Summary
Spatial control of mRNA translation is a well established mechanism for generating cellular asymmetries and for functional specialization of polarized cells like neurons. A requirement for the translational repressor Nanos (Nos) in the Drosophila larval peripheral nervous system (PNS) implicates translational control in dendrite morphogenesis [1]. Nos was first identified by its requirement in the posterior of the early embryo for abdomen formation [2]. Nos synthesis is targeted to the posterior pole of the oocyte and early embryo through translational repression of unlocalized nos mRNA coupled with translational activation of nos mRNA localized at the posterior pole [3, 4]. Mutations that abolish nos localization prevent abdominal development whereas de-repression of unlocalized nos mRNA suppresses head/thorax development, indicating that spatial regulation of nos is essential for anterior-posterior patterning [3, 5]. The observation that both loss and overexpression of Nos affect dendrite branching complexity in class IV dendritic arborization (da) neurons suggests that nos might also be regulated in these larval sensory neurons [1]. Here we show that localization and translational control of nos mRNA are essential for late stages of da neuron morphogenesis. RNA-protein interactions that regulate nos translation in the oocyte and early embryo also regulate nos in the PNS. Live imaging of nos mRNA shows that the cis-acting signal responsible for posterior localization in the oocyte/embryo mediates localization to the processes of class IV da neurons, but suggests a different transport mechanism. The need to target nos mRNA to the processes of da neurons may reflect a requirement for Nos protein in controlling translation locally within dendrites.
Results and Discussion
Nos is Required in Da Neurons to Maintain Dendrite Complexity
Da neurons, which innervate the larval epidermis, can be divided into four classes based on the complexity of their dendritic arbors, with class IV being the most highly branched [6]. These neurons elaborate primary and secondary branches during the first instar stage of larval development. By the second instar stage, higher order branches extend to completely cover the larval body wall [7]. Complete, nonredundant coverage or “tiling” of the epidermis by class IV da neurons is maintained throughout larval development [8]. Mutation of nos results in a reduction in the number of higher order branches of class IV da neurons without affecting the morphology of the main branches [1]. This decreased branching complexity could reflect an early role for nos in the initial elaboration of the dendritic branches or a later role in maintaining coverage of the receptive field during larval growth. To distinguish between these possibilities, we examined the morphology of nos mutant class IV da neurons at different larval stages. In these, and all subsequent experiments, class IV da neurons are marked by mCD8:GFP, expressed using the GAL4477 driver [9]. Branching complexity was monitored by quantitation of branch termini (see Supplemental Experimental Procedures and Figure 1 legend).
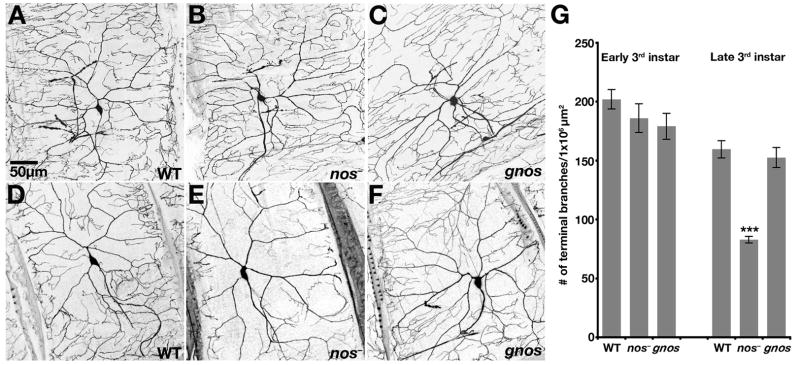
Figure 1. nos Plays a Role in Maintenance of Dendritic Branching.
Confocal Z-series projections of class IV da neurons in (A–C) early third instar larvae and (D–F) late third instar larvae. Da neurons are marked here and in all subsequent figures by using GAL4477 to drive expression of UAS-mcd8:GFP. (A,D) Neurons from wild-type larvae. (B,E) Neurons from nos mutant larvae. (C,F) Neurons from nos mutant larvae carrying the gnos rescue transgene. (G) Quantitation of total number of terminal branches within a 1 × 106 μm2 region of the dendritic tree of an individual neuron (see Experimental Procedures). Values are the average +/− standard error. One neuron per larva was analyzed from early third instar: wild-type (n=10 neurons); nos mutant (n=9 neurons); gnos (n=10 neurons); or late third instar: wild-type (n=15 neurons); nos mutant (n=10 neurons); gnos (n=10 neurons). Here and in all subsequent figures, P values were determined by the Student’s t-test and are labeled as *, **, and *** to denote p<0.05, p<0.01 and p<0.001 respectively.
From the first larval instar through the early third instar stage, da neurons in wild-type larvae, nos mutant larvae, and nos mutant larvae carrying a genomic nos transgene, gnos [3], show no significant difference in branching complexity (Figure 1A–C, G and data not shown). Morphological defects are first detected at the late third instar stage, when a significant reduction of higher order branching is observed in nos mutant da neurons as compared to wild-type neurons (Figure 1D,E). Whereas terminal branch density decreases slightly as body size increases from early to late third instar stages in wild-type larvae, the density of terminal branches decreases dramatically in nos mutant larvae (Fig. 1G). Wild-type branching is restored in nos mutant larvae by addition of gnos, which includes native transcriptional regulatory sequences required for nos expression and rescues all nos mutant embryonic phenotypes [3, 10] (Figure 1F,G). These results indicate that Nos is not required for the initial elaboration of dendritic branches but instead plays a role at later stages of development, possibly by maintaining existing branches or promoting new branch extension during larval growth.
nos is Localized to the Processes of Da Neurons
Localized translation of nos required for embryonic patterning is achieved through a combination of mRNA localization and translational control. To determine if nos is spatially regulated in da neurons, we analyzed the distribution of nos mRNA by modifying a fluorescent labeling method previously used to investigate the mechanism of nos mRNA localization during oogenesis [4]. In this method, a fusion between bacteriophage MS2 coat protein (MCP) and either GFP or RFP is tethered to nos mRNA bearing 6 stem-loop binding sites for MCP [nos-(ms2)6]. Here, we have improved detection of nos by introducing 18 MCP-binding stem-loops [nos-(ms2)18]. The nos-(ms2)18 transgene behaves indistinguishably from gnos and the previously described nos-(ms2)6 transgenes in the oocyte and early embryo (K. Forrest and E.R.G., unpublished). To label nos-(ms2)18 RNA specifically in da neurons, we expressed MCP-RFP under UAS control using GAL4477 in larvae carrying the nos-(ms2)18 transgene. In neurons from control larvae that express MCP-RFP without nos-(ms2)18 mRNA, RFP fluorescence is largely confined to the nucleus due to a nuclear localization signal engineered in MCP-RFP that targets unbound MCP-RFP to the nucleus (Fig. 2A). By contrast, RFP-labeled nos-(ms2)18 mRNA (nos*RFP) can be detected in the cell body and in particles that are distributed along the dendrites and axons of class IV da neurons (Fig. 2B). Analysis of nos*RFP throughout larval development showed that localization of nos to the processes of da neurons can first be detected early at the third instar stage (data not shown).
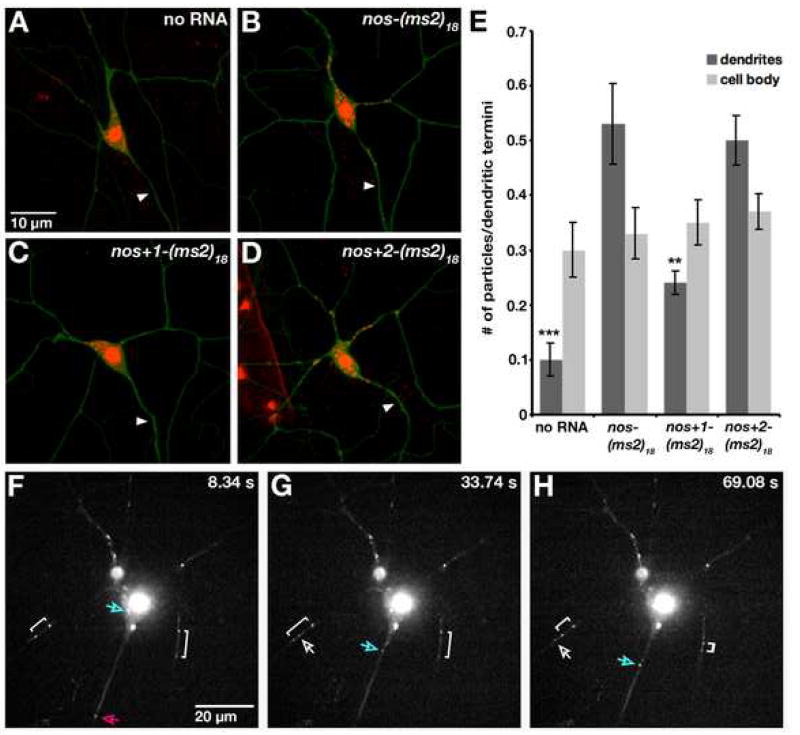
Figure 2. Localization of nos to the Processes of Da Neurons.
(A–D) Class IV da neurons in semi-intact third instar larvae expressing mCD8:GFP, MCP-RFP and (A) no ms2-tagged nos mRNA (control); (B) nos-(ms2)18 mRNA; (C) nos+1-(ms2)18 mRNA; (D) nos+2-(ms2)18 mRNA. MCP-RFP that is not bound to mRNA is sequestered in the nucleus due to an NLS in the MCP-RFP fusion protein. Arrowhead indicates the axon, as identified in lower power images. (E) Quantitation of nos*RFP particles in dendritic branches. All neurons were imaged using identical confocal settings. A merged image showing both green (mcd8:GFP) and red (nos*RFP or MCP-RFP alone) channels was enlarged and adjusted in Adobe Photoshop so that green channel was just visible. Red particles encompassed within the branches or cell body were counted and each total was normalized to the total number of dendritic termini within the field imaged (3.6×104 μm2). Two independent lines analyzed for each transgene produced similar results and one line for each is shown. For each genotype, values are the average +/− standard error for 10 neurons. (F–H) Time lapse sequence of RFP-labeled nos-(ms2)18 mRNA in a Class IV da neuron (only the red channel is shown). Each panel shows a single confocal section captured at the indicated time. See Supplemental Movie S2 for the complete 75 second time series. Examples of movement are indicated. Brackets illustrate movement toward and away from the cell body. Particles indicated by the bracket on the left move bidirectionally – first apart from each other, then toward each other. The pink arrow illustrates a particle that moves out of the frame. The white arrow shows a particle that crosses paths with one of the particles indicated by the bracket. The blue arrow marks a particle traveling from the cell body to a dendrite.
We have not been able to confirm localization of native nos mRNA in da neurons by in situ hybridization methods, most likely due to a combination of low transcript abundance and high background from the underlying muscle tissue. However, we have previously shown that fluorescently labeled nos mRNA is a valid proxy for native nos mRNA in the oocyte and embryo [4]. Moreover, the correlation between dendritic localization of nos*RFP and its ability to rescue the nos mutant dendritic branching defect, described below, gives us confidence that it recapitulates the distribution of native nos in these neurons.
The nos 3′UTR is Required for Efficient nos Localization in the PNS
Posterior localization of nos in the oocyte and early embryo is mediated by a complex cis-acting localization signal in the nos 3′ untranslated region (3′UTR) comprising multiple, partially functional localization elements [11]. To test whether the same sequences direct dendritic localization of nos in da neurons, we analyzed the distribution of RFP-labeled nos–(ms2)18 RNAs bearing 3′UTR deletions (Figure S1A). Deletion of the entire localization signal (nosΔLS) or three of the four localization elements (nos+1), respectively, abolishes or severely reduces posterior localization of nos in the oocyte and embryo [11]. Both deletions also compromise localization to the processes of da neurons (Figure 2C and data not shown). Quantitation of nos*RFP particles shows reduced accumulation in dendrites of larvae expressing nos+1-(ms2)18 mRNA relative to larvae expressing nos-(ms2)18 mRNA, whereas no significant difference is detected within the cell body (Figure 2E). In contrast, the distribution of RFP-labeled nos+2-(ms2)18 mRNA is similar to that of nos-(ms2)18mRNA (Figure 2D,E). This RNA lacks two of the four localization elements but retains the nos +2 element, which confers near wild-type localization in the embryo [11]. The nos+1-(ms2)18 and nos+2-(ms2)18 transcripts are present at comparable levels in da neurons as determined by RT-PCR (Figure S2) and similar results were obtained for two independent lines of each transgene (data not shown), indicating that the observed difference in localization to neuronal processes is not likely due to a difference in expression or stability. Thus, the same sequences that mediate posterior localization of nos at earlier developmental stages target nos to the processes of class IV da neurons. This result suggests that one or more factors that recognize this localization signal to mediate localization during oogenesis may be used again for dendritic localization.
Localization of nos is Required for its Function in the PNS
Since posterior localization of nos is essential for its function in embryonic development, we investigated whether dendritic localization of nos is also required for its function in the larval PNS. The nos-(ms2)18, nos+1-(ms2)18 and nos+2-(ms2)18 transgenes were introduced into nos mutant larvae and assayed for their ability to rescue the nos mutant dendritic defect. All three transgenes include sequences required for nos translational regulation (see below) and none of these transgenes on its own affects dendrite branching complexity (data not shown). Class IV da neurons in nos mutant larvae carrying either the nos-(ms2)18 or nos+2-(ms2)18 transgene exhibit nearly wild-type dendritic branching, indicating that both transgenes are able to rescue the nos mutant phenotype (Figure 3A–C, E). In contrast, the nos+1-(ms2)18 transgene fails to rescue, as larvae show reduced branching complexity (Figure 3D,E).
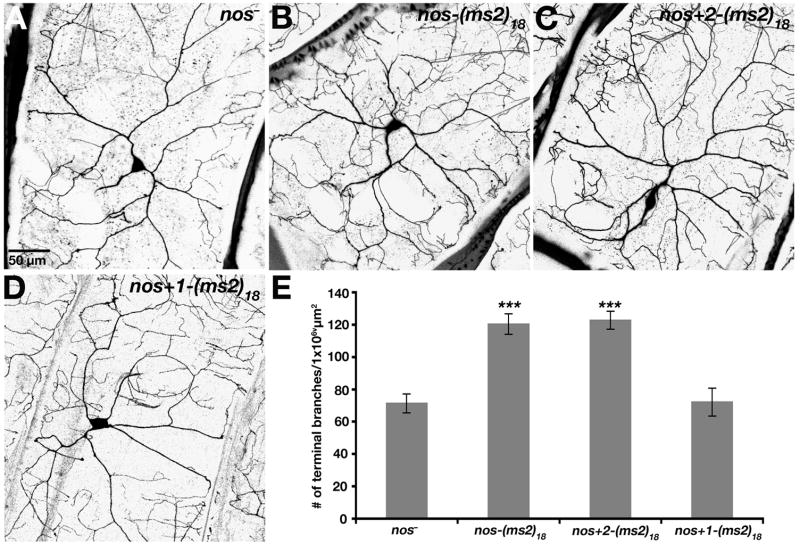
Figure 3. nos mRNA Localization is Required for Dendrite Morphogenesis.
The nos transgenes analyzed in Fig. 2 were tested for their ability to rescue the nos mutant defect in dendrite morphogenesis. (A–D) Confocal Z-series projections of class IV da neurons in third instar nos mutant larvae (A) or nos mutant larvae expressing nos-(ms2)18 (B), nos+2-(ms2)18 (C), or nos+1-(ms2)18 (D) transgenes. (E) Quantitation of dendritic terminal branches. Two independent lines for each transgene produced similar results and one line for each is shown. Values are the average +/− standard error for nos− (n=11 neurons); nos-(ms2)18 (n=11 neurons); nos+2-(ms2)18 (n=9 neurons); nos+1-(ms2)18 (n=11 neurons).
These results indicate that the localization of nos to class IV da neurons is required for nos function in dendrite morphogenesis. Because currently available anti-Nos antibodies are not adequate to detect Nos protein in da neurons (our data, also see Ref. [1]), we cannot show definitively that nos mRNA localization leads to local production of Nos protein. However, the correlation between the localization to neuronal processes and the ability to rescue the nos mutant branching defect revealed by the nos+1-(ms2)18 and nos+2-(ms2)18 mRNAs provides strong evidence that nos mRNA localization plays a critical role by targeting synthesis of Nos to dendrites.
Live imaging of nos mRNA Particle Movement in Da Neurons
Although most mRNAs are thought to be transported as particles along cytoskeletal elements by motor proteins, nos accumulates at the posterior of the oocyte by a passive diffusion and entrapment mechanism [4]. As a first step toward investigating the mechanism of nos localization in da neurons, we performed time lapse imaging of nos*RFP in da neurons of intact larvae at high magnification and time resolution. Control neurons expressing only MCP-RFP contain few RFP-labeled particles outside of the cell body and these particles rarely exhibit movement (Movie S1). In contrast, in neurons expressing nos*RFP, dynamic particles are readily detected in the cell body and processes (Figure 2F–H and Movie S2). Photobleaching of RFP that occurs at the requisite high image capture rates and the potential for tissue damage limits our time sequences to ≤90 sec. During these short periods, we observe particles traveling with linear trajectories, in both anterograde and retrograde directions along the neuronal processes, and in some cases, individual particles exhibit bi-directional movement (Figure 2F–H; Movie S2). By analyzing sustained particle runs in a single direction (average run distance = 4.1 μm; see Supplemental Experimental Procedures) within the dendrites of five neurons from three independent larvae, we calculated a mean average dendritic particle velocity of 0.56 μm/sec (range=0.21–0.97 μm/sec, n=40). This value is similar to those we have observed, using the same labeling method, for dynein-dependent transport of bicoid (bcd) mRNA in the Drosophila oocyte [12] and similar rates have been observed for microtubule-dependent transport of ribonucleoprotein particles in the dendrites of cultured hippocampal neurons [13–16]. Like nos, these dendritic RNA-containing particles exhibit bi-directional movement. The generation of brighter and more photostable MCP fusion proteins that permit visualization of particles over longer time periods and in various mutant backgrounds will enable us to determine how these complex particle dynamics lead to accumulation of RNA in dendrites.
Whereas localization of nos during oogenesis occurs by diffusion and entrapment [4], the trajectories and velocities exhibited by dendritic nos particles are characteristic of cytoskeletal-based transport. Analysis of microtubule polarity in da neurons indicates that the majority of microtubules are oriented with their minus ends distal to the cell body [17]. Although individual da neuron subclasses were not distinguished in this study, the observations suggest that transport of nos mRNA particles into dendrites utilizes dynein. nos RNA injected into blastoderm embryos exhibits microtubule-dependent apical localization characteristic of pair-rule transcripts, whose transport is dynein-mediated [18, 19]. Since endogenous nos mRNA is not apically localized, the significance of such transport has been unclear. Our results suggest that the ability of nos to engage dynein-dependent transport machinery is indeed relevant to its role in the PNS.
Regulation of Dendrite Morphogenesis by Glo and Smg
Translational activation of nos at the posterior pole is tightly coupled to translational repression of unlocalized nos mRNA to prevent accumulation of Nos in the anterior of the embryo, where Nos suppresses anterior development [5]. Since nos localization during oogenesis is inefficient, this linkage is essential to silence nos mRNA that remains distributed throughout the bulk cytoplasm [20]. Translational repression of nos mRNA is mediated by a structural motif, the translational control element (TCE), within the nos 3′UTR [21–23] (Figure S1B). TCE function requires the formation of two stem-loops, designated as II and III, that have temporally distinct activities [24, 25]. Whereas stem-loop III mediates repression of nos during oogenesis, through its interaction with Glorund (Glo), stem-loop II is responsible for repression of nos in the early embryo, through its interaction with a different repressor, Smaug (Smg) [26–28].
Replacement of the nos 3′UTR by α-tubulin 3′UTR sequences (nos-tub3′UTR) abolishes nos localization and translational repression, leading to unrestricted synthesis of Nos and defects in anterior development [5]. GAL4 mediated overexpression of a UAS-nos-tub3′UTR transgene in class IV da neurons is also deleterious, causing decreased branching complexity. This overexpression phenotype is ameliorated by reinsertion of the nos TCE [1]. The observation that both loss and overexpression of nos cause similar defects indicates that although nos is required for dendrite morphogenesis, the level of Nos protein must be carefully modulated in da neurons. Moreover, the ability of the TCE to suppress the toxicity of nos mRNA overexpression in da neurons suggests that it may normally function to control Nos levels in the PNS. We therefore sought to determine whether endogenous nos is regulated by the TCE in da neurons.
Ectopic expression studies have identified several additional somatic cell types where the TCE can repress translation, including neuroendocrine cells and the dorsal pouch epithelium [29, 30]. However, TCE function in the dorsal pouch does not depend on the Glo or Smg binding sites, but requires a distinct sequence motif with homology to the Bearded (Brd) box [29]. Mutation of the Brd box-like motif does not abrogate the ability of the TCE to suppress excess nos activity in da neurons (data not shown). Consequently, to determine whether endogenous nos mRNA might be regulated by the TCE, we first analyzed da neurons in glo and smg mutant larvae.
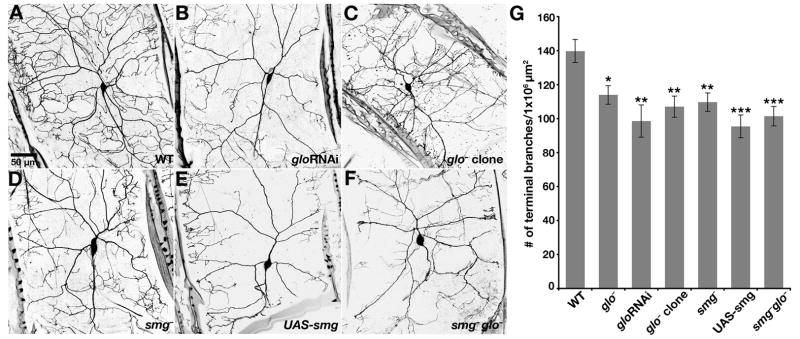
Larvae mutant for glo or smg survive until third instar stage, permitting us to examine the effect of eliminating either repressor on dendrite morphology of da neurons. Compared to wild-type class IV da neurons, glo mutant larvae show a significant decrease in the number of higher order dendritic branches as reflected by a decreased number of terminal dendritic processes (Figure 4G). Because glo mutant larvae exhibit additional defects (J.L.B. and E.R.G., unpublished), we disrupted glo function specifically in class IV da neurons either by using GAL4477 to express a UAS-gloRNAi transgene or by using the MARCM method [31] to generate mosaic animals. In both cases, glo mutant da neurons show decreased branching complexity (Figure 4A–C,G). Mutation of smg or GAL4477-mediated overexpression of a UAS-smg transgene also causes loss of high order branches (Figure 4D,E,G). Larvae doubly mutant for glo and smg do not show a more severe phenotype than larvae mutant for either gene alone (Figure 4F,G), suggesting that each repressor contributes independently. Thus, defects due to loss or overexpression of the repressors are consistent with defects caused by loss or overexpression of nos. Due to the aforementioned inadequacy of anti-Nos antibodies, we have not been able to monitor changes in Nos protein levels in glo and smg mutant da neurons. However, when combined with the analysis of Glo and Smg binding site mutations presented below, these results strongly support a role for glo and smg in regulation of nos for dendrite morphogenesis.
Figure 4. Glo and Smg are Required for Dendrite Development.
(A–F) Confocal Z-series projections of class IV da neurons in third instar larvae. (A) Class IV da neuron from a wild-type larva. (B) Neuron from a glo RNAi larva. (C) glo mutant neuron generated by MARCM. (D) Neuron from a smg mutant larva. (E) Neuron from larva overexpressing smg (UAS-smg). (F) Neuron from larva doubly mutant for glo and smg. (G) Quantitation of total number of terminal branches within a 1 × 106 μm2 region of the dendritic tree of an individual neuron. Values are the average +/− standard error for wild-type (n=15 neurons), glo− (n=10 neurons), gloRNAi (n=10 neurons), glo− MARCM clone (n=5 neurons), smg− (n=10 neurons), UAS-smg (n=9 neurons) and smg−glo− (n=10 neurons).
TCE-dependent Translational Regulation of nos is Required for Dendrite Development
In the oocyte, Glo binds specifically to the distal double-stranded helix of TCE stem-loop III (the Glo Recognition Helix or GRH; Figure S1B) [27]. In the embryo, Smg interacts with nos TCE stem-loop II, via nucleotides within the loop designated as the Smg Recognition Element (SRE; Figure S1B) [22, 24]. A second SRE located downstream of the TCE in the nos 3′UTR appears to act redundantly [22, 23]. To determine whether the defects observed in glo and smg mutant da neurons are due to loss of TCE-mediated repression, we tested whether mutation of the nos GRH or SREs produces a similar phenotype. Mutations that disrupt both SREs (SREs−), the binding site for Glo (GRH−), or the SREs and GRH (SREs−GRH−) together (Figure S1B) were introduced into the gnos transgene. The resulting gnosSREs−, gnosGRH−, and gnosSREs−GRH− transgenes all produce mRNAs that show wild-type localization in the early embryo but whose translation is not restricted to the posterior pole (Ref [10]; E.R.G., unpublished). When compared to larvae expressing the wild-type gnos transgene, branching complexity is significantly reduced in da neurons of larvae expressing gnosSREs−, gnosGRH−, and gnosSREs−GRH− transgenes (Figure 5). Moreover, each of these transgenes behaves similarly to the gnos-tub3′UTR transgene, which lacks the entire nos 3′UTR, indicating that mutation of the GRH and/or SREs is sufficient to disrupt nos regulation in the PNS. Together, these results show that TCE-mediated regulation of nos in da neurons is essential for dendrite morphogenesis. Furthermore, the finding that the same phenotype is produced by either eliminating the repressors or mutating their binding sites provides strong evidence that this regulation is mediated by Glo and Smg.
Figure 5. Effect of TCE Mutations on nos Regulation in Da Neurons.
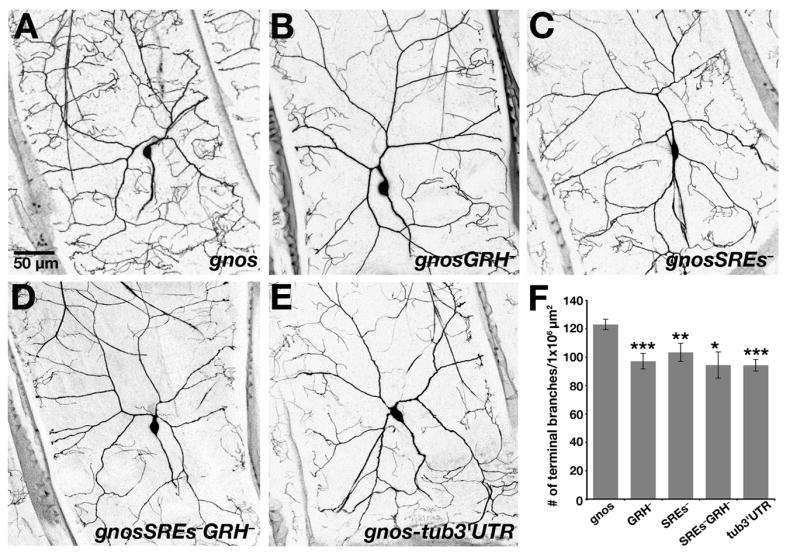
(A–E) Confocal Z-series projections of class IV da neurons in third instar larvae expressing the (A) gnos, (B) gnosGRH−, (C) gnosSREs−, (D) gnosSREs−GRH−, and (E) gnos-tub3′UTR transgenes. (F) Quantitation of dendritic terminal branches. Similar results were obtained from analysis of three independent lines for each transgene and data obtained from one line for each is shown. For each transgene, values are the average +/− standard error: gnos (n=9 neurons); gnosGRH− (n= 10 neurons); gnosSREs− (n=9 neurons); gnosSREs−GRH− (n=8 neurons); gnos-tub3′UTR (n=10 neurons).
In many cell types, protein synthesis is spatially regulated through the transport of translationally silent mRNAs and activation of these mRNAs at the target destination. Linkage of translation and localization serves not only to prevent premature accumulation of nos during transit to the oocyte posterior, but to silence the large pool of nos that remains unlocalized due to inefficient posterior localization [20]. We cannot yet distinguish whether localization of nos in da neurons is similarly inefficient or whether translational repression of nos serves primarily to repress translation during transport. However, the deleterious effect on dendrite morphogenesis caused by mutations that disrupt TCE function show that, as for maternally synthesized nos mRNA, localization alone is not sufficient to modulate its activity.
Conclusions
nos plays an important role in dendrite morphogenesis and we show that nos function in da neurons requires spatial regulation of nos mRNA. Cis-acting sequences and two cognate factors that control nos mRNA localization and/or translation in the oocyte and early embryo are redeployed during larval stages to regulate localization and translation of nos in da neurons. Localization of nos mRNA to the processes of class IV da neurons is essential for dendritic branching. For the first time, we observe movement of RNA particles in neurons of intact animals and analysis of nos mRNA particle movement suggests that nos localization occurs by different mechanisms depending on cellular context. Taken together, our results support a role for Nos as a local regulator of translation in the PNS.
In the early embryo, Nos functions in a complex with the RNA-binding protein Pumilio (Pum) to repress hunchback mRNA translation, thereby promoting abdominal development [32, 33]. Whereas Pum is produced throughout the embryo [33, 34], restriction of Nos synthesis to the posterior limits the spatial domain of the repressor complex. Mutations in nos and pum produce similar defects in dendrite morphogenesis, suggesting that Nos and Pum also act together to repress translation in da neurons [1]. Thus, spatial regulation of nos may serve a similar function in the PNS as it does in the early embryo, by restricting the activity of the Nos/Pum repressor complex to dendrites.
Supplementary Material
Acknowledgments
We are grateful to K. Forrest for initiating this project by generating and characterizing the nos-(ms2)18 and UAS-MCP-RFP transgenic lines, T. Weil for assistance with particle movement analysis and preparation of the Supplemental Movies, A. Becalska for generation of nos+2-(ms2)18 transgenic lines, and N. Jeurkar for generation of UASp-smg transgenic lines. We also thank Y.N. Jan and R. Wharton for fly stocks, B. Ye, U. Mayor, and W. Grueber for technical advice, J. Goodhouse (Princeton) and A. Sossick (Gurdon Institute) for microscopy assistance, and B. Ye, T. Weil, and I. Clark for comments on the manuscript. E.R.G. thanks her sabbatical host A. Brand and members of the Brand lab for hospitality during this work. This work was supported initially by the NSF (IOB-0344728) and subsequently by the NIH (R01 GM061107, R01 GM067758).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. nanos and pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 3.Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- 4.Forrest KM, Gavis ER. Live imaging of endogenous mRNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 5.Gavis ER, Lehmann R. Translational regulation of nanos by RNA localization. Nature. 1994;369:315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 6.Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- 7.Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- 9.Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein Cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- 10.Gavis ER, Chatterjee S, Ford NR, Wolff LJ. Dispensability of nanos mRNA localization for abdominal patterning but not for germ cell development. Mech Dev. 2008;125:81–90. doi: 10.1016/j.mod.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 12.Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 17.Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, Doe CQ. Polarity and intracellular compartmentalization of Drosophila neurons. Neural Develop. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- 19.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 20.Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126:659–669. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- 21.Gavis ER, Lunsford L, Bergsten SE, Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- 22.Smibert CA, Wilson JE, Kerr K, Macdonald PM. Smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- 23.Dahanukar A, Wharton RP. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 1996;10:2610–2620. doi: 10.1101/gad.10.20.2610. [DOI] [PubMed] [Google Scholar]
- 24.Crucs S, Chatterjee S, Gavis ER. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol. Cell. 2000;5:457–467. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- 25.Forrest KM, Clark IE, Jain RA, Gavis ER. Temporal complexity within a translational control element in the nanos mRNA. Development. 2004;131:5849–5857. doi: 10.1242/dev.01460. [DOI] [PubMed] [Google Scholar]
- 26.Smibert CA, Lie YS, Shillingaw W, Henzel WJ, Macdonald PM. Smaug, a novel and conserved protein contributes to repression of nanos mRNA translation in vitro. RNA. 1999;5:1535–1547. doi: 10.1017/s1355838299991392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Glorund, an hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev. Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- 29.Duchow HK, Brechbiel JL, Chatterjee S, Gavis ER. The nanos translational control element represses translation in somatic cells by a Bearded box-like motif. Dev Biol. 2005;282:207–217. doi: 10.1016/j.ydbio.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 30.Clark IE, Duchow HK, Vlasak AN, Gavis ER. A common translational control mechanism functions in axial patterning and neuroendocrine signaling in Drosophila. Development. 2002;129:3325–3334. doi: 10.1242/dev.129.14.3325. [DOI] [PubMed] [Google Scholar]
- 31.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 32.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992;6:2312–2326. doi: 10.1101/gad.6.12a.2312. [DOI] [PubMed] [Google Scholar]
- 34.Macdonald PM. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development. 1992;114:221–232. doi: 10.1242/dev.114.1.221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.