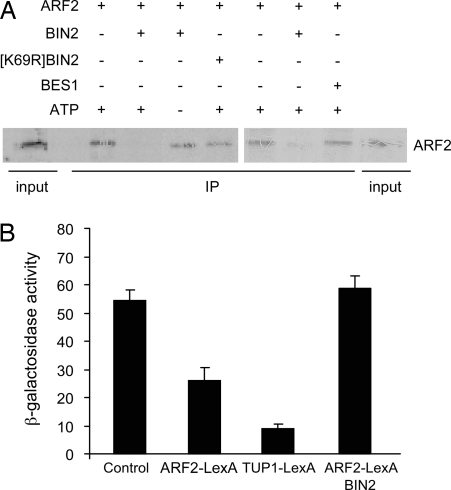
To test whether phosphorylation by BIN2 alters ARF2 DNA binding, we used in vitro DNA-binding experiments with the promoter of SAUR-15 gene (previously called SAUR-AC1) as a target. SAUR-15 is among the earliest detectable auxin- and BR-induced transcripts, and BES1 binding to the SAUR-15 promoter has been shown by chromatin immunoprecipitation (13) and in vitro binding studies (17). The target region has five E-boxes, known to be required for BES1 activation (13), one canonical AuxRE (TGTCTC), and 11 core TGTC elements. As has been reported previously (17), BIN2 showed no DNA binding ability on its own, and BIN2 phosphorylation of BES1 reduced BES1 binding to the promoter of SAUR-15 (Fig. S3). Using the same conditions, ARF2 could bind to the SAUR-15 promoter, and preincubation with BIN2 greatly reduced DNA binding activity (Fig. 3A). The addition of BES1 did not alter ARF2 interaction with DNA (Fig. 3A). BIN2-dependent inhibition of ARF2 DNA binding required ATP and functional kinase activity as shown by the kinase-dead [K69R] BIN2 control (32). This finding suggests that BIN2 effects on ARF2 occurred via phosphorylation.
Fig. 3.
Phosphorylation alleviates ARF2 DNA binding and repressor activity. (A) DNA:protein pull-down experiments with biotinylated DNA fragments from SAUR-15 promoter with ARF2 translated in vitro. ARF2 protein was incubated with BIN2 or kinase-dead [K69R] BIN2 before pull-down experiments with DNA-bound streptavidin beads. Two representative examples from seven similar experiments are shown. BES1 does not interfere with ARF2 binding to DNA. Purified recombinant MBP-BES1 was added where indicated. (B) Yeast repression assay. Repression of the transcription of a LexAop-CYC1::LacZ reporter by a ARF2-LexA fusion in the presence or absence of BIN2 was assessed by β-galactosidase activity. Results are presented as mean of two separate assays of three independent transformants. Error bars indicate standard error.
To address whether BIN2-mediated phosphorylation of ARF2 modulates its repressor activity, we tested ARF2 repression activity in yeast. A LacZ reporter gene was fused downstream from the constitutive CYC1 promoter fused to 4xLexA operators. Reporter expression was then measured in the presence or absence of candidate repressors fused to the LexA binding domain. Reduced β-galactosidase activity, indicating repression, was clearly detected in the presence of the control, yeast repressor TUP1 (33) fused to LexA (TUP1-LexA), or in the presence of ARF2-LexA. Thus, ARF2 acted as a repressor in our system. Coexpression of BIN2 with ARF2-LexA completely alleviated ARF2 transcriptional repression activity (Fig. 3B). These results provide strong evidence that BIN2 regulates both DNA binding and transcriptional activity of ARF2.
ARF2 Is Required for Normal BR Response.
Previous studies of loss-of-function arf2 mutants found a range of growth-related defects (31, 34–36). Some of these phenotypes are similar to those observed in mutants with moderate increases in BR sensitivity, such as plants overexpressing the transcription factor BEE1 (37). There are conflicting results regarding ARF2 effects on known auxin response genes, but the large size of the ARF family may complicate analysis of individual loss-of-function mutants. To evaluate the involvement of ARF2 in BR signaling, hypocotyl elongation of dark-grown arf2 seedlings was assayed on a range of concentrations of BRZ, a BR biosynthesis inhibitor. Mutations in arf2 resulted in substantial resistance to BRZ, suggesting that ARF2 is a negative regulator of the BR pathway (Fig. 4A). No difference in response to exogenous BRs could be detected (Fig. 4B), perhaps as a result of redundancy with or compensation by other players in the BR or auxin pathways.
Fig. 4.
ARF2 is a negative regulator of BR responses. Hypocotyl elongation in response to BRZ (A) and BL (B) is shown. Four-day-old dark-grown wild-type (filled bars) and arf2 (open bars) seedlings were grown on plates with different concentrations of BRZ or BL. Results are presented as mean of two independent experiments ± SD (n = 25).
To further examine the relationship between ARF2 and the BR pathway, global transcriptome analysis was performed comparing BRZ response in arf2 and wild-type seedlings. In both genotypes, BRZ treatments induced expression of genes encoding BR biosynthetic enzymes, including DWARF4 (At3g50660), AtBR6OX1 (At5g38970), and AtBR6OX2 (At3g30180) (Tables S2–S4). In wild-type plants, BRZ also increased expression of CPD (At5g05690), encoding an additional BR biosynthetic enzyme, and decreased levels of CYP72C1 (At1g17060), encoding a BR-inactivating enzyme. Interestingly, arf2 mutants did not show significant changes in expression of these additional genes, perhaps indicating an ARF2-dependent feedback loop.
A number of genes involved in auxin metabolism, transport, or response were misregulated in an arf2 mutant (Tables S5 and S6). Although SAUR-15 does not show altered expression in arf2 mutants in these conditions, several closely related SAUR genes, as well as members of the Aux/IAA and GH3 early auxin response families, are altered in arf2. Nearly 40% of the genes differentially regulated in the arf2 background were responsive to BRZ (355 of 896 genes; Table S4), suggesting that ARF2 may act to integrate auxin and BR pathways. This result also implies that BR-regulated activators are required for full expression of ARF2-regulated genes, because the loss of BRs acts to restore normal gene expression in an arf2 mutant. Whereas most differentially expressed genes are up-regulated in arf2, there are genes with reduced expression, likely reflecting secondary effects.
A significant overrepresentation of genes involved in tryptophan (and auxin) biosynthesis are up-regulated in the arf2 background (P < 0.01, BioMaps: http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/virtualplant.cgi; Tables S5 and S7). Linear modeling generated a list of genes that had a differential response to BRZ depending on genotype (g*t genes; Table S8). This list included the tryptophan biosynthesis genes. Although no effect of BRZ could be detected in a wild-type background, BRZ treatment returned gene expression to wild-type levels in arf2 plants (Tables S7 and S8). If these changes lead to increased auxin levels, they could explain the increased resistance to BRZ seen in arf2 mutants. Growing plants at reduced temperature completely suppressed the arf2 BRZ resistance phenotype (Fig. S4), perhaps reflecting the temperature sensitivity of the auxin biosynthesis pathway (38).
Discussion
Previous physiological and transcriptome studies highlighted the close relationship between auxin and BRs, including evidence that promoter regions of BR-responsive genes are enriched in ARF-binding sites (4, 6). In the present study, we have further characterized the relationship between auxin and BRs and shown a molecular link connecting the two signaling pathways. We provide strong evidence that ARF2 mediates some BR responses and that the BR-regulated kinase BIN2 regulates ARF2 by phosphorylation.
Our data suggest a model where BRs and auxin coordinately regulate gene expression through ARF-binding sites. It is well established that the transcriptional activation activity of activator ARFs is modulated by auxin-mediated degradation of Aux/IAA corepressors (9). We propose that repressor ARFs, such as ARF2, may compete for binding with activator ARFs at AuxREs in the promoters of some genes. Phosphorylation of ARF2 by BIN2 would result in removal of ARF2 from DNA and the loss of ARF2 repression activity. In effect, BRs would release a brake from such coregulated genes, and auxin would provide gas to drive increased expression through release of repression on the activator ARFs. Addition of both hormones would result in elevated and potentially prolonged expression of target genes. This model is consistent with recent studies showing the requirement of activator ARF activity for expression of multimerized ARF-binding site reporters and target genes (39). Increased levels of activator ARFs would induce expression by outcompeting repressor ARFs for DNA binding, thus mimicking the effect of adding BRs.
This mode of coregulation is likely to represent only part of the combinatorial control mechanism of auxin and BRs. Binding sites for the BR-regulated activator BES1 are also overrepresented in genes regulated by both hormones (6), suggesting potential additional layers of control on promoters of coregulated genes. In addition, BRs have been shown to promote auxin transport (40, 41), and recent studies have connected auxin signaling and BR biosynthesis (42). In the root, auxin increases BR levels through induction of CPD. BR levels in turn regulate the magnitude of auxin effect. A multilevel interaction between auxin and BRs is further supported by our data suggesting that ARF2 links BR and auxin biosynthetic pathways. arf2 mutants show elevated expression of genes required for normal auxin biosynthesis. BRZ treatment suppresses the expression of these genes to near wild-type levels, suggesting that auxin production may be partially dependent on BRs. In addition, some components of the BR homeostasis mechanisms appear to require ARF2 function.
The role of the repressor ARFs in auxin signal transduction has remained elusive. Here we present evidence that one of these proteins, ARF2, acts as a point of interaction with the BR pathway. Other studies have shown that ARF2 protein levels are antagonistically regulated by ethylene and light (31). Together these findings suggest that ARF2, and perhaps other repressor ARFs, may act as integrators of multiple pathways regulating photomorphogenesis.
Methods
Hypocotyl Length Measurements.
Seeds were sterilized for 15 min in 70% ethanol and 0.01% Triton X-100, followed by 10 min of 95% ethanol. After sterilization, seeds were suspended in 0.1% low-melting-point agarose and spotted on plates containing 0.5× Murashige minimal organics medium (Gibco/BRL) and 0.8% phytagar (Gibco/BRL). Seeds on plates were then stratified in the dark at 4°C for 4 days. Plants were maintained in the dark but transferred to 22°C. Hypocotyl lengths from 10–14 seedlings were measured on day 4 with NIH Image 1.62. All dose–response experiments were performed at least in duplicate.
Quantitative PCR.
Nine-day-old, light-grown seedlings were immersed in mock, 1 μM BR, 1 μM auxin (IAA), or both hormones in 0.5× Murashige minimal organics medium (Invitrogen) for 1, 3, or 6 h before they were harvested. Total RNA was extracted by using a Qiagen RNAeasy kit, and first-strand cDNA was synthesized by using an Invitrogen SuperScript First-Strand cDNA Synthesis kit. cDNAs were diluted 20-fold and combined with SYBR master mix (Bio-Rad). Three independent biological replicates were used and analyzed with a Bio-Rad iCycler. A standard curve was constructed for each primer by using an equal mixture of all cDNAs.
Two-Hybrid Screening.
The entire BIN2 ORF was cloned into the pDBLeu vector, carrying the GAL4 DNA binding domain, and transformed into yeast AH109 cells. Yeast cells carrying the bait vector were then transformed with the plasmid DNAs of the CD4–22 Arabidopsis cDNA library prepared from 3-day-old etiolated seedlings. Putative BIN2 interactors were screened by growth on synthetic medium lacking Ade and His and confirmed by blue color on 5-bromo-4-chloro-3-indolyl-α-d-galactoside-containing medium. To confirm the interaction, prey plasmids were rescued and retransformed in AH109 cells expressing the GAL4 DNA binding domain or the BIN2-GAL4 DNA binding domain fusion protein. The resulting yeast cells were assayed for the activation of the ADE2, HIS3, and LacZ reporter genes. Full-length ARF2 and BES1 ORFs were cloned in pEXP-AD502 and assayed for interaction with BIN2 as described above.
Recombinant Protein Preparation.
BIN2 and [K69R]BIN2 were cloned into pGEX-5X-1 to generate GST fusions. ARF2 was cloned in pMALc2x to create MBP fusions. Recombinant proteins were prepared according to the manufacturer's suggestion (Amersham Pharmacia and New England Biolabs, respectively).
In Vitro Phosphorylation Assay.
MBP-ARF2 was incubated with GST-BIN2 in 20 μl of kinase buffer (20 mM Tris, pH 7.5/100 mM NaCl/12 mM MgCl2/100 μM ATP) and 10 μCi of [γ-32P]ATP. After incubation at 30°C for 20 min, the reactions were stopped by adding 20 μl of 2× SDS buffer and boiling at 94°C for 5 min. Proteins were resolved on a SDS polyacrylamide gel, and phosphorylation was detected by exposing the dried gel to x-ray film.
Yeast Repression Assay.
The GAL4 DNA binding domain sequence of pBridge was replaced by the LexA gene. ARF2 was subsequently cloned in the modified pBridge, in frame with LexA, under the control of ADH1 promoter. Finally, BIN2 was cloned in the second multiple cloning site of the modified version of pBridge, under the control of MET25 promoter. MaV203 yeast cells were sequentially transformed with pJK1621, carrying 4xLexA operators upstream of UAS and promoter of CYC1 fused to bacterial LacZ gene as a reporter (33), and the modified pBridge carrying the LexA-ARF2 gene and BIN2. β-Galactosidase assays were performed on yeast protein extracts using ONPG as a substrate, as described previously (MATCHMAKER two-hybrid system; Clontech).
DNA-Binding Experiments.
ARF2 in the pCMX-pL1 vector was subjected to 35S-Met T7 in vitro transcription/translation with a reticulocyte system (Promega) and then incubated with GST-BIN2 or GST-[K69R]BIN2 in the presence/absence of ATP. PCR-generated biotinylated pSAUR15 DNA fragments were bound to streptavidin-coated paramagnetic beads (Invitrogen) and washed three times in IP100 buffer (100 mM potassium glutamate/50 mM Tris·HCl, pH 7.6/2 mM MgCl2/0.05% Nonidet P-40). Radiolabeled ARF2 was added to DNA-bound beads and rotated in a 4°C cold room for 2 h. Beads were washed three times with IP100 buffer; proteins were removed by boiling with 2× SDS buffer and subjected to SDS/PAGE.
Microarray Analysis.
Total RNA was extracted from 4-day-old, etiolated Arabidopsis seedlings grown on 0.5 μM BRZ or mock treatments and used to probe ATH1 microarrays (Affymetrix), according to the manufacturer's protocols. All experiments used at least two independent biological replicates. Data analysis was performed in R (43). We performed standard Affymetrix quality-control procedures using the BioConductor package simpleaffy (44). Expression was normalized and estimated by using the gcRMA package of BioConductor (45) and subsequently analyzed by using linear models and empirical Bayes analysis (limma package) (46). To be considered differentially expressed, genes were required to have a false discovery rate adjusted P value of <0.05 and an empirical Bayes log odds value of >0.
Supplementary Material
Acknowledgments.
We thank Hai Li and Joseph Ecker (The Salk Institute) for arf2-6 seeds and anti-ARF2 antibody, Zach Smith and Jeff Long (The Salk Institute) for the yeast repression system, Jianming Li (University of Michigan, Ann Arbor) for BIN2 constructs, Alison Crowe (University of Washington, Seattle) for consultations and equipment needed for in vitro DNA binding assays, Fangxin Hong for assistance with microarray analysis, and Kris Lawrence and Rachel Denney for technical assistance. This work was supported by National Science Foundation Grant IOS-0649389, a Human Frontier Science Program Organization long-term fellowship (to G.V.), and funds from the University of Washington. J.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE11216).
This article contains supporting information online at www.pnas.org/cgi/content/full/0803996105/DCSupplemental.
References
- 1.De Smet I, Jurgens G. Patterning the axis in plants—Auxin in control. Curr Opin Genet Dev. 2007;17:337–343. doi: 10.1016/j.gde.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Friml J. Auxin transport—shaping the plant. Curr Opin Plant Biol. 2003;6:7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- 3.Halliday KJ. Plant hormones: The interplay of brassinosteroids and auxin. Curr Biol. 2004;14:R1008–R1010. doi: 10.1016/j.cub.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mussig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129:1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 8.Kepinski S. The anatomy of auxin perception. BioEssays. 2007;29:953–956. doi: 10.1002/bies.20657. [DOI] [PubMed] [Google Scholar]
- 9.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Goda H, et al. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura A, et al. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 2003;133:1843–1853. doi: 10.1104/pp.103.030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 14.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 15.He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mora-Garcia S, et al. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 18.Zhao J, et al. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gampala SS, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu H, et al. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;19:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 22.Nozue K, Maloof JN. Diurnal regulation of plant growth. Plant Cell Environ. 2006;29:396–408. doi: 10.1111/j.1365-3040.2005.01489.x. [DOI] [PubMed] [Google Scholar]
- 23.Hardtke CS. Transcriptional auxin-brassinosteroid crosstalk: Who's talking? BioEssays. 2007;29:1115–1123. doi: 10.1002/bies.20653. [DOI] [PubMed] [Google Scholar]
- 24.Delarue M, Prinsen E, Onckelen HV, Caboche M, Bellini C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998;14:603–611. doi: 10.1046/j.1365-313x.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- 25.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 26.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 27.Zenser N, Dreher KA, Edwards SR, Callis J. Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J. 2003;35:285–294. doi: 10.1046/j.1365-313x.2003.01801.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 30.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell. 2004;7:193–204. doi: 10.1016/j.devcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 33.Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 34.Ellis CM, et al. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132:4563–4574. doi: 10.1242/dev.02012. [DOI] [PubMed] [Google Scholar]
- 35.Okushima Y, Mitina I, Quach HL, Theologis A. AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant J. 2005;43:29–46. doi: 10.1111/j.1365-313X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- 36.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 37.Friedrichsen DM, et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics. 2002;162:1445–1456. doi: 10.1093/genetics/162.3.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S, Tiwari SB, Hagen G, Guilfoyle TJ. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell. 2005;17:1979–1993. doi: 10.1105/tpc.105.031096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao F, et al. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004;134:1624–1631. doi: 10.1104/pp.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L, Xu J, Xu ZH, Xue HW. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell. 2005;17:2738–2753. doi: 10.1105/tpc.105.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mouchel CF, Osmont KS, Hardtke CS. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature. 2006;443:458–461. doi: 10.1038/nature05130. [DOI] [PubMed] [Google Scholar]
- 43.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 44.Wilson CL, Miller CJ. Simpleaffy: A BioConductor package for Affymetrix Quality Control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- 45.Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. Department of Biostatistics Working Papers, Johns Hopkins University. Working Paper 1. 2004 Available at http://www.bepress.com/jhubiostat/paper1. [Google Scholar]
- 46.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. State Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.