Abstract
Rhizobia can infect roots of host legume plants and induce new organs called nodules, in which they fix atmospheric nitrogen. Infection generally starts with root hair curling, then proceeds inside newly formed, intracellular tubular structures called infection threads. A successful symbiotic interaction relies on infection threads advancing rapidly at their tips by polar growth through successive cell layers of the root toward developing nodule primordia. To identify a plant component that controls this tip growth process, we characterized a symbiotic mutant of Medicago truncatula, called rpg for rhizobium-directed polar growth. In this mutant, nitrogen-fixing nodules were rarely formed due to abnormally thick and slowly progressing infection threads. Root hair curling was also abnormal, indicating that the RPG gene fulfils an essential function in the process whereby rhizobia manage to dominate the process of induced tip growth for root hair infection. Map-based cloning of RPG revealed a member of a previously unknown plant-specific gene family encoding putative long coiled-coil proteins we have called RRPs (RPG-related proteins) and characterized by an “RRP domain” specific to this family. RPG expression was strongly associated with rhizobial infection, and the RPG protein showed a nuclear localization, indicating that this symbiotic gene constitutes an important component of symbiotic signaling.
Keywords: genetics, symbiosis, coiled-coil
In the symbiotic interaction between legumes and soil bacteria called rhizobia, nitrogen-fixing nodules are formed that allow plant growth to be independent of added combined nitrogen, and the plant provides rhizobia with a carbon source derived from photosynthesis. During initial signal exchange in the rhizosphere, rhizobia respond to plant flavonoids by producing lipochito-oligosaccharidic molecules called Nod factors (NFs). Host-specific recognition of NFs triggers a controlled infection leading to rhizobial internalization, and the induction of a new plant organ, the nodule, in which nitrogen fixation occurs (1).
Rhizobial infection of host legumes is generally via root hairs (RHs) that undergo marked curling. Compared with normal RH tip growth in which vesicles, containing cell wall and membrane material, travel in an actin- and microtubule-dependent fashion to the RH tip, where they fuse with the cell membrane (2), this rhizobium-induced growth reorientation involves alterations to the plant cytoskeleton and the redirection of vesicle traffic away from the RH tip to a new site (3, 4). Inside a closed chamber formed by root hair curling (RHC), the plant cell wall is locally degraded and the plasma membrane becomes invaginated. Rhizobia enter a newly formed, plant-derived structure, the infection thread (IT), that undergoes inward tip growth within the RH. Underlying outer cortical cells change into highly polarized pre-IT cells, which guide IT passage to the nodule primordium formed, in M. truncatula, in the inner root cortex (3). Here, bacteria are released into plant cells and differentiate into nitrogen-fixing bacteroids.
Whereas the establishment of nodulation probably results from the concerted interplay of hundreds of plant genes, only a subset is likely to be uniquely implicated in the symbiotic process. In M. truncatula, NFP and LYK3 encode putative NF receptors, whereas DMI1, DMI2, DMI3, NSP1, and NSP2 control early steps of NF signal transduction (5), and DMI1 and DMI2 are also necessary for the formation of a high-affinity NF binding site (6). Activation of the NFP–DMI–NSP signaling pathway precedes infection, whereas LYK3 is more specifically involved in infection and controls polarization of epidermal and cortical cells, and NIN functions downstream of NF signaling to control nodule formation (7–9). In addition to these genes that control RHC and IT formation, others intervene later. M. truncatula bit1, lin, nip, api, latd, and itd mutants, crinkle mutants of Lotus japonicus, and pea sym7, sym34, sym37, and sym38 mutants all show arrested infection (reviewed in ref. 9; ref. 30). Nonsymbiotic phenotypes are described for latd, nip, and crinkle mutants, supporting the idea that IT formation relies largely on endogenous cellular functions (4).
To identify a plant component with a specific function to control and/or steer the polar growth process of IT formation, we characterized the rpg mutant of M. truncatula. RHC and IT formation were aberrant in this mutant, and map-based cloning showed that the RPG (Rhizobium-directed polar growth) gene encodes a putative long coiled-coil protein.
Results
The Medicago truncatula rpg Mutant Shows Abnormal Infection Threads and Root Hair Curls.
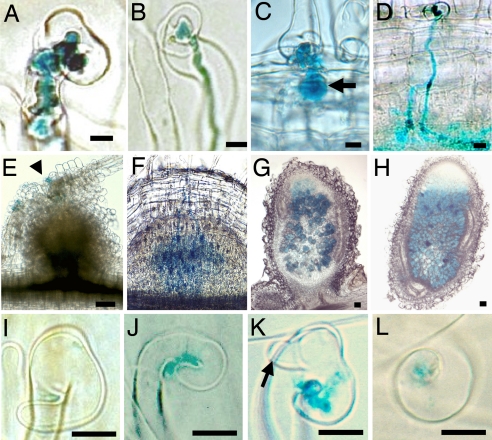
The rpg mutant was found in a screen for nodulation-deficient ethyl methanesulfonate-induced mutants of M. truncatula (10). rpg roots inoculated with Sinorhizobium meliloti always showed unusually thick ITs with bulbous protrusions, quite distinct from the thin and straight wild-type (WT) structures, and 3–4 times wider (Fig. 1 A and B). Mutant ITs progressed slowly and mostly remained within RH cells, but sometimes progressed into the cortex as balloon-shaped structures, whereas WT ITs progressed straight down through the root cortex (Fig. 1 C and D). ITs were at least as numerous on mutant roots compared with WT. Twenty-one days post inoculation (dpi), rpg roots showed uninfected nodule-like structures, whereas on WT roots, infected nodule primordia were formed within 4–5 dpi (Fig. 1 E and F) and pink (nitrogen-fixing), elongated nodules formed 7 dpi. Rare pink nodules formed on rpg roots (on average a single nodule on 1 of 20 plants, 21 dpi), with a normal structure, but more patchy infection compared with WT nodules (Fig. 1 G and H).
Fig. 1.
ITs, nodules, and RHC in response to S. meliloti expressing lacZ, 5 (A–D and F), 21 (E, G, and H), and 3 (I–L) dpi. (A and B) RH infection in the rpg mutant (A) and in WT (B). (C and D) Cortical infection in the rpg mutant, showing an enlarged sac-like structure (arrow) (C) and in WT (D). (E) Nodule-like structure on the rpg mutant showing infection (blue coloration) limited to the epidermis (arrowhead). (F) An infected nodule primordium in WT. (G and H) Nodule sections in the rpg mutant (G) and WT (H). (I–L) RHC [loose and incomplete with RH outgrowth (arrow)] in the rpg mutant (I–K) and (tight) in WT (L). [Scale bars: 10 μm (A–D and I–L) and 100 μm (E–H).]
The rpg mutant showed a delay of ≈24 h in the formation of bacterial chambers in RH curls compared with WT roots, whereas there was no subsequent delay in the initiation of ITs. Rhizobial entrapment by RHC in rpg roots was the result of loose curling, which was frequently incomplete, with the RH growing straight again, and often associated with one or more new poles of growth (Fig. 1 I–K). Tight and complete RHC around bacteria, which is typical of WT plants (Fig. 1L), was never seen in the rpg mutant, whereas RHC in WT plants was not accompanied by new poles of RH growth as in the rpg mutant.
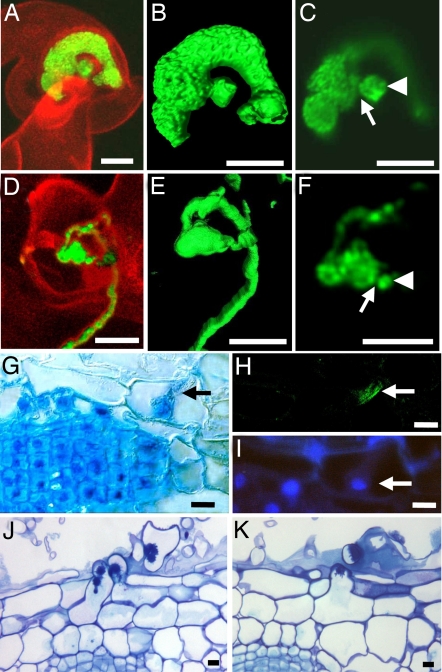
Compared with WT, rpg ITs contained much larger numbers of bacteria, which were densely packed, indicating active bacterial multiplication (Fig. 2A, B, D, and E). Confocal sections indicated that each rpg IT initiated from a narrow entry site, comparable to the WT situation (Fig. 2 C and F), although more detailed studies are necessary to confirm this. Normal local degradation of the plant cell wall at the onset of IT formation would be consistent with the absence of any delay in the formation of rpg ITs after RHC.
Fig. 2.
RH infection and cortical cell responses. (A–F) Confocal images of S. meliloti expressing GFP. ITs in the rpg mutant (A–C) and WT (D–F) are shown in 3D representations (B and E), and confocal sections (C and F) show bacterial chambers (arrowheads) and probable restricted entry sites (arrows). (G–K) Root sections. (G–I) Cytoplasmic bridge (arrow in G) in an outer cortical cell overlying cell divisions in the rpg mutant, 4 dpi. (H) Immunolocalization of α-tubulin showing parallel microtubules (arrow). (I) Nucleus (arrow) visualized by using DAPI. (J and K) Infected cortical RHs 7 dpi, with toluidine blue coloration, in rpg inoculated with S. meliloti WT (J) and in WT inoculated with S. meliloti exoA (K). (Scale bars: 10 μm.)
Nod Factor Responsiveness and Other Phenotypes Are Normal in the rpg Mutant.
Given that NFs are required for infection, we tested whether the rpg infection defect could be overcome with a strain of S. meliloti that constitutively overproduces NFs (GMI6390) (10). Normal IT formation was not restored. NF responsiveness was further tested by using serial 10-fold dilutions of purified S. meliloti NF (10−8 to 10−11 M). RH branching and MtENOD11 expression, detected by using an rpg mutant line carrying a pMtENOD11::GUS fusion, were indistinguishable from WT phenotypes (data not shown).
In rpg plants, the morphology of plant structures exhibiting polar growth (RHs, pollen tubes, and trichomes) was no different from WT, and a normal mycorrhization phenotype was observed (data not shown), indicating that rpg plants are defective specifically for the Rhizobium–legume symbiosis.
Outer Cortical Cells Become Polarized in the rpg Mutant.
Because the majority of rpg ITs remained within RH cells, we asked whether underlying outer cortical cells prepare normally for infection by looking for pre-ITs (PITs). Four days after spot inoculation, outer cortical cells located over division foci (seen on 75% of inoculations), displayed central cytoplasmic bridges joining the inner and outer periclinal cell walls (Fig. 2G). Immunolocalization of α-tubulin coupled to DAPI coloration indicated that these structures contained microtubules organized in parallel strands, and nuclei were positioned against the inner periclinal wall (Fig. 2 H and I). These features are typical of PITs (3).
Bulges were often seen where the cytoplasmic bridge was connected to the outer periclinal wall. These bulges gave rise to cortical RHs that emerged between two epidermal cells and often subsequently became infected with thick ITs characteristic of rpg epidermal RH cells (Fig. 2J). WT roots inoculated with the same WT rhizobial strain did not form cortical RHs, whereas infected and uninfected cortical RHs were observed on WT roots after inoculation with an S. meliloti exoA mutant that induces abortive ITs (11) (Fig. 2K).
Map-Based Cloning of the RPG Gene.
Nodulation phenotypes of F1 (6 Nod+/0 Nod−) and F2 (73 Nod+/25 Nod−) plants indicated that the rpg Nod− character is controlled by a single recessive locus, named RPG, for Rhizobium-directed polar growth. This locus was mapped to chromosome 1 between markers DSI and MTIC370 by using 300 Nod− F2 mapping individuals [supporting information (SI) Fig. S1].
Phenotyping and genotyping of 3,000 F2 mapping individuals placed the RPG locus 12 and 23 recombinants distant from MTIC370 and DSI, respectively (Fig. S1A). From these starting points a contig of 14 BACs (≈1,300 kb) was built. Within this, the RPG region was restricted to ≈50 kb within the BAC mth2–53B15, which was sequenced (Fig. S1B). Among the five predicted proteins (Fig. S1C), two are represented as ESTs from nodules: a globin and a protein with moderate identity with myosin heavy chains. Globins function in nitrogen fixation, so we sequenced the myosin-like gene in the rpg mutant and revealed a mutation leading to a premature stop codon in the third exon. This gene and its putative promoter region were cloned and transformed into mutant roots. Such roots nodulated (46 of 60 plants with three nodules per plant on average), and showed thin straight ITs (Fig. S2), indicating that the candidate gene corresponds to RPG.
RPG Is a Putative Coiled-Coil Protein That Shows a Nuclear Localization in Nicotiana benthamiana Leaves.
RPG is a 5.8-kb gene composed of seven predicted exons (Fig. S3A). cDNA sequencing confirmed the 3,768-bp ORF, encoding a putative protein of 1,255 aa (Fig. S3B). We predicted potential phosphorylation sites, a potential nuclear localization signal (NLS), and three different regions in the protein sequence (Fig. S3B). First, a 140-aa region of short α-helices and β-sheets and low solvent accessibility is predicted. The second region (170 aa long), containing the rpg mutation (Fig. S3B), is characterized by hydrophilic and charged amino acid residues, particularly serines. This region is predicted to be “disordered” with no secondary structure. The third part of RPG is predicted to be α-helical, and it shows sequence conservation (≈20% identity, 40% similarity) to coiled-coil proteins such as myosin heavy chains and vesicular tethering proteins. The prediction of coiled-coil structures is supported by the periodic pattern of leucine, isoleucine, or other hydrophobic residues at 7-aa intervals (Fig. S3B), a commonly observed heptad repeat structure in coiled-coil proteins. Proline residues, which usually prevent α-helix formation, were found in predicted hinge sequences, supporting our delimitation of four coiled-coil domains.
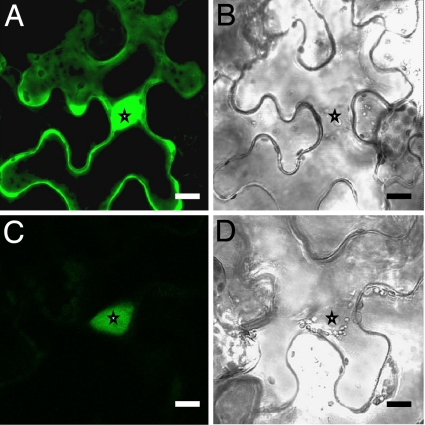
The subcellular localization of RPG was studied by using a translational GFP fusion expressed transiently in Nicotiana benthamiana leaves. This showed a clear nuclear signal and significantly reduced signal within the cytoplasm, distinct from the control construct expressing GFP alone, for which both nuclear and cytoplasmic regions showed strong signals (Fig. 3).
Fig. 3.
Subcellular localization of RPG in Nicotiana benthamiana leaf epidermal cells. (A and B) The p35S::GFP control showing nuclear and cytoplasmic GFP. (C and D) p35S::GFP::RPG showing nuclear localization of GFP. Stars indicate nuclei. (Scale bars: 20 μm.)
RPG Has a Largely Symbiosis-Specific Expression Pattern, Strongly Associated with the Infection Process.
Quantitative RT-PCR showed a 60- ± 20-fold induction of RPG expression in nodules compared with uninoculated roots, and very low expression in flowers, leaves, and stems (Table 1). At 1 and 3 dpi with S. meliloti, RPG expression was induced 10- and 30-fold, respectively (Table 2). In two early infection mutants, B56 and D8, affected in the HCL and LIN genes, respectively (ref. 7, data not shown), RPG expression was induced only to 13% of the level found in WT roots at 3 dpi (Table 2).
Table 1.
Expression analysis of RPG and M. truncatula RRP genes by quantitative RT-PCR
| Gene | Expression levels |
||||
|---|---|---|---|---|---|
| Root | Nodule | Leaf | Stem | Flower | |
| RPG | 0.02 ± 0.00 | 1 ± 0.35* | 0.05 ± 0.00 | 0.04 ± 0.01 | 0.01 ± 0.00 |
| RRP1 | 1* | 0.06 ± 0.01 | 0.84 ± 0.03* | 0.70 ± 0.29* | 0.06 ± 0.01 |
| RRP2 | 0.61 ± 0.13* | 0.04 ± 0.01 | 1 ± 0.32* | 0.53 ± 0.04* | 0.13 ± 0.01 |
| RRP3 | 0.36 ± 0.00* | 0.40 ± 0.01* | 1 ± 0.15* | 0.55 ± 0.08* | 0.40 ± 0.02* |
| RRP4 | 1 ± 0.12* | 0.51 ± 0.20* | 0.88 ± 0.23* | 0.87 ± 0.28* | 0.82 ± 0.06* |
| RRP5 | 0.68 ± 0.19* | 0.80 ± 0.05* | 0.62 ± 0.03* | 0.81 ± 0.25* | 1 ± 0.17* |
Values are ratios relative to the sample having the highest expression level, ± SEMs.
*Preferential expression pattern.
Table 2.
Expression analysis of RPG by quantitative RT-PCR at early time points after rhizobial inoculation of wild-type (WT) and M. truncatula hcl and lin mutants
| Plant | Expression levels |
||
|---|---|---|---|
| 0 dpi | 1 dpi | 3 dpi | |
| WT | 0.03 ± 0.00 | 0.34 ± 0.14 | 1 ± 0.18 |
| hcl | 0.03 ± 0.01 | ND | 0.14 ± 0.01 |
| lin | 0.01 ± 0.00 | ND | 0.13 ± 0.01 |
Values are ratios relative to the sample having the highest expression level, ±SEMs.
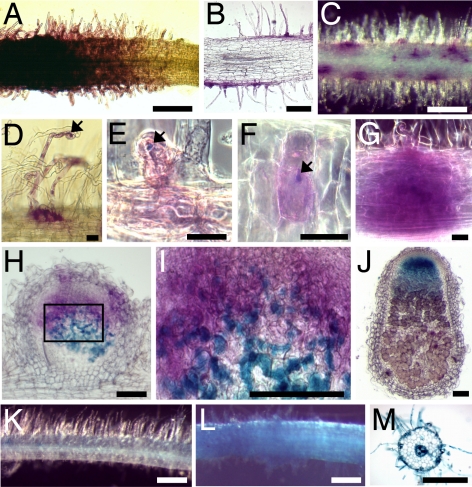
The spatiotemporal expression pattern of RPG was analyzed by using a promoter-GUS fusion containing the same promoter region used for functional complementation. Noninoculated roots showed weak GUS activity in RH cells of lateral roots and in the vascular system (data not shown). At 2–3 dpi, strong GUS activity was detected in growing and recently matured RHs (Fig. 4 A and B). From 3 dpi, when ITs were first visible, GUS activity progressively localized to infected RH cells (Fig. 4 C–F). GUS staining was associated with all cell layers of developing nodule primordia at 5 dpi (Fig. 4G), but localized to the preinfection and infection zones in young and mature nodules (Fig. 4 H–J). When DAPI coloration was used to distinguish meristematic nodule cells (characteristically small and square with a large, round, and central nucleus), GUS staining was not associated with such cells, but was strongest in directly adjacent cell layers (data not shown). Expression was lower in the inter zone II/III and not detectable in the central nitrogen-fixing zone of nodules (Fig. 4J). An S. meliloti NodA strain unable to produce NFs (GMI6702) (12) induced no increase in GUS activity (data not shown), whereas pure S. meliloti NFs at 10−8 M induced strong GUS activity in growing and recently matured RHs, and also in the vascular system of roots (Fig. 4 K–M).
Fig. 4.
Spatiotemporal analysis of RPG gene expression by using an RPG promoter-GUS fusion. Shown is GUS coloration of whole roots (A, C, D, K, and L) or sections (B, E–J, and M), after inoculation with S. meliloti lacZ (A–J) or after NF/control treatment (K–M). GUS coloration is magenta (A–I) or blue (J–M), and bacteria are colored blue (E–I) or magenta (J). (A and B) RH cells at a lateral root apex, 2 dpi. (C–F) Deformed and infected (arrows) RHs, 3 dpi. Sectioning shows GUS expression limited to the infected cell (E and F). (G) A developing nodule primoridium, 5 dpi. (H and I) Young nodule section 8 dpi (I is a zoom of H). (J) Section of a mature nodule 18 dpi, showing GUS coloration in the infection zone. (K–M) Without (K) or with (L and M) 10−8 M NF treatment. Sectioning shows that NF induction is localized in RH cells and the central vascular system (M). [Scale bars: 300 μm (A, C, and J–M), 100 μm (B, H, and I), 10 μm (D–G).]
RPG Is a Member of a Small, Uncharacterized Plant-Specific Gene Family.
We identified four homologous, but uncharacterized, genes in both A. thaliana and rice. In M. truncatula, EST sequences for five homologous genes were identified, and called MtRRP1-MtRRP5 (RPG-related proteins) (Table S1). MtRRP1 and MtRRP2 were expressed mostly in leaves, stems, and roots, whereas MtRRP3, MtRRP4, and MtRRP5 were ubiquitously expressed (Table 1). These profiles correspond well with in silico expression data, which also show that MtRRP3 and MtRRP5 are expressed during pathogen attack and mycorrhization (Table S1). The gene most similar to RPG, MtRRP1, was fully sequenced, and homologous genes were identified in other plant species, but not in animals, fungi, or bacteria. RPG, RRP1, and family members from A. thaliana and rice have a conserved domain structure (Fig. 5), with a highly conserved first RRP domain (Fig. S4). The second region, always serine-rich and predicted to be disordered, displays low conservation of sequence except that residues are predominantly hydrophilic and charged. The third region is always an α-helix, with variable numbers of predicted coiled-coil motifs. Sequence conservation is generally good at both ends of this third region. Certain proteins have unique predicted features; a putative, N-terminal NLS in RPG, a putative inner NLS in Os10g36060, a signal peptide and a transmembrane domain in Os10g21940, and an inner sequence repetition within the α-helical domain of Os03g01710. The generally well conserved C-terminal region is not well conserved in RRP1 and is absent from RPG.
Fig. 5.
Predicted secondary structure of RPG homologs. Proteins are drawn to scale and aa lengths are given. Sequence conservation is given as percentage identity values. RRP, RRP domains (gray); D, disordered regions (striped); α-helix, coiled-coil regions (black). Because of low sequence conservation, At1g22060 and Os10g21940 (a) and RRP1 (b) were excluded from the analysis of sequence conservation. 1NLS sequence at the start of RPG. 2Signal peptide and transmembrane domain at the start of Os10g21940. 3NLS sequence within the RRP domain of Os10g36060.
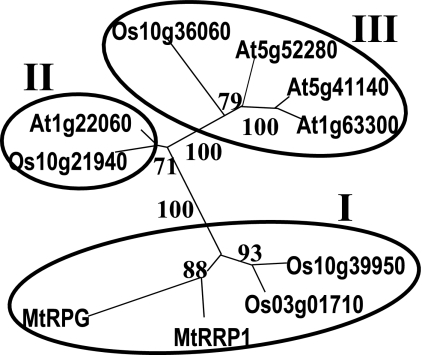
Phylogenetic analysis using either the RRP domain or whole proteins gave the same phylogenetic tree containing three clades (Fig. 6). Analysis of intron positions revealed a high conservation for genes whose proteins grouped in the same clade (Table S2). However, RPG differs from other proteins in clade I by modification of the exonic composition at the 5′ and 3′ extremities, in accordance with distinctive features at each end of the RPG protein (Table S2).
Fig. 6.
Unrooted phylogenetic tree of RPG homologs based on the RRP domain. One thousand bootstrap replicates were performed and percentage bootstrap supports are given. Family clades are circled.
Discussion
RPG Is a Symbiotic Gene of M. truncatula That Controls Rhizobial Infection.
The rpg rhizobial infection phenotype was characterized by delayed and abnormal RHC and ITs. Despite this, ITs formed with a normal, if not increased, frequency, in agreement with reports that rhizobial infection does not necessarily have to initiate from tight RHCs (12, 13). In rare cases rpg ITs grew sufficiently to give rise to apparently functional nodules, indicating that the mutation affects progression of ITs, but neither completely blocks their development nor affects bacterial release into nodule cells. The rpg mutation specifically affects the rhizobial infection process, because no apparent alterations in mycorrhization and nonsymbiotic phenotypes were observed, and the rpg mutation is likely to be null (the premature stop codon should be detected by the nonsense-mediated mRNA decay mechanism).
The rpg infection defect was associated with the formation of infectable cortical RHs that derived from PIT cells. Normally, PITs accommodate and guide growing ITs through the cortex, and cell polarization for PIT formation is mediated by NFs (14), indicating that this NF signaling pathway is functional in the rpg mutant. The great majority of rpg ITs do not reach the cortex. Therefore, in the absence of cortical infection, NF signaling for the induction of polarity changes might continue and result in the induction of cortical RHs. In favor of this hypothesis, infectable cortical RHs also formed when WT plants were inoculated with an infection-defective, but not with a WT, S. meliloti strain (this work), in Vicia sativa in response to EPS or cellulose-deficient mutants of Rhizobium leguminosarum (15), and on the RH-less L. japonicus rhl1 mutant when inoculated with Mesorhizobium loti (16).
Compared with nfp, dmi, and nsp NF signaling mutants of M. truncatula (5), the rpg mutant was not altered for NF-induced RH branching and MtENOD11 expression. The presence of RHC, ITs, and PITs distinguished rpg from hcl mutants of M. truncatula (7). rpg also differs from M. truncatula api, nip, and latd mutants, which have abnormal RH or root development, from M. truncatula lin and bit1 mutants, and from itd mutants of L. japonicus that show reduced and abortive infection (9, 30). This, together with genetic analysis, indicates that RPG is a previously unknown symbiotic M. truncatula gene controlling early steps of rhizobial infection, but not early steps of NF signaling nor polarization of the outer cortex.
RPG Controls Rhizobium-Induced Polar Growth During the Infection Process.
The two infection steps altered in the rpg mutant (RHC and IT growth) are not well characterized, but are known to be deviations from normal polar RH growth that are dependent on NF-producing rhizobia. Redirection of tip growth during RHC involves break-up of the actin network at the RH tip, followed by its reassembly at an off-center site that is stabilized, or selected, by the microtubule network (3, 4). Then ITs form by inward polar growth, with an actively streaming column of cytoplasm present between the RH nucleus and the IT tip, which is the site of cell wall and membrane deposition (4).
rpg RHs do not respond to rhizobia by sustained polar growth changes, resulting in both loose, rather than tight RHCs, and thick, slowly progressing, rather than narrow, fast-growing ITs. This indicates that a common molecular mechanism underlies these two steps. van Batenberg et al. (17) proposed that dominant rhizobially induced tip growth can explain tight RHC and IT growth. For curls, a single rhizobial microcolony would become dominant and dictate that RHC occurs continuously around it by the sustained redirection of tip growth. The loose RHC and formation of new poles of tip growth in the rpg mutant suggest a defect for maintenance of this dominance. For normal ITs, only rhizobia present at the tip divide and induce growth, thus producing narrow and rapidly advancing structures (4). The wide and bumpy nature of rpg ITs suggests that bacteria are not just multiplying in a narrow zone, and that they are either passively filling ITs that are not extending normally in a polar fashion, or they are all stimulating IT growth. This all suggests that RPG fulfils an essential function in the process whereby rhizobia dominate the process of induced tip growth for RHC and IT growth.
NFs are directly implicated in the polar changes that accompany RH infection because NFs stimulate cytoplasmic polarity, affect RH cytoskeleton organization, and induce a new growth axis and new polar growth (18). The mechanism underlying these changes is also probably responsible for infection, with bacteria bound to the RH surface acting as a point source of NF (4). NF recognition is clearly necessary for RHC and for proper development of ITs (8, 19–21). Consistent with normal responsiveness to NFs in the rpg mutant, RPG may control RHC downstream of the NFP-DMI-NSP NF-signaling pathway, providing the first known link between NF signaling and redirection of endogenous RH growth for infection.
RPG Expression Is Strongly Associated with the Infection Process.
Initial, widespread GUS expression driven by the RPG promoter region was NF dependent and inducible by pure NFs. This suggests that RPG induction depends on the NFP/DMI/NSP pathway that mediates all known NF-induced transcriptional responses and potentially places RPG as a response element of this pathway under the control of recently identified transcription factors (9). This possibility is supported by preliminary data indicating that NF-induced RPG induction is DMI3 dependent (data not shown).
During infection, the RPG promoter was down-regulated in all epidermal cells except those containing ITs, where the RPG promoter was more strongly induced. Such an NF-mediated, preinfection-, and then infection-related expression pattern is similar to that of MtENOD11 (22), but different from NFP and DMI2, which are expressed ahead of ITs in nodule primordia (20, 23). Analysis of hcl and lin mutants that both uncouple NF responsiveness (normal) from infection (blocked), suggests that after the early, NF-mediated induction of RPG gene expression, sustained expression depends on the infection process and functional HCL and LIN genes. The strong reduction in RPG gene expression in hcl plants, mutated in the NF entry receptor gene LYK3, might partly explain the defects in polar growth of ITs in LYK3 knock-down and mutant lines (8, 24).
RPG Is Likely to Be an Important Component of Symbiotic Signaling.
Features of the predicted structure of RPG, together with its nuclear localization, give clues about the potential function of the protein. First, RPG has a region predicted to be intrinsically unstructured. Typically, such disordered domains mediate protein–protein interactions, becoming structured on binding to their targets and concomitantly altering the action of their partner to facilitate recruiting of further components and complex functioning. Such domains are found notably in “assemblers” or “hubs,” which assemble multiprotein complexes (25). The major part of RPG consists of coiled-coil regions that are widespread oligomerization motifs. Long coiled-coil proteins can form filaments or networks that have structural roles or can act as scaffolds (26). Coiled-coil proteins are found in the nucleus, and disordered proteins often have functions in the regulation of transcription.
Taken together, these features are consistent with RPG fulfilling a regulatory role in controlling gene expression. For example, RPG might be recruited as a transcriptional activator for the regulation of genes involved in spatial subcellular reorganizations that lead to the localized deposition of cell wall and membrane material to new polar growth sites during tip growth for RHC and IT growth. A nuclear localization for RPG is interesting in light of the symbiotic nucleoporin genes, NUP133 and NUP85, of L. japonicus and the nuclear localization of several NF signaling proteins and NF-induced calcium spiking, presumably all involved in a major reprogramming of transcription (9). It will now be interesting to know how RPG is regulated during the symbiotic interaction, and with which proteins RPG interacts.
The RRP Family Defines a Class of Plant-Specific Long Coiled-Coil Proteins.
To our knowledge, RPG is the founding member of a new family of plant-specific proteins, the RRPs, including four proteins each for A. thaliana and rice, and six proteins so far predicted for M. truncatula. Characterization of the rpg mutant therefore has allowed an RRP gene to be assigned to a precise biological process. All RRPs are putative long coiled-coil proteins with a conserved domain structure, of which the RRP domain can be considered the signature of the family. Given the predicted structural features of RRPs, they might mediate protein-protein interactions in signaling processes.
The rice RRP Os10g36060 binds to a cyclin and is thus implicated in cell cycle regulation (27). A role in polar growth for RRPs is supported by the fact that two rice RRP genes, one in the RPG clade, are preferentially expressed in mature pollen (MPSS database, accessed August 1, 2006). Sequence conservation in the RRP domain, and at both ends of the α-helical domain, suggests common properties for RRPs. However, sequence variation in the disordered domain and the variable numbers of coiled-coils suggest functional or partner specificity.
Among M. truncatula RRPs, the unique features and expression pattern of RPG probably reflect specialization for a role during rhizobial infection. The nuclear localization of RPG in N. benthamiana leaves provides a starting point to determine the precise molecular function of RPG in this process, and further characterization of the plant-specific RRP family will elucidate in which other cellular processes this family of proteins is involved.
Methods
Plant Material and Growth Conditions.
The M. truncatula rpg mutant (originally called B99) is an ethyl methanesulfonate mutant isolated and grown as described (10).
Microscopic Methods.
Infection events by S. meliloti were observed as described (10), by histochemical staining for β-galactosidase activity expressed by pXLGD4 (28), or by confocal studies for GFP from pHC60 (24), and immunolocalization of microtubules was done as described (7). Other methods are described in SI Text.
Positional Cloning of RPG.
M. truncatula genetic markers (www.medicago.org; ref. 29) for all chromosomes were tested on F2 plants from a cross between rpg and the M. truncatula accession DZA315.16, for segregation with the RPG locus. Chromosome walking generated a BAC contig covering the RPG region. CAPs markers (SI Text) were generated from BAC end sequences and analyzed on the mapping population. For complementation, an 11-kb SalI fragment, carrying RPG (coding sequence and 1.5 kb upstream) and a potential globin gene, was cloned into pCAMBIA2201 (www.cambia.org). The globin gene was deleted by StuI–PacI digestion followed by plasmid religation. Agrobacterium rhizogenes Arqua1 was used for hairy root transformation (24).
Sequence Analysis.
Genes and ESTs were identified by using the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/), FGENESH (www.softberry.com), and the Medicago EST Navigation System (MENS) database (http://medicago.toulouse.inra.fr/MENS). RRP1 was fully sequenced by using the BACs mth2–26G04 and mth2–60F19. Domain structure, alignments, and phylogeny were analyzed as described in SI Text.
Expression Analysis.
Plant growth, RNA extractions, quantitative RT-PCRs, and result standardization using ACTIN2 were performed as described (SI Text and ref. 20). For the RPG promoter-GUS fusion, 1.5 kb upstream of the RPG start codon was amplified (SI Text) and cloned in the binary vector pLP100 (31) by using SalI–KpnI digestion. Expression analysis was performed as described in ref. 20.
Subcellular Localization of RPG.
cDNA of RPG was amplified (SI Text) and cloned into the Gateway vector pGWB6 (Clontech) to generate a translational GFP fusion of RPG controlled by the cauliflower mosaic virus 35S promoter. The p35S::GFP construct was used as a control. After infiltration into 4-week-old Nicotiana benthamiana plants for transient protein expression (SI Text), observations by confocal laser scanning microscopy (Leica SP2) were made 5–6 days later (SI Text).
Supplementary Material
Acknowledgments.
We are grateful to S. Bensmihen, F. Debellé, and J. Dénarié for critical reading of the manuscript, F. Maillet for NFs, F. Debellé and S. Bensmihen for cloning help, the GENOSCOPE (Evry, France) for BAC sequencing, T. Huguet for genetic markers, T. Vernié for M. truncatula RNA, P. Rougé for help in sequence analysis, and V. Bayle for confocal observations. We thank the Plate-Forme Génomique of the Toulouse Génopole for use of robotics facilities. J.F.A. and O.G. received doctoral grants from the French Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank databank [accession nos. DQ854741 (RPG), DQ854742 (RRP1), EF222477 (GB), EF222478 (GR), EF222479 (HS), and EF222480 (KH)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0710273105/DCSupplemental.
References
- 1.Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campanoni P, Blatt MR. Membrane trafficking and polar growth in root hairs and pollen tubes. J Exp Bot. 2007;58:65–74. doi: 10.1093/jxb/erl059. [DOI] [PubMed] [Google Scholar]
- 3.Timmers AC, Auriac MC, Truchet G. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development. 1999;126:3617–3628. doi: 10.1242/dev.126.16.3617. [DOI] [PubMed] [Google Scholar]
- 4.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geurts R, Fedorova E, Bisseling T. Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr Opin Plant Biol. 2005;8:346–352. doi: 10.1016/j.pbi.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Hogg BV, Cullimore JV, Ranjeva R, Bono JJ. The DMI1 and DMI2 early symbiotic genes of Medicago truncatula are required for a high-affinity nodulation factor-binding site associated to a particulate fraction of roots. Plant Physiol. 2006;140:365–373. doi: 10.1104/pp.105.068981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catoira R, et al. The HCL gene of Medicago truncatula controls Rhizobium-induced root hair curling. Development. 2001;128:1507–1518. doi: 10.1242/dev.128.9.1507. [DOI] [PubMed] [Google Scholar]
- 8.Smit P, et al. Medicago LYK3, an entry receptor in rhizobial Nod factor signaling. Plant Phys. 2007;145:183–191. doi: 10.1104/pp.107.100495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with rhizobial infection. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 10.Catoira R, et al. Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell. 2000;12:1647–1666. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callaham DA, Torrey JG. The structural basis for infection of root hairs of Trifolium repens by Rhizobium. Can J Bot. 1981;59:1647–1664. [Google Scholar]
- 13.Bauer WD. Infection of legumes by Rhizobia. Annu Rev Plant Physiol. 1981;32:407–449. [Google Scholar]
- 14.van Brussel AA, et al. Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- 15.Laus MC, van Brussel AAN, Kijne JW. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol Plant–Microbe Interact. 2005;18:533–538. doi: 10.1094/MPMI-18-0533. [DOI] [PubMed] [Google Scholar]
- 16.Karas B, et al. Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs. Plant Physiol. 2005;137:1331–1344. doi: 10.1104/pp.104.057513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Batenberg FHD, Jonker R, Kijne JW. Rhizobium induces marked root hair curling by redirection of tip growth: A computer simulation. Physiol Plant. 1986;66:476–480. [Google Scholar]
- 18.Timmers ACJ. The role of the plant cytoskeleton in the interaction between legumes and rhizobia. J Microscopy. 2008 doi: 10.1111/j.1365-2818.2008.02040.x. in press. [DOI] [PubMed] [Google Scholar]
- 19.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 20.Arrighi JF, et al. The Medicago truncatula LysM-receptor kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Den Herder J, et al. Nod factor perception during infection thread growth fine-tunes nodulation. Mol Plant–Microbe Interact. 2007;20:129–137. doi: 10.1094/MPMI-20-2-0129. [DOI] [PubMed] [Google Scholar]
- 22.Andriankaja A, et al. AP2-ERF transcription factors mediate Nod factor-dependent MtENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell. 2007;19:2866–2885. doi: 10.1105/tpc.107.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bersoult A, et al. Expression of the Medicago truncatula DM12 gene suggests roles of the symbiotic nodulation receptor kinase in nodules and during early nodule development. Mol Plant–Microbe Interact. 2005;18:869–876. doi: 10.1094/MPMI-18-0869. [DOI] [PubMed] [Google Scholar]
- 24.Limpens E, et al. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- 25.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 26.Rose A, Meier I. Scaffolds, levers, rods and springs: Diverse cellular functions of long coiled-coil proteins. Cell Mol Life Sci. 2004;61:1996–2009. doi: 10.1007/s00018-004-4039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper B, et al. Identification of rice (Oryza sativa) proteins linked to the cyclin-mediated regulation of the cell cycle. Plant Mol Biol. 2003;53:273–279. doi: 10.1023/b:plan.0000007001.30865.0f. [DOI] [PubMed] [Google Scholar]
- 28.Leong SA, Williams PH, Ditta GS. Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 1985;13:5965–5976. doi: 10.1093/nar/13.16.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoquet P, et al. The molecular genetic linkage map of the model legume Medicago truncatula: An essential tool for comparative legume genomics and the isolation of agronomically important genes. BMC Plant Biol. 2002;2:1. doi: 10.1186/1471-2229-2-1. Available at www.biomedcentral.com/bmcplantbiol/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teillet A, et al. api, a novel Medicago trunctula symbiotic mutant impaired in nodule primordium invasion. Mol Plant–Microbe Interact. 2008;21:535–546. doi: 10.1094/MPMI-21-5-0535. [DOI] [PubMed] [Google Scholar]
- 31.Szabados L, Charrier B, Kondorosi A, de Bruijn FJ, Ratet P. New plant promoter and enhancer testing vectors. Mol Breeding. 1995;1:419–423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.