Abstract
A missense mutation in the αB-crystallin (CryAB) gene triggers a severe form of desmin-related cardiomyopathy (DRCM) characterized by accumulation of misfolded proteins. We hypothesized that autophagy increases in response to protein aggregates and that this autophagic activity is adaptive. Mutant CryAB (CryABR120G) triggered a >2-fold increase in cardiomyocyte autophagic activity, and blunting autophagy increased the rate of aggregate accumulation and the abundance of insoluble CryABR120G-associated aggregates. Cardiomyocyte-restricted overexpression of CryABR120G in mice induced intracellular aggregate accumulation and systolic heart failure by 12 months. As early as 2 months (well before the earliest declines in cardiac function), we detected robust autophagic activity. To test the functional significance of autophagic activation, we crossed CryABR120G mice with animals harboring heterozygous inactivation of beclin 1, a gene required for autophagy. Blunting autophagy in vivo dramatically hastened heart failure progression with a 3-fold increase in interstitial fibrosis, greater accumulation of polyubiquitinated proteins, larger and more extensive intracellular aggregates, accelerated ventricular dysfunction, and early mortality. This study reports activation of autophagy in DRCM. Further, our findings point to autophagy as an adaptive response in this proteotoxic form of heart disease.
Keywords: protein aggregation, remodeling
Protein conformation disease, characterized by toxic aggregations of misfolded proteins, is a growing family of human disorders, which includes Alzheimer's disease, Parkinsonism, amyotrophic lateral sclerosis, and both polyglutamine and polyalanine expansion disorders (1). A common feature of these diseases is the formation of intracellular aggregates of toxic proteins. In muscle, where myofibrillar architecture is maintained by desmin and other intermediate filaments, intracellular protein aggregates contain desmin and perturbation of the desmin cytoarchitecture is a major feature of disease (leading to the designation desmin-related myopathy). [With the discovery that a number of other proteins are present in these intracellular inclusions, the more generic term myofibrillar myopathy is often used (2).] In all of these muscle disorders, cardiomyopathy is a major cause of mortality.
Cumulative pathologic stress on the heart elicits a syndrome of failure (3), a major source of morbidity and mortality and a significant drain on health-care resources worldwide. Desmin-related cardiomyopathies (DRCMs) are particularly severe and progressive forms of heart failure for which there are currently no effective treatments. This class of disease arises from mutations in several different proteins, including desmin, myotilin, and dystrophin (4). In a subset, disease is caused by a failed interaction between desmin and αB-crystallin (CryAB), a small heat shock protein (5). CryAB associates with desmin and functions as a molecular chaperone, preventing aggregation of desmin and thus maintaining myofibrillar structure (6). Mutations that disrupt the interaction between desmin and CryAB produce a phenotype of protein aggregation, myofibrillar disarray, cardiac dysfunction, and sudden cardiac death (4, 7).
Clinical presentation of the index family with DRCM caused by a missense (CryABR120G) mutation was characterized by early-onset cataracts, proximal and distal muscle weakness, and severe cardiomyopathy (7). CryABR120G-associated DRCM has now been replicated in two independently derived transgenic mouse models (8, 9). The CryABR120G mutation results in protein aggregation and aggresome formation (10), mitochondrial toxicity (11), disruption of proteasome function (12), and induces a state of “reductive stress” (9). However, whereas CryABR120G-induced pathogenesis has been well characterized, we have limited understanding of the adaptive cellular pathways that function to protect cardiomyocytes from CryABR120G-induced proteotoxicity.
Autophagy is increasingly appreciated as a cellular stress response involved in a variety of disease states (13). Best characterized as a mechanism of lysosome-mediated proteolysis, work by our group and others has demonstrated that cardiomyocyte autophagy is activated in heart by pressure overload and ischemia/reperfusion (14). Given that excessive protein aggregation is central to CryABR120G pathology, we postulated that autophagy, a process of bulk-protein degradation, could be a mechanism through which the heart protects itself in the setting of DRCM.
A number of characteristics associated with CryABR120G-induced cardiomyopathy led us to hypothesize that, in this setting, autophagy functions in a protective manner. CryABR120G-associated DRCM results from the chronic expression of an aggregate-inducing protein in terminally differentiated, nondividing cells. This paradigm is very similar to neurodegenerative diseases, where the abnormal deposition of proteins within intracellular aggregates is a prominent pathological feature. The prevailing theory in this field is that autophagic pathways facilitate removal of aggregates too large for efficient proteasome-mediated clearance, thus acting in a salutary fashion (15). Similarly, we previously reported that pressure overload or pharmacologically induced protein aggregation are sufficient to induce robust cardiomyocyte autophagy, which then functions to attenuate the accumulation of protein aggregates and aggresome formation (16). Based on the prominent role of protein aggregation in these diseases, we hypothesized that autophagy is up-regulated in DRCM, decreases the accumulation of toxic protein aggregates, and thereby attenuates disease progression.
Results
Mutant CryABR120G Increases the Abundance of Autophagosomes in Cardiomyocytes.
We hypothesized that the presence of aggregate-prone protein would induce autophagic activity in cardiac myocytes. To test this hypothesis, neonatal rat ventricular myocytes (NRVMs) were infected with virus expressing WT human CryAB (Ad-CryABWT), virus expressing mutant human CryABR120G (Ad-CryABR120G), or empty virus (control). Infected cells were cultured for 5 days and then processed for analysis by EM to evaluate for double membrane-bound vacuoles consistent with autophagosomes.
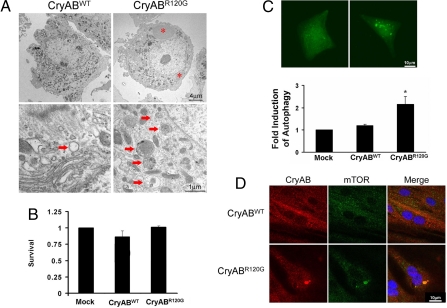
As expected, and consistent with the known “housekeeping” function of constitutive autophagy in many cells, modest numbers of autophagosomes were detected in both control and Ad-CryABWT-expressing cells (Fig. 1A). These autophagosomes were relatively uniform in morphology, appearing as clearly discernable double-membrane vacuoles, >0.5 μm in diameter, and containing heterogeneous proteinaceous material. In striking contrast, 5 days of Ad-CryABR120G expression triggered the appearance of large, perinuclear structures (Fig. 1A, *). The appearance of these structures was suggestive of aggresomes, intermediate filament-encaged collections of damaged proteins localized in a microtubule-dependent manner at the microtubule organizing center (MTOC) (10). Interestingly, we detected increased numbers of autophagosomes in the perinuclear region of aggresome-positive cells, with morphology distinct from the autophagosomes seen in healthy control cells. CryABR120G-induced autophagosomes were heterogeneous in morphology, multilamellar (in contrast to just one double membrane), contained high-density proteinaceous material, and showed evidence of mitochondrial sequestration (Fig. 1A, arrows). Despite the presence of large perinuclear aggresomes and accumulated autophagosomes, cell viability was not altered through the 5-day experimental period (Fig. 1B).
Fig. 1.
CryABR120G expression is a potent activator of cardiomyocyte autophagy. (A) Representative low (×5,000; Upper) and high (×20,000; Lower) magnification images of NRVMs 5 days after expression of CryABWT or CryABR120G. Aggresomes are evident in CryABR120G-expressing cells (asterisks) as are extensive perinuclear autophagosomes (arrows). (B) Despite extensive induction of autophagy, there is no appreciable change in cell viability 5 days postinfection. (C) NRVMs were transiently transfected with a GFP-LC3 construct and then infected with WT or mutant CryAB. Twenty-four hours later, autophagy was quantified as the number of punctate-positive cells divided by the total number of GFP+ cells. (D) Representative images of NRVMs (two examples of each) infected with CryAB and processed for mTOR immunocytochemistry. mTOR is distributed throughout the cytoplasm in NRVMs expressing WT CryAB. In contrast, CryABR120G triggered formation of perinuclear aggregates that stain for both mTOR and crystallin.
To quantify the increase in autophagic activity in response to Ad-CryABR120G expression, NRVMs were transfected with a GFP-LC3 autophagy-reporter construct, followed by infection with control adenovirus, Ad-CryABWT, or Ad-CryABR120G. LC3 is an intermembrane component of the early autophagosome, and its redistribution from a diffuse cytosolic signal to punctate dots is a sensitive and specific indicator of autophagy (17). After 24 h of infection, the abundance of autophagic vesicles was measured in live cells, quantified as the number of GFP-LC3 punctate-positive cells divided by the total number of GFP-positive cells. In these experiments, autophagic activity was increased >2-fold (P < 0.05) in NRVMs expressing Ad-CryABR120G, whereas infection with Ad-CryABWT had no effect (Fig. 1C).
Aggresomes invariably recruit cytoplasmic components, including chaperones and elements of the ubiquitin and proteasome pathways. In certain polyglutamine-expansion disorders, intracellular protein aggregates sequester mTOR (mammalian target of rapamycin), a well established inhibitor of autophagy, reducing levels of the soluble protein with a consequent increase in autophagy (18). Given our findings of autophagic activation in association with protein aggregate accumulation, we evaluated mTOR localization in NRVMs expressing CryABR120G. Consistent with findings reported in Huntington's disease, we detected a perinuclear coalescence of mTOR in cells expressing mutant crystallin (Fig. 1D). In contrast, mTOR remained freely distributed through the cytoplasm of cells expressing WT CryAB (Fig. 1D). These data, then, lend credence to the notion that protein aggregates induced by mutant CryAB trigger an autophagic response in cardiac myocytes.
Inhibiting Autophagy Increases the Abundance and Size of CryABR120G-Induced Aggregates.
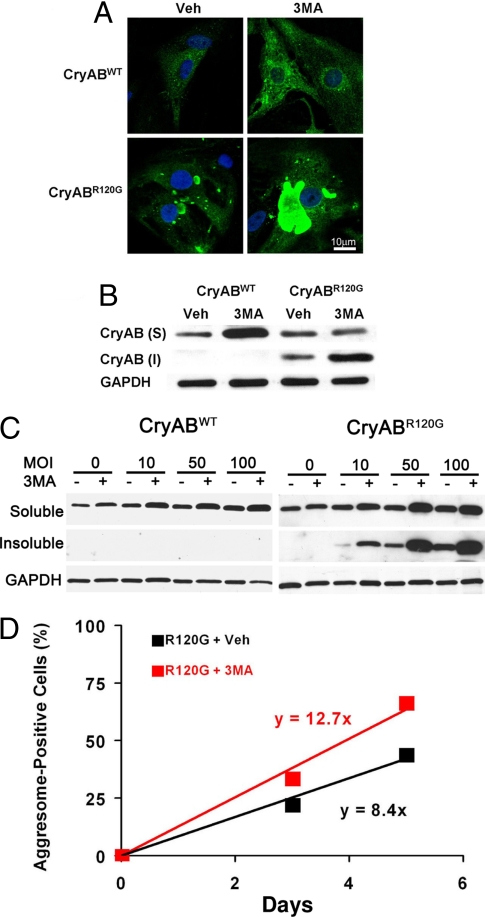
To test whether autophagosomes help clear CryABR120G–induced protein aggregates, NRVMs were infected with either Ad-CryABWT or mutant Ad-CryABR120G. After infection, the cells were treated daily with 5 mM 3-methyladenine (3MA), an inhibitor of class III phosphoinositide-3-kinase (PI3K), an enzyme required for initiation of autophagosome formation (19). After 5 days in culture, immunocytochemical analysis revealed that WT CryAB protein was distributed diffusely throughout the cytoplasm of vehicle-treated NRVMs infected with Ad-CryABWT (Fig. 2A). Inhibition of autophagy with 3MA increased the intensity of the CryAB-like immunoreactivity signal, but the immunoreactivity remained diffusely distributed throughout the cytoplasm with no evidence of protein aggregation or formation of aggresomes (Fig. 2A).
Fig. 2.
Autophagic activity serves to clear CryABR120G-induced protein aggregates. (A) Representative images depicting NRVMs subjected to 5 days of CryABR120G expression, demonstrating robust perinuclear aggresome formation. Blunting of autophagy with 3MA (5 mM) dramatically increased aggresome size. (B) Blunting autophagy increased the amount of insoluble CryAB in cells expressing mutant, but not WT, protein. (C) Dose–response relationship between CryABWT or CryABR120G expression and accumulation of soluble and insoluble protein. (D) CryABR120G-induced aggregate accumulation is accelerated in the setting of blunted autophagy, suggesting that autophagic pathways function to clear the aggregates.
Consistent with our EM studies, CryAB-associated perinuclear aggregates were detected in Ad-CryABR120G-infected NRVMs (Fig. 2A). Disruption of microtubules with nocodazole (10 μM) was sufficient to prevent perinuclear coalescence, supporting the idea that they are aggresomes [supporting information (SI) Fig. S1A]. Further, γ-tubulin, a structural component of the MTOC was found to colocalize with the structures (Fig. S1B). Together, these findings confirm that these intracellular structures are aggresomes.
As autophagic mechanisms are activated in neurodegenerative disease to clear protein aggregates, we tested the role of autophagy in aggresome formation in cardiac myocytes by blunting autophagy with 3MA. Here, we observed a dramatic increase in aggresome size (Fig. 2A). To evaluate for changes in the distribution of CryAB proteins, Nonidet P-40-soluble and insoluble proteins were fractionated from CryAB-overexpressing cells. In CryABWT-expressing cells, CryAB was detected only in the detergent-soluble fraction, regardless of the addition of 3MA to block autophagy (Fig. 2B). In contrast, CryAB was found in both the soluble and insoluble fractions of Ad-CryABR120G-infected cells, and blunting of autophagy with 3MA elicited a significant increase in the levels of insoluble CryAB (Fig. 2B). Similar findings were observed across a range of doses of CryABWT and CryABR120G (Fig. 2C). These results, then, suggest that autophagy plays a role in the clearance of CryABR120G protein, which may serve to protect myocytes from the accumulation of misfolded aggregates and subsequent incorporation into aggresomes.
The prevalence of CryAB-positive aggregates was also increased in response to 3MA. To examine the underlying kinetics, we quantified the number of NRVM with detectable aggresomes at 3 and 5 days postinfection. Three days after Ad-CryABR120G infection, 23.1% (±7.1) of all NRVMs (n = 479) were positive for aggresomes. Treatment with 3MA significantly increased the number of aggresome-positive cells to 33.5% (±9.3, n = 455, P < 0.05). Five days after infection, 48.8% (±14.4) of vehicle-treated cells (n = 274) were aggresome-positive, whereas in cells exposed to 3MA (n = 317), the prevalence increased to 66.3% (±10.1, P < 0.05) (Fig. 2D). Logistic regression analysis revealed that the rate of aggresome accumulation in 3MA-treated cells was accelerated (P = 0.03). Collectively, these findings demonstrate that expression of CryABR120G protein is sufficient to trigger robust autophagic activity in cardiac myocytes. Further, these data suggest that autophagic activity functions as a clearance pathway for these potentially toxic protein aggregates.
Hearts Expressing Mutant CryABR120G Display Increased Autophagic Activity.
To evaluate the relationship between autophagy and DRCM, we studied transgenic mice expressing cardiomyocyte-restricted hCryABR120G (αMHC-CryABR120G), a model of DRCM where disease progression is remarkably similar to that of humans carrying a mutant allele (9). The αMHC-CryABR120G transgene, expressed at levels comparable to our in vitro studies (Fig. S2), triggers a late-onset cardiomyopathy with severe pathological remodeling, protein aggregation, and death caused by heart failure at 10–12 months of age (9). Initial analysis was performed on 2-month-old animals, a time point that precedes ventricular remodeling or detectable reductions in cardiac function.
First, consistent with a prior report (20), we detected significant colocalization of crystallin and desmin proteins in the αCryABR120G mice (Fig. S3). Further, consistent with our in vitro studies, αMHC-CryABR120G-expressing cardiomyocytes contained perinuclear aggregates detectable by anti-CryAB immunohistochemistry (Fig. S4), even at this early, preclinical stage of disease (see below). Ultrastructural analysis also revealed numerous protein aggregates (Fig. S4B). By contrast, normal cellular architecture, with tightly packed arrangements of sarcomeres and mitochondria, was seen in WT littermates (Fig. S4B). Again, we detected an abundance of autophagosomes immediately adjacent to the aggregates in αMHC-CryABR120G-expressing cardiomyocytes, suggesting a functional relationship between aggregated proteins and autophagy (Fig. S4B).
Because metabolic stress and starvation are capable of activating autophagy, we tested whether increases in autophagic activity were the result of CryABR120G expressed within myocytes or a secondary consequence of systemic illness. To do this, we studied skeletal muscle from each animal in tandem with the heart samples. Ultrastructural analysis of skeletal muscle (soleus) from αMHC-CryABR120G mice revealed an absence of aggregates, no sarcomeric disarray, and no increase in autophagic activity (Fig. S4B). Indeed, skeletal muscle from these transgenic mice was identical on EM to WT littermates, suggesting that systemic factors do not account for the observed activation of autophagic pathways.
Blunting Autophagy Accelerates Pathological Remodeling in αMHC-CryABR120G Hearts.
To evaluate the effects of autophagic activity on the structural remodeling of DRCM, we studied hearts from αMHC-CryABR120G mice crossed into a beclin 1 haploinsufficient background. Beclin 1, the mammalian homologue of the proautophagic gene ATG6 in yeast, is required for recruitment of Atg12–Atg5 conjugates to preautophagosomal membranes (21), and hence is required for autophagic vesicle formation. Although beclin 1−/− mice are not viable, it has been demonstrated that heterozygous beclin 1+/− animals have an ≈50% reduction in autophagic capacity (22, 23). Hearts were studied at 9 months of age, a time point where severe structural abnormalities exist in the αMHC-CryABR120G model (Table S1), and yet mortality was not yet excessive. At this age, percent fractional shortening (%FS), a measure of ventricular systolic function, averages ≈40%; values below this seen at later time points are indicative of terminal heart failure with high risk of sudden cardiac death (24). Echocardiographic determination of posterior wall thickness and necropsy evaluation of heart mass both revealed similar degrees of hypertrophic growth in αMHC-CryABR120G and αMHC-CryABR120G;beclin 1+/− mice (Table S1).
Hearts were stained with Masson's trichrome stain (Fig. S5A) or picrosirius red (data not shown) for interstitial fibrosis, a hallmark of pathological myocardial remodeling and an indirect marker of cell death. In WT and beclin 1+/− mice, we detected no signs of interstitial fibrosis at 9 months. In contrast, hearts from 9-month-old αMHC-CryABR120G animals manifested extensive intracellular aggregates, but only minor increases in fibrosis were detected. Similar to αMHC-CryABR120G animals and consistent with our in vitro findings, numerous intracellular protein aggregates were detected in hearts from αMHC-CryABR120G;beclin 1+/− mice. However, in contrast with the other three genotypes, hearts from αMHC-CryABR120G;beclin 1+/− mice manifested signs of severe pathological remodeling characterized by extensive (3-fold increased) deposition of interstitial fibrosis (Fig. S5A).
Additional qualitative differences were detected on ultrastructural analysis in CryABR120G hearts where autophagy was blunted by beclin 1 haploinsufficiency. Although both αMHC-CryABR120G and αMHC-CryABR120G;beclin 1+/− hearts contained protein aggregates, the aggregates were both more prevalent and larger in αMHC-CryABR120G;beclin 1+/− mice (Fig. S5B). Additional evidence for protein aggregate accumulation was obtained on immunoblot by evaluating for high molecular weight polyubiquitinated proteins in ventricular lysates. The abundance of these high molecular weight proteins was similar in WT and beclin 1+/− animals (Fig. S5C). Consistent with the induction of aggregates containing degraded protein, αMHC-CryABR120G hearts contained increased levels of high molecular weight polyubiquitinated proteins. And consistent with a role for autophagic pathways in the elimination of these proteins, their abundance was even greater in αMHC-CryABR120G;beclin 1+/− animals (Fig. S5C).
A Reduction in Autophagy Does Not Increase Apoptosis.
Significant interactions exist between autophagic and apoptotic signaling pathways (25, 26). For example, similar types of stress can induce either apoptosis or autophagy depending on cellular context, and when cells are induced to undergo apoptosis while the activation of caspases is prevented, the cells die via caspase-independent mechanisms (27). In a different transgenic model of cardiac-restricted expression of mouse CryABR120G, a model where animals die very early (≈5 months), increases in apoptosis have been reported (11). To test for apoptosis as a contributor to heart failure in our model of moderately overexpressed (≈6-fold) hCryABR120G, paraffin-fixed sections of hearts from 5- and 9-month-old animals were processed for TUNEL staining. Cardiac size and performance in αMHC-CryABR120G mice are both normal at 5 months, whereas signs of heart failure are emerging at 9 months (see below). Hearts from WT and beclin 1+/− mice manifested very low levels of TUNEL-positive cells at both time points (Fig. S6A). At 5 months, the prevalence of TUNEL-positive cells was only modestly increased in both αMHC-CryABR120G and αMHC-CryABR120G;beclin 1+/− hearts (Fig. S6B), and the increases were similar (P, not significant) in both genotypes. Similar findings were seen at 9 months of age, a time point where significant pathological remodeling is evident. Indeed, we detected a decrease in apoptotic signal in αMHC-CryABR120G;beclin 1+/− heart at 9 months as compared with αMHC-CryABR120G (Fig. S6B). Additional evidence for a lack of up-regulated apoptotic activity was obtained on Western blot, where no changes in caspase-3 cleavage (Fig. S6C) or Bcl-2 phosphorylation (data not shown) were detected across all four genotypes.
Attenuation of Autophagy Accelerates Heart Failure Progression in αMHC-CryABR120G Mice.
Findings on necropsy at 9 months were consistent with the notion that down-regulated autophagy promotes accumulation of polyubiquitinated proteins, aggresome formation, and pathological remodeling. To determine whether these increases in autophagic activity are adaptive or maladaptive, αMHC-CryABR120G mice were crossed with beclin 1 haploinsufficient mice. We took advantage of the diminished autophagic response in these mice to titrate the cellular response to CryABR120G, and we evaluated cardiac structure and function by serial echocardiography, followed by necropsy analysis.
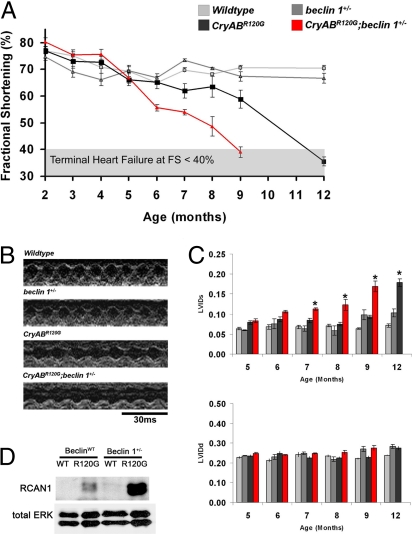
WT and beclin 1+/− mice manifested no signs of compromise of cardiac performance over the 12-month period of study (Fig. 3A). %FS was constant throughout at 70–80%, consistent with a previous report (23). As expected, αMHC-CryABR120G mice developed a progressive decline in %FS that reached statistical significance compared with WT at 9 months of age (Fig. 3A). By 12 months of age, these mice manifested overt heart failure (%FS <40%).
Fig. 3.
Accelerated heart failure and early mortality in αMHC-CryABR120G;beclin 1+/− mice. (A) Cardiac function was monitored by serial echocardiography in nonsedated animals. αMHC-CryABR120G animals developed late-onset heart failure with the first signs of functional decline appearing at 9 months and development of heart failure by 12 months of age. In contrast, αMHC-CryABR120G;beclin 1+/− mice manifested an accelerated disease course, with early signs of functional decline apparent at 6 months and terminal heart failure at 9 months. (B) Representative M-mode echocardiograms recorded in 9-month-old animals. (C) Declines in systolic performance are caused primarily by progressive increases in left ventricular internal dimension at end systole (LVIDs), with little change seen in end-diastolic diameter (LVIDd). By 12 months of age, the CryABR120G;Beclin 1+/− animals had experienced 100% mortality. (D) Accumulation of RCAN1, indicative of calcineurin activation, as observed in αMHC-CryABR120G hearts at 9 months of age. RCAN1 levels were substantially higher in αMHC-CryABR120G;beclin 1+/ − hearts.
To test the role of autophagic activity in αMHC-CryABR120G mice, we studied αMHC-CryABR120G;beclin 1+/− mice. Intriguingly, declines in cardiac performance were greatly accelerated in these mice (Fig. 3A). Indeed, these animals developed significant declines in cardiac function by 6 months with severe heart failure by 9 months of age, a rate of disease progression that was statistically significantly accelerated relative to αMHC-CryABR120G (P = 0.007 by log-rank statistic) (Fig. 3 A and B). Mortality was significantly increased (P < 0.05) in αMHC-CryABR120G;beclin 1+/− mice, as well: CryABR120G;beclin 1+/− mice manifested 25% mortality (two of eight animals) by 9 months of age, whereas no mortality was seen in WT (zero of seven), beclin 1+/− (zero of nine), or α-MHC-CryABR120G (zero of eight) animals at that time point. Echocardiographic determination of posterior wall thickness and necropsy evaluation of heart mass both revealed similar degrees of hypertrophic growth in αMHC-CryABR120G and αMHC-CryABR120G;beclin 1+/− mice (Fig. 3B and Table S1). Interestingly, in both α-MHC-CryABR120G and CryABR120G;beclin 1+/− mice, heart failure was systolic in nature, with significant increases developing in end-systolic diameter but with no significant changes in end-diastolic dimensions (Fig. 3C).
Finally, to probe the role of intracellular signaling mechanisms, we measured steady-state levels of RCAN1 (Regulator of Calcineurin), a downstream target of the pathological signaling molecule calcineurin. Accumulation of RCAN1, indicative of calcineurin activation, was observed in αMHC-CryABR120G hearts (Fig. 3D), consistent with the cardiomyopathic phenotype. RCAN1 levels were substantially higher still in αMHC-CryABR120G;beclin 1+/− hearts, consistent with additional activation of calcineurin in the setting of blunted autophagy.
Discussion
Misfolding and aggregation of mutant or damaged proteins underlies the pathogenesis of multiple neurodegenerative diseases, skeletal myopathies, and heart failure. In these disorders, cellular mechanisms responsible for recognizing and disposing of aggregating proteins are overwhelmed, leading to accumulation of soluble and insoluble isoforms of toxic protein. The present study implicates autophagic activity in DRCM and demonstrates that autophagic activity in this disorder has a protective role by serving to clear toxic aggregates.
Proteinopathy.
Abnormal protein aggregation and accumulation of ubiquitinated proteins in the cytosol have been detected in human hearts with idiopathic or ischemic cardiomyopathies (28, 29). Recent work from our group has demonstrated accumulation of protein aggregates and aggresomes in a very common form of acquired heart disease, namely load-induced heart failure (16). In the case of the desmin-related myopathies, severe cardiomyopathy and early death are thought to be caused, at least in part, by disruption of the desmin architecture within the cell, leading to contractile dysfunction and cell damage. In other cases of proteinopathy, the mechanism whereby a mutated protein is toxic to the cell is less clear. Although some mechanisms are likely disease-specific, related to loss of function of the mutated or misfolded protein, there is general agreement that early, still-soluble aggregates are potentially toxic (30). Indeed, evidence suggests that it is the soluble preamyloid aggregates that are the most toxic in neurons (31). Also, in many instances, the mutations responsible for proteinopathies confer a toxic gain of function to the relevant protein.
Molecular chaperones, such as HSP70, monitor protein quality and either facilitate refolding of the misfolded protein or promote degradation via the proteasome. Excess misfolded proteins that escape this quality-control mechanism begin to aggregate. The presence of protein aggregates, in turn, overwhelms and inhibits proteasome activity, potentially disrupting other important proteasome functions (32). Autophagy can relieve proteasome inhibition by removing aggregates that have escaped proteasome clearance. In parallel, unprocessed protein aggregates are directed toward sequestration in the perinuclear aggresome.
It is likely that the low level of apoptotic activity that we observe contributes in part to the cardiac pathology seen in αMHC-CryABR120G mice, but it is unlikely to be the sole cause of cardiac failure. Further, we did not detect increased levels of myocyte apoptosis in αMHC-CryABR120G;beclin 1+/− mice as compared with αMHC-CryABR120G animals, despite the marked acceleration of heart failure progression in the former. This observation contrasts with prior studies in another model of DRCM where disease progression is markedly accelerated relative to our model (20). It suggests that differences exist between cellular responses stemming from increases in the expression of aggregating protein and the processing of those aggregates via autophagic clearance. It is interesting to note that our line of αMHC-CryABR120G mice develop systolic failure with no evidence of chamber dilation, whereas the αMHC-CryABR120G mice developed by Robbins and colleagues (20), which manifest high levels of apoptosis, develop systolic dysfunction with ventricular dilation. This apparent discrepancy raises the intriguing possibility that these different transgenic lines may highlight distinct, stage-specific features of DRCM.
Autophagy and Myofibrillar Myopathy.
In the context of nutrient deprivation, autophagic activity is adaptive in that degradation of cytosolic components releases substrates for intermediary metabolism. Autophagy is also a mechanism for eliminating damaged proteins and organelles that might otherwise be toxic or trigger apoptotic death. Some evidence suggests that autophagy can efficiently target species that are not in aggregates large enough to be seen on light microscopy (33). The observation of inclusion formation in the neurons of mice with neuron-restricted inactivation of autophagy genes is consistent with the ability of autophagy to clear soluble and oligomeric aggregate precursors (34, 35). In general, it seems that the capacity to aggregate, rather than the protein aggregates themselves, is correlated with toxicity. In both brain and heart, however, little is known regarding whether these intracellular inclusions are toxic themselves, or whether they represent a compensatory mechanism that sequesters harmful, soluble proteins within the cytoplasm. However, a model has been proposed in which increased autophagic activity in neurodegenerative disorders does not directly clear aggregates themselves but clears aggregate precursors, shifting the equilibrium away from aggregate formation (30).
Diseases caused by polyglutamine-expansion mutations, such as Huntington's disease, as well as Parkinson's disease and other late-onset neurodegenerative diseases, strongly depend on macroautophagy pathways for clearance of intracellular protein aggregates (30). Each disease is caused by the expression of a dominant-negative, aggregate-prone protein in terminally differentiated postmitotic cells. In that context, mechanisms capable of removing damaged proteins are particularly important, because of limited capacity for replacement of defective cells. Here, we describe an increase in the abundance of autophagic vesicles in cardiac myocytes in response to expression of a mutant protein causing DRCM. Whereas autophagic activity has been shown to be protective in cell and fly models of aggregate-prone diseases (18), our study demonstrates such in a mammalian model of myopathy.
Autophagy in Heart Disease.
Recent studies have uncovered a role for autophagic activity in ischemia/reperfusion injury and heart failure. Depending on the stressor and disease context, these studies point to either adaptive or maladaptive roles of this protein clearance pathway (14). Here, we report that cardiomyocyte autophagy triggered by abnormal aggregation of intracellular proteins is beneficial, which is consistent with observations made in neurodegenerative diseases. Indeed, a growing body of evidence implicates autophagy as a protective response in genetic diseases associated with cytoplasmic aggregation-prone proteins (36), which we extend here to heart disease triggered by defective chaperone function. Indeed, it has been suggested that autophagy may have two distinct beneficial effects in protein conformation disease. First, this pathway functions to clear the primary toxin causing these diseases. Second, enhanced autophagy attenuates apoptotic responses to various insults, rendering the cell resistant to programmed cell death. Importantly, recent studies demonstrating that pharmacological up-regulation of autophagy is protective in a wide variety of disease models associated with intracellular protein aggregation raise the exciting prospect of autophagic activation as a novel therapeutic strategy.
There is some evidence to suggest that activation of autophagy involving an increase in Beclin 1 expression is indicative of damaging autophagic activity (23, 37). Significantly, we see no elevation in Beclin 1 levels in the αMHC-CryABR120G mice (data not shown), consistent with autophagy being beneficial in this setting.
It is important to recognize that the increases in autophagic vesicle abundance could be caused either by an increase in vesicle formation or a block in the clearance of vesicles. Our evidence suggests that αMHC-CryABR120G protein stimulates vesicle formation (rather than inhibiting their clearance), because decreasing autophagic capacity, either pharmacologically or genetically, increased the abundance of aggregated proteins. This fact, then, demonstrates that the protein aggregates caused by CryABR120G can indeed be removed through autophagic pathways. That said, it remains possible that CryABR120G-associated aggregates eventually overwhelm and inhibit autophagy the same way they have been shown to inhibit proteasomal function. In either scenario, our finding that DRCM is more severe and progresses more rapidly in animals with reduced autophagic capacity suggests that increasing autophagic capacity may be beneficial in patients with this and related disorders.
Perspective.
Desmin-related cardiomyopathy is a severe and progressive disease for which there are limited therapeutic options. In this article, we identify autophagy as a robust cellular response to CryABR120G-associated proteinopathy and further demonstrate that this response plays a significant role in attenuating disease progression. These findings are clinically relevant for a number of reasons: (i) they indicate that in DRCM, autophagy is a pathway suitable for consideration as a therapeutic intervention, (ii) they suggest that interindividual variations in autophagic capacity or responsiveness may account for the heterogeneous presentation of disease, and (iii) they may serve as a paradigm for myofibrillar myopathy of diverse molecular etiology. Finally, given that drugs that alter the process of autophagy are already in clinical use, advances in this field are all the more urgent.
Materials and Methods
Primary Culture of Neonatal Rat Ventricular Myocytes.
Cardiomyocytes were isolated from the ventricles of 1- to 2-day-old Sprague–Dawley rat pups as described (23).
CryAB Transgenic Lines.
Transgenic mice expressing mutant human CryAB (CryABR120G) were engineered as described (9). Only CryABR120GHigh animals were used.
Additional materials and methods are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Christopher Gilpin, Tom C. Januszewski, and Laurie M. Meuller in the Imaging Facility at UT Southwestern for support and David Leonard for statistical advice. This work was supported by National Institutes of Health Grants HL-075173, HL-080144, HL-006296, HL-063834, and HL-072016 and American Heart Association Grants 0640084N and 0655202Y.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706802105/DCSupplemental.
References
- 1.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 2.Goebel HH, Warlo IA. Surplus protein myopathies. Neuromuscul Disord. 2001;11:3–6. doi: 10.1016/s0960-8966(00)00165-6. [DOI] [PubMed] [Google Scholar]
- 3.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 4.Omary MB, Coulombe PA, McLean WH. Intermediate filament proteins and their associated diseases. N Engl J Med. 2004;351:2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- 5.Dalakas MC, et al. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. N Engl J Med. 2000;342:770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- 6.Perng MD, Wen SF, van den IP, Prescott AR, Quinlan RA. Desmin aggregate formation by R120G αB-crystallin is caused by altered filament interactions and is dependent upon network status in cells. Mol Biol Cell. 2004;15:2335–2346. doi: 10.1091/mbc.E03-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vicart P, et al. A missense mutation in the αB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, et al. Mouse model of desmin-related cardiomyopathy. Circulation. 2001;103:2402–2407. doi: 10.1161/01.cir.103.19.2402. [DOI] [PubMed] [Google Scholar]
- 9.Rajasekaran NS, et al. Human αB-crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell. 2007;130:427–439. doi: 10.1016/j.cell.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanbe A, et al. Desmin-related cardiomyopathy in transgenic mice: A cardiac amyloidosis. Proc Natl Acad Sci USA. 2004;101:10132–10136. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maloyan A, et al. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in αB-crystallin desmin-related cardiomyopathy. Circulation. 2005;112:3451–3461. doi: 10.1161/CIRCULATIONAHA.105.572552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res. 2005;97:1018–1026. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 13.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothermel BA, Hill JA. Myocyte autophagy in heart disease: Friend or foe? Autophagy. 2007;3:632–634. doi: 10.4161/auto.4913. [DOI] [PubMed] [Google Scholar]
- 15.Williams A, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: Therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 16.Tannous P, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 19.Petiot A, Ogier-Denis E, Blommaart EFC, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, et al. Expression of R120G-αB-crystallin causes aberrant desmin and αB-crystallin aggregation and cardiomyopathy in mice. Circ Res. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 21.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothermel BA, et al. Differential activation of stress-response signaling in load-induced cardiac hypertrophy and failure. Physiol Genomics. 2005;23:18–27. doi: 10.1152/physiolgenomics.00061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit beclin-1-dependent autophagy. Cell. 2005;122:927–933. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 27.Lang-Rollin ICJ, Rideout HJ, Noticewala M, Stefanis L. Mechanisms of caspase-independent neuronal death: Energy depletion and free radical generation. J Neurosci. 2003;23:11015–11025. doi: 10.1523/JNEUROSCI.23-35-11015.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heling A, et al. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circ Res. 2000;86:846–853. doi: 10.1161/01.res.86.8.846. [DOI] [PubMed] [Google Scholar]
- 29.Kostin S, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 30.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 31.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol. 2006;40:451–454. doi: 10.1016/j.yjmcc.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. α-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 34.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 35.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 36.Rubinsztein DC, et al. Autophagy and its possible roles in nervous system diseases, damage, and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- 37.Matsui Y, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.