Abstract
Mitochondria generate reactive oxygen species, whose downstream lipid peroxidation products, such as 4-hydroxynonenal, induce uncoupling of oxidative phosphorylation by increasing proton leak through mitochondrial inner membrane proteins such as the uncoupling proteins and adenine nucleotide translocase. Using mitochondria from rat liver, which lack uncoupling proteins, in the present study we show that energization (specifically, high membrane potential) is required for 4-hydroxynonenal to activate proton conductance mediated by adenine nucleotide translocase. Prolonging the time at high membrane potential promotes greater uncoupling. 4-Hydroxynonenal-induced uncoupling via adenine nucleotide translocase is prevented but not readily reversed by addition of carboxyatractylate, suggesting a permanent change (such as adduct formation) that renders the translocase leaky to protons. In contrast with the irreversibility of proton conductance, carboxyatractylate added after 4-hydroxynonenal still inhibits nucleotide translocation, implying that the proton conductance and nucleotide translocation pathways are different. We propose a model to relate adenine nucleotide translocase conformation to proton conductance in the presence or absence of 4-hydroxynonenal and/or carboxyatractylate.
Keywords: carboxyatractylate (CAtr), 4-hydroxynonenal (HNE), proton leak, rat liver mitochondrion, trypsin
Abbreviations: ANT, adenine nucleotide translocase; CAtr, carboxyatractylate; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; HNE, 4-hydroxynonenal; ROS, reactive oxygen species; TPMP, triphenylmethylphosphonium; UCP, uncoupling protein
INTRODUCTION
Excessive ROS (reactive oxygen species) production is deleterious to cells, and is implicated in the aetiology and/or progression of a wide variety of pathologies including cardiovascular and neurodegenerative diseases [1–3]. However, ROS also play a role in signal transduction [4], and previously Echtay et al. [5,6] proposed that ROS and their downstream lipid peroxidation products such as HNE (4-hydroxynonenal) are not merely cytotoxic, but act as signals that result in mitochondrial uncoupling via the UCPs (uncoupling proteins) and ANT (adenine nucleotide translocase). The mitochondrial respiratory chain is a significant producer of ROS [7,8], particularly at high mitochondrial membrane potential [9]. Echtay and others have suggested that ROS-induced mild uncoupling via UCPs/ANT effectively acts as a negative-feedback loop by decreasing mitochondrial membrane potential, thereby attenuating further ROS production [6,10].
The proteins that are thought to contribute to proton leak in mitochondria belong to the mitochondrial solute carrier family [6,11–13]. Despite extensive research into uncoupling pathways via the UCPs and ANT, the molecular mechanisms by which activators induce proton conductance remain contentious. For example, fatty acids have long been known to activate ANT [11,12] and UCPs [14,15], but there is no consensus on how this activation is mediated. The main suggested mechanisms are the flip-flop model [16], the cofactor model [17] and the functional competition model [18–20]. The flip-flop model is generally invoked to explain uncoupling of ANT by fatty acids. All three models are discussed for UCPs, and should also be considered for ANT (see [21]). In the flip-flop model, a protonated fatty acid crosses the mitochondrial inner membrane into the matrix, dissociates, and the fatty acid anion is translocated back out of the matrix by the protein. Cycling of the fatty acid anion in its protonated and unprotonated forms results in net flux of protons into the matrix, thereby causing uncoupling. In the cofactor model, fatty acid carboxyl groups introduce a proton-conducting pathway. Attempts to clarify whether activation occurs via the flip-flop model or the cofactor model have included using ‘unflippable’ lipids such as glucose-O-ω-palmitate. However, results from different laboratories appear to conflict (reviewed in [22]). In the functional competition model, fatty acids are not directly involved in the transport mechanism, but competitively overcome inhibition of a native proton conductance by ATP without displacing the nucleotide from the protein.
Alkenal induction of proton leak is even less well characterized [23]. The main proposal is that alkenals covalently modify target proteins, thereby increasing their proton conductance [24]. Numerous studies show that alkenals, such as HNE, form adducts with ANT [25–27], but this has not formally been linked to uncoupling. Others have suggested that alkenals only stimulate uncoupling if they are first converted into a fatty acid [28].
Following the finding that activation of uncoupling by HNE in skeletal muscle mitochondria requires energization [29], and given our inability to discriminate between UCP3 and ANT using inhibitors [30], in the present study we chose to characterize the factors involved in this phenomenon using a UCP-free system. In the present study, we use the interaction of HNE with ANT in rat liver mitochondria as a model for defining the factors involved in activation of uncoupling by alkenals. The rat liver system is ideal for characterizing HNE-induced uncoupling because of the abundance of ANT compared with UCPs (which simplifies data interpretation) and the availability of a tight-binding ANT inhibitor such as CAtr (carboxyatractylate) [31] (which allows ANT-specific activity to be calculated). In the present study we demonstrate that in the presence of HNE, energization (specifically high membrane potential) is required to activate proton leak through ANT, that this uncoupling is not reversible by CAtr (in contrast with fatty-acid-induced proton leak through ANT, which has been shown to be both preventable and reversible by CAtr [12,14]), and that the proton conductance and nucleotide translocation pathways of ANT are behaviourally distinct.
EXPERIMENTAL
Animals
Female Wistar rats (Charles River Laboratories, U.K.) were housed at 21±2 °C, humidity 57±5% with a 12 h light/dark cycle. Food and water were available ad libitum. Home Office Guidelines for the Care and Use of Laboratory Animals (UK) were followed.
Tissue collection and mitochondrial isolation
Liver was dissected from rats after killing by stunning and cervical dislocation. Mitochondria were prepared by the method previously described in [32]. The following steps were carried out at 4 °C. Tissue was placed in STE buffer [250 mM sucrose, 5 mM Trizma base and 2 mM EGTA (pH 7.4)], minced, homogenized and centrifuged at 1047 g for 3 min. The supernatant was decanted and centrifuged at 11621 g for 10 min. The pellet was resuspended in STE buffer and centrifuged at 11621 g for 10 min a further two times. The final pellet was resuspended in approx. 1–1.2 ml of STE buffer and stored on ice for up to 6 h. The protein concentration was determined by the biuret method.
Respiratory control ratios
To estimate the functional quality of mitochondria, the respiratory control ratio (state 3/state 4oligomycin) was measured for each preparation (see below for oxygen consumption measurements). State 3 respiration was induced in the presence of 4 mM succinate by 600 μM ADP and terminated to state 4 by the addition of 1 μg/ml oligomycin. All mitochondria used had a respiratory control ratio of at least four.
Proton leak kinetics
Oxygen consumption and membrane potential were measured simultaneously at 37 °C, using a Clark-type oxygen electrode and an electrode sensitive to the potential-dependent cation, TPMP (triphenylmethylphosphonium) [33]. Mitochondria were incubated at 0.5 mg of protein/ml in air-saturated KPHEB medium [120 mM KCl, 5 mM KH2PO4, 3 mM Hepes and 1 mM EGTA (pH 7.2), supplemented with 0.3% (w/v) defatted BSA]. Additions of 5 μM rotenone (a complex I inhibitor, to prevent oxidation of endogenous NAD-linked substrates), 1 μg/ml oligomycin (to prevent ATP synthesis) and 80 ng/ml nigericin (to abolish the pH gradient) were made, in the presence or absence of 2.5 μM CAtr (Calbiochem) and/or 35 μM HNE (Cayman).
The chamber was sealed, avoiding air bubbles, and the TPMP electrode was calibrated with sequential additions of 0.5 μM TPMP to a final concentration of 2.5 μM. To calculate membrane potentials, data were corrected for TPMP binding to mitochondria using a binding correction of 0.4 (μl/mg of protein)−1 [33]. Proton leak rates were calculated by multiplying respiration rates by the H+/O ratio of 6 for succinate oxidation. Mitochondria were energized with 4 mM succinate, followed by titration with malonate or KCN (up to 2.3 mM or 0.2 mM respectively) to vary post-energization membrane potential and respiration rate for determination of proton leak kinetics. To vary the membrane potential during energization, pre-additions of malonate or cyanide were made before succinate. After each run, 0.3 μM FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone) was added to release TPMP for baseline correction.
ANT-specific proton leak was calculated as total proton leak inhibitable by pre-addition of 2.5 μM CAtr. CAtr may also (indirectly) prevent proton leak through UCPs [30], and UCP2 can be expressed in liver mitochondria under certain conditions [34–37]; however, immunoblotting of UCP2 calibrated using recombinant rat UCP2 standards showed UCP2 content to be negligible, constituting 0.00002±0.00002% of total mitochondrial protein (results not shown), hundreds fold less than in lung and spleen [38].
Measurement of ANT content by CAtr titration
Given the high specificity and tight binding of CAtr to ANT [31], the minimum concentration of CAtr required to lower state 3 respiration rates to state 4 levels equals the ANT content. Respiration rates were measured as above, without oligomycin or nigericin. After a 5 min energization with 4 mM succinate in the presence or absence of 35 μM HNE, excess ADP (600 μM) was added to establish state 3 respiration. This was then successively inhibited by CAtr additions (up to 3 nmol/mg of mitochondrial protein) until state 4 was well established. ANT content was calculated as the point at which the gradients in state 3 and state 4 intersected when the respiration rate was plotted as a function of the amount of CAtr added.
Trypsin digestion assay
Mitochondria were treated as for proton leak assays, but to terminate succinate oxidation after 5 min, 0.29 μM myxothiazol (a complex III inhibitor) was added. The whole incubation (KPHEB medium containing 0.5 mg/ml mitochondria, 5 μM rotenone, 1 μg/ml oligomycin, 80 ng/ml nigericin, 2.5 μM TPMP, ±35 μM HNE, ±2.5 μM CAtr, 4 mM succinate and 0.29 μM myxothiazol) was frozen (−20 °C) and thawed (37 °C) to rupture membranes, and subsequently treated with 0.25 mg/ml trypsin at 37 °C. At various times, aliquots were treated with 0.5 mg/ml soybean trypsin inhibitor to stop trypsin digestion, and centrifuged at 16000 g for 5 min. The supernatant was removed. The mitochondrial pellet was resuspended in gel-loading buffer [10% (w/v) SDS, 250 mM Tris/HCl (pH 6.8), 5 mM EDTA, 50% (v/v) glycerol, 5% (v/v) 2-mercaptoethanol and 0.05% (w/v) Bromophenol Blue], boiled for 5 min and vortex mixed vigorously.
Immunoblotting
Proteins from the trypsin digestion assay were separated by SDS/PAGE (12% gel) [39], transferred on to a Protran nitrocellulose membrane (Schleicher and Schuell), and probed with 0.8 μg/ml of an anti-human ANT goat polyclonal IgG antibody (sc-9300, Santa Cruz Biotechnology). The secondary antibody was a HRP (horseradish peroxidase)-conjugated donkey anti-goat IgG used at 0.08 μg/ml (sc-2020, Santa Cruz Biotechnology). The immunoblot was developed using a Lumigen® ECL® (enhanced chemiluminescence) Plus Western Blotting Detection system (Amersham Biosciences). Protein was quantified by analysing band intensities using ImageJ software (http://rsb.info.nih.gov/ij/). Membranes were stained with Gelcode Blue Stain Reagent (Pierce) to verify digestion of mitochondrial proteins by trypsin.
Chemicals
All chemicals were from Sigma–Aldrich and BDH except where otherwise stated.
Data analysis
Data are quoted as means±S.E.M. unless otherwise stated. Significance was tested by single factor ANOVA with Tukey's honestly significant difference post-hoc analysis. Values of *P<0.05 and **P<0.01 were considered significant.
RESULTS
Time-dependent uncoupling through ANT in energized rat liver mitochondria
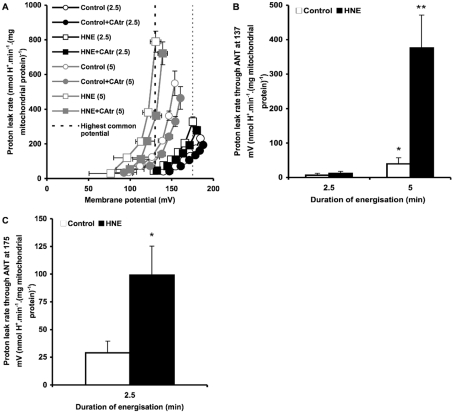
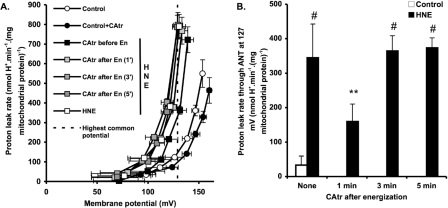
CAtr is a tight-binding inhibitor of ANT [31], so the CAtr-sensitive portion of uncoupling was used as a measure of proton leak mediated by ANT. Initial measurements of proton leak kinetics revealed that ANT-mediated proton conductance increased progressively after the mitochondria were energized by addition of succinate. This is shown in Figure 1(A), where at any given driving force (membrane potential) the proton leak rate under a variety of conditions was higher when mitochondria were energized with succinate for 5 min compared with for 2.5 min. Proton leak under these conditions at the highest common membrane potential (137 mV) is shown in Figure 1(B). ANT-mediated proton conductance increased more prominently in the presence of 35 μM HNE compared with in its absence (control). To show the effect of HNE at shorter times more clearly, Figure 1(C) shows the analysis at the highest membrane potential common to the 2.5 min data points (175 mV). Even with only 2.5 min of energization, proton leak through ANT was significantly greater in the presence of HNE than in the control. Subsequent experiments also using 2.5 min energization (Figures 2 and 3) gave proton conductances through ANT in the presence of HNE that were not significantly different from Figure 1(C).
Figure 1. Proton leak kinetics in rat liver mitochondria: energization-dependent activation.
(A) Rat liver mitochondria incubated in KPHEB medium plus 5 μM rotenone, 1 μg/ml oligomycin and 80 ng/ml nigericin ±2.5 μM CAtr ±35 μM HNE were energized with 4 mM succinate for 2.5 min (black symbols) or 5 min (grey symbols). To determine proton leak kinetics, oxygen consumption and membrane potential were measured as succinate oxidation was progressively inhibited by titration with malonate (up to 2.3 mM). (B) Proton leak rates through ANT at the highest potential common to all data sets [thick broken line in (A)]. Data are means±S.E.M. of at least five independent experiments. *P<0.05 and **P<0.01 compared with 2.5 min. (C) Proton leak rates through ANT at the highest common potential at 2.5 min of energization [fine broken line in (A)]. Data are means±S.E.M. of 11 independent experiments. *P<0.05 compared with control.
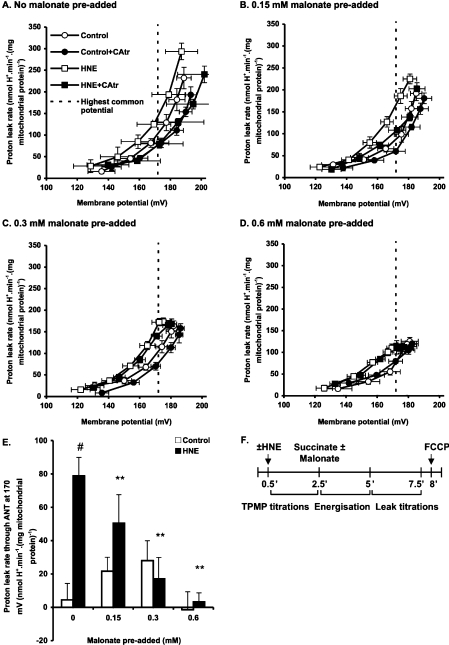
Figure 2. Effect of malonate pre-addition on ANT-mediated proton leak.
Rat liver mitochondria incubated in KPHEB plus 5 μM rotenone, 1 μg/ml oligomycin and 80 ng/ml nigericin ±2.5 μM CAtr ±35 μM HNE were energized with 4 mM succinate after pre-addition of different concentrations of malonate: (A) none added, (B) 0.15 mM, (C) 0.3 mM and (D) 0.6 mM added. After 2.5 min of energization, proton leak kinetics were determined by malonate titration (up to 2.3 mM). Data are means±S.E.M. of four independent experiments. (E) Proton leak through ANT at the highest common potential. Values are calculated as each condition minus its ‘plus CAtr’ condition for each independent experiment and subsequently averaged to give means±S.E.M. of four independent experiments. **P<0.01 compared with #. (F) Experiment time course. Zero time is the addition of 0.5 mg/ml mitochondria to KPHEB medium.
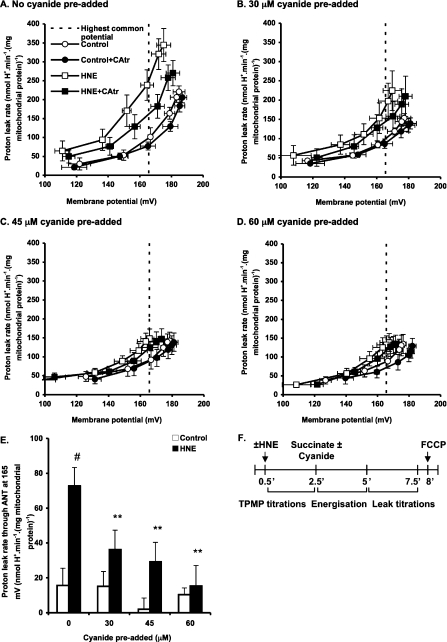
Figure 3. Effect of cyanide pre-addition on ANT-mediated proton leak.
Rat liver mitochondria incubated in KPHEB plus 5 μM rotenone, 1 μg/ml oligomycin and 80 ng/ml nigericin ±2.5 μM CAtr ±35 μM HNE were energized with 4 mM succinate after pre-addition of different concentrations of cyanide: (A) none added, (B) 30 μM, (C) 45 μM and (D) 60 μM added. After 2.5 min of energization, proton leak kinetics were determined by cyanide titration (up to 0.2 mM). Data are means±S.E.M. of four independent experiments. (E) Proton leak through ANT at the highest common potential. Data are calculated as each condition minus its ‘plus CAtr’ condition for each independent experiment and subsequently averaged to give means±S.E.M. of four independent experiments. **P<0.01 compared with #. (F) Experiment time course. Zero time is the addition of 0.5 mg/ml mitochondria to KPHEB medium.
It has previously been shown that the uncoupling effect of HNE is observable even at 1 μM HNE, but that 35 μM is required in the presence of BSA [6]. Although perhaps more physiologically relevant, the effect at 1 μM would be too small to allow investigation of its characteristics, particularly in the presence of BSA to minimize fatty-acid-mediated uncoupling through ANT, therefore HNE was used at 35 μM in all of the experiments in the present study.
The activation of ANT-mediated proton conductance was dependent upon the time between the initiation of energization and assay of proton leak, rather than on the total incubation time of mitochondria with or without HNE (results not shown). The effect of longer energization times on control proton leak are dealt with in more detail in [30], which also shows that increased proton leak is an energization-dependent effect and not a result of damage to mitochondria that are stirred for longer periods.
Although CAtr stabilizes the c-conformation of ANT, which is thought to promote opening of the mitochondrial permeability transition pore [40], our medium contained excess EGTA, which inhibits pore opening. In the present study, CAtr did not increase proton leak through ANT as would be the case with pore opening, but inhibited it, so proton leak did not involve pore opening in the results presented.
Thus the proton conductance of ANT increases with time of energization, particularly when HNE is present.
Membrane-potential-dependent uncoupling through ANT in energized rat liver mitochondria
To determine whether membrane potential was responsible for the energization-dependent activation of proton leak through ANT in a UCP-free model, we progressively decreased the state 4 membrane potential during energization by pre-addition of different concentrations of the succinate dehydrogenase inhibitor, malonate (Figures 2A–2D). Analysis at the highest common potential (170 mV) shows that in the presence of HNE, proton leak through ANT decreased at higher concentrations of malonate (Figure 2E). This was not the case in the absence of HNE (Figure 2E).
Since malonate additions not only lower membrane potential but also progressively oxidize the electron transport chain, we tested whether changes in redox state of the respiratory chain were responsible for the energization-dependent activation of proton leak through ANT. We decreased the membrane potential during energization by pre-addition of cyanide, a complex IV inhibitor that progressively reduces the electron transport chain (Figures 3A–3D). Cyanide pre-additions resulted in decreased proton conductance through ANT in the presence, but not the absence of HNE (Figure 3E), showing that the redox state of the electron transport chain was not an important factor.
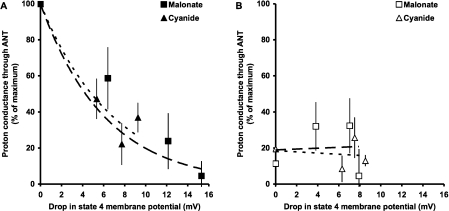
To make direct comparisons between the data in Figures 2 and 3, proton leak rates were normalized to the maximum rate on each day, then averaged. ANT-mediated proton leak rates at a fixed membrane potential were then plotted as a function of the drop in state 4 membrane potential during energization (Figure 4). The data sets followed the same pattern, suggesting that it is indeed membrane potential that is important for HNE-induced proton leak through ANT (Figure 4A). The loss of membrane potential did not affect the control (endogenous) proton conductance at fixed energization duration (Figure 4B), implying that the HNE-activated and endogenous proton leak pathways through ANT differ.
Figure 4. Relationship between ANT-mediated proton leak (at fixed membrane potential) and drop in state 4 membrane potential caused by pre-addition of malonate or cyanide.
Data from experiments shown in Figures 2 and 3 were first normalized to the maximum proton leak on each day, then averaged. (A) The presence of 35 μM HNE. Separate regression lines forced through 100% were fitted for malonate and cyanide. The parameters of the regressions were not significantly different between the data sets. Exponential fits were used on empirical grounds; linear fits also showed no difference. (B) HNE absent (control). Separate linear regression lines were fitted for malonate and cyanide. The parameters of the regressions were not significantly different between the data sets.
Thus the activation of proton conductance after 2.5 min through ANT caused by addition of HNE requires the presence of a high membrane potential, whereas that of the endogenous pathway does not.
Irreversibility of HNE-induced uncoupling through ANT
Figure 5 shows the reversibility of HNE-induced activation of ANT. CAtr was either added before energization with succinate, to prevent activation, or at 1, 3 or 5 min after energization, to test whether activation could be reversed. Proton leak kinetics were then measured 5 min after succinate addition (Figure 5A). Figure 5(B) shows ANT-dependent proton leak rates measured at the highest common potential of 127 mV, which were calculated by subtracting the rate with CAtr added at the start of the experiment. CAtr added 3 min or 5 min after energization with succinate did not reverse the HNE-induced activation of proton conductance through ANT, since the rates were the same as when no CAtr was added after energization. CAtr added 1 min after energization was partially inhibitory, presumably because it prevented further time-dependent activation by HNE rather than reversing activation that had already occurred.
Figure 5. Effect of CAtr on HNE-induced ANT activation.
(A) Rat liver mitochondria incubated in KPHEB plus 5 μM rotenone, 1 μg/ml oligomycin and 80 ng/ml nigericin ±35 μM HNE were energized with 4 mM succinate. Where indicated, 2.5 μM CAtr was added at the start of the experiment (CAtr before En), or 1 min, 3 min or 5 min after succinate addition (CAtr after En). After 5 min of energization (En), proton leak kinetics were determined by malonate titration (up to 2.3 mM). Values are means±S.E.M. of four independent experiments. (B) Proton leak rates through ANT at the highest common potential. Data are calculated as each condition minus its ‘plus CAtr (and before HNE)’ condition for each independent experiment and subsequently averaged to give means±S.E.M. of four independent experiments. For HNE data, **P<0.01 compared with #.
Thus the time-dependent increase in proton conductance of ANT caused by addition of HNE is prevented by pre-incubation with CAtr but is not reversed by subsequent addition of CAtr after 3 min of energization.
ANT conformation during activation of HNE-induced uncoupling through ANT
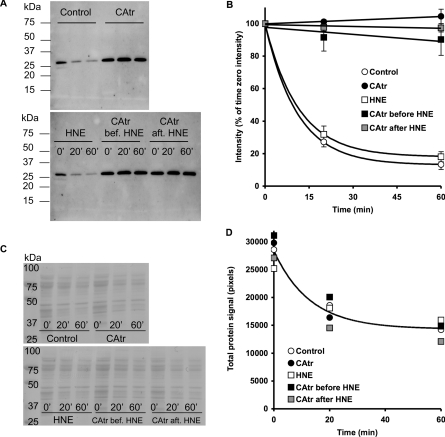
The lack of reversibility by CAtr of the effect of HNE on ANT-mediated proton leak raises two possibilities: either HNE modifies ANT conformation sufficiently to prevent CAtr binding, or CAtr still binds to ANT in the presence of HNE but no longer prevents proton leak through it. We investigated whether HNE modifies ANT conformation by using trypsin digestion (whereby the susceptibility of ANT trypsin digestion sites is affected by the conformation that ANT assumes under given conditions). Figures 6(A) and 6(B) show that exogenous trypsin rapidly degrades ANT in mitochondria, and CAtr is strongly protective [presumably by binding tightly and altering ANT conformation, since Coomassie Blue staining of membranes revealed that CAtr had no effect on the ability of trypsin to digest other mitochondrial proteins (Figures 6C and 6D)]. Addition of HNE did not alter the susceptibility of ANT to digestion by trypsin, and did not affect the ability of CAtr to protect (Figures 6A and 6B), suggesting that HNE does not modify ANT conformation sufficiently to prevent CAtr binding.
Figure 6. Effect of CAtr and HNE on ANT conformation as probed by digestion with trypsin.
Rat liver mitochondria were treated as for proton leak experiments; where indicated, 2.5 μM CAtr and 35 μM HNE were added. CAtr was added at the start of the experiment (CAtr and CAtr bef. HNE) or after HNE, 5 min after addition of succinate (CAtr aft. HNE). At the point where malonate titration would be carried out, mitochondria were inhibited with myxothiazol, and subsequently freeze–thawed to rupture membranes. Thawed mitochondria in KPHEB medium were treated with 0.25 mg/ml trypsin at 37 °C. At 0, 20 and 60 min after trypsin addition, aliquots were treated with 0.5 mg/ml trypsin inhibitor. Mitochondria were pelleted and proteins were separated by SDS/PAGE (12% gel), then immunoblotted for ANT. (A) Typical immunoblot of trypsin digestion experiment. Molecular mass markers (in kDa) are shown on the left-hand side of two typical immunoblots of samples from different incubation conditions. Times (0, 20 or 60 min) after trypsin addition are indicated. (B) Trypsin digestion profiles for different conditions. Data are means±S.E.M. of four independent experiments. Exponential regressions were fitted to control and HNE data sets (not significantly different from each other), and linear regression lines to the remaining data sets (not significantly different from each other), all forced through 100%. (C) Nitrocelluose membranes treated with Gelcode Blue Stain Reagent after immunoblotting in (A). (D) Densitometric analysis of all bands in each lane in (C). Exponential regression analysis showed that the parameters of the curves for different conditions were not significantly different from each other.
Thus treatment with HNE does not prevent CAtr from protecting ANT against degradation by trypsin, suggesting that CAtr still binds to ANT in the presence of HNE even though it no longer prevents proton leak through ANT.
No effect of HNE on state 3 respiration rate or CAtr titre
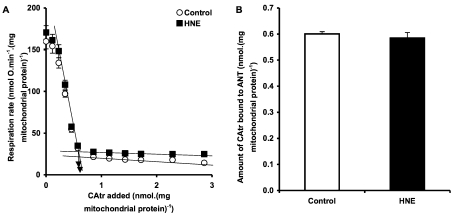
To investigate further whether CAtr still binds to ANT in the presence of HNE, we titrated state 3 respiration with CAtr, with or without HNE pre-treatment (Figure 7). HNE did not affect the ability of low concentrations of CAtr to inhibit state 3 respiration (Figure 7A), suggesting that HNE did not alter binding of CAtr. This conclusion was confirmed by assaying the amount of CAtr required to fully inhibit state 3 to the state 4 rate: CAtr titre was unchanged in HNE-treated mitochondria, showing that CAtr still binds tightly and inhibits adenine nucleotide transport after pre-treatment with HNE (Figure 7B).
Figure 7. CAtr titre of respiration.
(A) Rat liver mitochondria incubated in KPHEB plus 5 μM rotenone ±35 μM HNE were energized with 4 mM succinate for 5 min, then state 3 respiration was achieved by addition of 600 μM ADP. State 3 was titrated by successive additions of CAtr to bring mitochondria into a CAtr -inhibited state 4. (B) Amount of CAtr required to fully titrate state 3 to state 4, calculated from the intersection of linear regressions as shown in (A). Data are means±S.E.M. of four independent experiments. Data sets were not significantly different.
Thus treatment with HNE does not prevent CAtr from binding tightly to ANT and inhibiting adenine nucleotide transport, even though treatment with HNE prevents CAtr from inhibiting proton leak through ANT. This suggests that the proton conductance and nucleotide translocation pathways in ANT are behaviourally and perhaps physically distinct.
DISCUSSION
In the present study, we used ANT as a model for understanding the factors involved in HNE-induced mitochondrial uncoupling. The results may also be relevant to the activation of UCPs by HNE [6]. Our results show that energization, by generating a high membrane potential, plays an important role in the ability of HNE to activate ANT-mediated proton conductance, and that increased time at high membrane potential results in greater uncoupling. This time-dependent activation appears to be irreversible over the short term by subsequent CAtr addition. The results of the present study also imply that the proton conductance pathway is distinct from the nucleotide translocation pathway in ANT.
We find that after 2.5 min energization, the HNE-induced pathway behaves differently from the endogenous pathway in being membrane-potential-dependent. In light of results in [30], this may be because the endogenous pathway has not been sufficiently activated at 2.5 min in liver, or it may be explained by tissue-specific differences. In liver, time at high membrane potential in the presence of HNE results in a striking increase in proton conductance, which may be interpreted in two ways. First, HNE may slowly form covalent adducts with ANT, progressively converting ANT into a form with high proton conductance, or secondly, HNE metabolism may progressively form fatty acids [41], which then activate proton conductance [28]. The lack of reversibility of HNE-induced proton conductance by CAtr (Figure 5) is unlike fatty-acid-mediated activation, which is known to be both prevented and reversed by CAtr [11,12]; this shows that the effect of HNE is not mediated indirectly by fatty acids. Indeed, it implies that HNE modifies ANT permanently, perhaps by formation of covalent adducts [25–27]. HNE is known to form Michael adducts with sulfhydryl groups and lysine or histidine residues of proteins [42]. We therefore propose that high membrane potential influences the conformation of ANT or its lipid milieu [43,44], allowing HNE to interact with ANT and activate ANT-mediated proton conductance. Conformational changes may involve a transition of ANT from the c-state to the m-state, which unmasks the sulfhydryl groups [45] and lysine residues [46–48] of ANT, rendering them susceptible to attack by HNE. Work by Aquila et al. [49] on purified and reconstituted ANT shows that the carrier is protected from trypsin digestion by CAtr addition, but rendered susceptible by bongkrekate addition. Their ANT digestion profiles are consistent with those that we see in mitochondria that are treated (or not) with CAtr. Furthermore, Vignais and Vignais [50] report that the uncoupler FCCP prevents alkylation of ANT sulfhydryl groups, presumably by preventing a c-state to m-state transition of the carrier. In much the same way, decreasing membrane potential during energization (Figure 4) might prevent HNE interaction with these groups. Similarly, pre-incubation with CAtr would fix the c-state and prevent HNE adduct formation.
Klingenberg et al. [31] show that pre-treatment with bongkrekate induces the m-state and prevents binding of both atractylate and CAtr, but when added subsequently it only removes atractylate. In contrast, the finding in the present study that CAtr can still bind to ANT after HNE treatment indicates that HNE does not behave like bongkrekate to permanently stabilize the m-conformation, but allows the carrier to revert to the c-state. Also, in contrast with the study by Vignais and Vignais [50] in which sulfhydryl group alkylation prevented nucleotide translocation and atractylate binding, and the study by Chen et al. [26] in which HNE inhibited the nucleotide translocation activity of ANT by sulfhydryl group alkylation, HNE affected neither parameter in the system used in the present study (Figure 7). It is likely that the extent of sulfhydryl group modification in our system is considerably less than in [50] and [26], which used a potent alkylating agent (N-ethylmaleimide) and millimolar concentrations of HNE respectively.
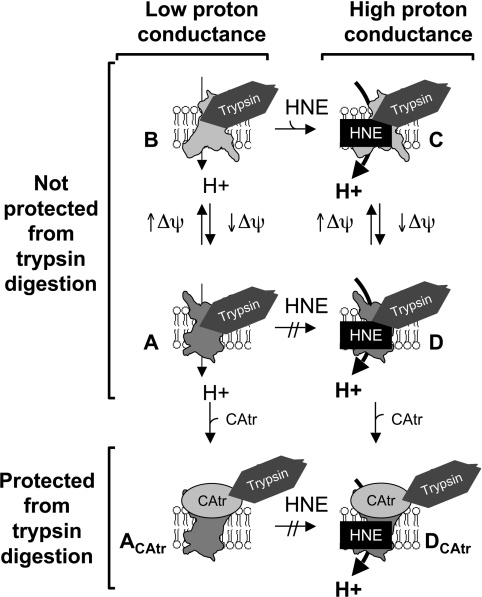
The simplest model to explain the observations in the present study (Figure 8) is that high membrane potential favours the m-state of ANT, which is required for HNE to react. Once HNE has reacted, proton translocation is permanently switched on and insensitive to later CAtr addition, but the carrier still translocates nucleotides, cycles between the c-state and the m-state, and can be trapped in the c-state by CAtr, preventing further nucleotide transport but permitting proton transport.
Figure 8. Proposed ANT conformation under different conditions.
At high membrane potential (Δψ), the c-like state (A) transitions towards an m-like state (B) thereby exposing reactive sulfydryl groups. If HNE is present, the m-like state can slowly be modified by adduct formation to a form with a higher proton conductance (C). ANT–HNE can then cycle between m-like (C) and c-like (D) states, allowing normal nucleotide transport. The c-like states (A and D) may still be trypsin-sensitive as de-energized mitochondria are degraded with similar profiles as shown in Figure 6 (V. Azzu and M. D. Brand, unpublished work). CAtr binding induces a conformational change in ANT (ACAtr, DCAtr), which protects it from trypsin digestion and prevents adenine nucleotide translocation. The fixed c-state (ACAtr) does not react with HNE but can accommodate pre-reacted HNE (DCAtr), in which case it retains high proton conductance. For further details see the Results and Discussion sections.
An alternative hypothesis is that ANT is required for activation of uncoupling, but the HNE-induced proton conductance pathway does not subsequently require ANT. This may explain why pre-addition of CAtr prevents proton leak induction, whereas its addition after HNE makes no difference to proton leak even though it can still inhibit nucleotide translocation by the carrier.
Acknowledgments
Support from the Medical Research Council, the Wellcome Trust (grants 065326/Z/01/Z and 066750/B/01/Z) (N. P., M. D. B.), and the School of Clinical Medicine, University of Cambridge, Cambridge, U.K. (to V. A.) is gratefully acknowledged.
References
- 1.Dalle-Donne I., Aldini G., Carini M., Colombo R., Rossi R., Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell Mol. Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry G., Castellani R. J., Hirai K., Smith M. A. Reactive oxygen species mediate cellular damage in Alzheimers disease. J. Alzheimers Dis. 1998;1:45–55. doi: 10.3233/jad-1998-1103. [DOI] [PubMed] [Google Scholar]
- 3.Jeremy J. Y., Shukla N., Muzaffar S., Handley A., Angelini G. D. Reactive oxygen species, vascular disease and cardiovascular surgery. Curr. Vasc. Pharmacol. 2004;2:229–236. doi: 10.2174/1570161043385691. [DOI] [PubMed] [Google Scholar]
- 4.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 5.Echtay K. S., Roussel D., St-Pierre J., Jekabsons M. B., Cadenas S., Stuart J. A., Harper J. A., Roebuck S. J., Morrison A., Pickering S., et al. Superoxide activates mitochondrial uncoupling proteins. Nature. 2002;415:96–99. doi: 10.1038/415096a. [DOI] [PubMed] [Google Scholar]
- 6.Echtay K. S., Esteves T. C., Pakay J. L., Jekabsons M. B., Lambert A. J., Portero-Otin M., Pamplona R., Vidal-Puig A. J., Wang S., Roebuck S. J., Brand M. D. A signalling role for 4-hydroxy-2-nonenal in regulation of mitochondrial uncoupling. EMBO J. 2003;22:4103–4110. doi: 10.1093/emboj/cdg412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLennan H. R., Degli Esposti M. The contribution of mitochondrial respiratory complexes to the production of reactive oxygen species. J. Bioenerg. Biomembr. 2000;32:153–162. doi: 10.1023/a:1005507913372. [DOI] [PubMed] [Google Scholar]
- 8.Lenaz G. The mitochondrial production of reactive oxygen species: mechanisms and implications in human pathology. IUBMB Life. 2001;52:159–164. doi: 10.1080/15216540152845957. [DOI] [PubMed] [Google Scholar]
- 9.Korshunov S. S., Skulachev V. P., Starkov A. A. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 10.Skulachev V. P. Role of uncoupled and non-coupled oxidations in maintenance of safely low levels of oxygen and its one-electron reductants. Q. Rev. Biophys. 1996;29:169–202. doi: 10.1017/s0033583500005795. [DOI] [PubMed] [Google Scholar]
- 11.Andreyev A., Bondareva T. O., Dedukhova V. I., Mokhova E. N., Skulachev V. P., Tsofina L. M., Volkov N. I., Vygodina T. V. The ATP/ADP-antiporter is involved in the uncoupling effect of fatty acids on mitochondria. Eur. J. Biochem. 1989;182:585–592. doi: 10.1111/j.1432-1033.1989.tb14867.x. [DOI] [PubMed] [Google Scholar]
- 12.Andreyev A., Bondareva T. O., Dedukhova V. I., Mokhova E. N., Skulachev V. P., Volkov N. I. Carboxyatractylate inhibits the uncoupling effect of free fatty acids. FEBS Lett. 1988;226:265–269. doi: 10.1016/0014-5793(88)81436-4. [DOI] [PubMed] [Google Scholar]
- 13.Samartsev V. N., Smirnov A. V., Zeldi I. P., Markova O. V., Mokhova E. N., Skulachev V. P. Involvement of aspartate/glutamate antiporter in fatty acid-induced uncoupling of liver mitochondria. Biochim. Biophys. Acta. 1997;1319:251–257. doi: 10.1016/s0005-2728(96)00166-1. [DOI] [PubMed] [Google Scholar]
- 14.Locke R. M., Rial E., Scott I. D., Nicholls D. G. Fatty acids as acute regulators of the proton conductance of hamster brown-fat mitochondria. Eur. J. Biochem. 1982;129:373–380. doi: 10.1111/j.1432-1033.1982.tb07060.x. [DOI] [PubMed] [Google Scholar]
- 15.Wojtczak L., Schonfeld P. Effect of fatty acids on energy coupling processes in mitochondria. Biochim. Biophys. Acta. 1993;1183:41–57. doi: 10.1016/0005-2728(93)90004-y. [DOI] [PubMed] [Google Scholar]
- 16.Skulachev V. P. Fatty acid circuit as a physiological mechanism of uncoupling of oxidative phosphorylation. FEBS Lett. 1991;294:158–162. doi: 10.1016/0014-5793(91)80658-p. [DOI] [PubMed] [Google Scholar]
- 17.Winkler E., Klingenberg M. Effect of fatty acids on H+ transport activity of the reconstituted uncoupling protein. J. Biol. Chem. 1994;269:2508–2515. [PubMed] [Google Scholar]
- 18.Rial E., Poustie A., Nicholls D. G. Brown-adipose-tissue mitochondria: the regulation of the 32000-Mr uncoupling protein by fatty acids and purine nucleotides. Eur. J. Biochem. 1983;137:197–203. doi: 10.1111/j.1432-1033.1983.tb07815.x. [DOI] [PubMed] [Google Scholar]
- 19.Shabalina I. G., Jacobsson A., Cannon B., Nedergaard J. Native UCP1 displays simple competitive kinetics between the regulators purine nucleotides and fatty acids. J. Biol. Chem. 2004;279:38236–38248. doi: 10.1074/jbc.M402375200. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Barroso M. M., Fleury C., Bouillaud F., Nicholls D. G., Rial E. The uncoupling protein UCP1 does not increase the proton conductance of the inner mitochondrial membrane by functioning as a fatty acid anion transporter. J. Biol. Chem. 1998;273:15528–15532. doi: 10.1074/jbc.273.25.15528. [DOI] [PubMed] [Google Scholar]
- 21.Lou P. H., Hansen B. S., Olsen P. H., Tullin S., Murphy M. P., Brand M. D. Mitochondrial uncouplers with an extraordinary dynamic range. Biochem. J. 2007;407:129–140. doi: 10.1042/BJ20070606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porter R. K. A new look at UCP 1. Biochim. Biophys. Acta. 2006;1757:474–479. doi: 10.1016/j.bbabio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Cannon B., Shabalina I. G., Kramarova T. V., Petrovic N., Nedergaard J. Uncoupling proteins: a role in protection against reactive oxygen species-or not? Biochim. Biophys. Acta. 2006;1757:449–458. doi: 10.1016/j.bbabio.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Brand M. D., Affourtit C., Esteves T. C., Green K., Lambert A. J., Miwa S., Pakay J. L., Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radical Biol. Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 25.Yan L. J., Sohal R. S. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12896–12901. doi: 10.1073/pnas.95.22.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J. J., Bertrand H., Yu B. P. Inhibition of adenine nucleotide translocator by lipid peroxidation products. Free Radical Biol. Med. 1995;19:583–590. doi: 10.1016/0891-5849(95)00066-7. [DOI] [PubMed] [Google Scholar]
- 27.Choksi K. B., Boylston W. H., Rabek J. P., Widger W. R., Papaconstantinou J. Oxidatively damaged proteins of heart mitochondrial electron transport complexes. Biochim. Biophys. Acta. 2004;1688:95–101. doi: 10.1016/j.bbadis.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Shabalina I. G., Petrovic N., Kramarova T. V., Hoeks J., Cannon B., Nedergaard J. UCP1 and defense against oxidative stress. 4-Hydroxy-2-nonenal effects on brown fat mitochondria are uncoupling protein 1-independent. J. Biol. Chem. 2006;281:13882–13893. doi: 10.1074/jbc.M601387200. [DOI] [PubMed] [Google Scholar]
- 29.Parker N., Vidal-Puig A. J., Brand M. D. Stimulation of mitochondrial proton conductance by hydroxynonenal requires a high membrane potential. Biosci. Rep. 2008;28:83–88. doi: 10.1042/BSR20080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker N., Affourtit C., Vidal-Puig A. J., Brand M. D. Energization-dependent endogenous activation of proton conductance in skeletal muscle mitochondria. Biochem. J. 2008;412:131–139. doi: 10.1042/BJ20080006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klingenberg M., Grebe K., Scherer B. The binding of atractylate and carboxy-atractylate to mitochondria. Eur. J. Biochem. 1975;52:351–363. doi: 10.1111/j.1432-1033.1975.tb04003.x. [DOI] [PubMed] [Google Scholar]
- 32.Chappell J., Hansford R. Preparation of mitochondria from animal tissue and yeast. In: Birnie G., editor. Subcellular components: preparation and fractionation. London: Butterworths; 1972. pp. 77–91. [Google Scholar]
- 33.Brand M. D. Measurement of mitochondrial protonmotive force. Bioenergetics. In: Brown G. C., Cooper C. E., editors. A practical approach. Oxford: IRL Press; 1995. pp. 39–62. [Google Scholar]
- 34.Larrouy D., Laharrague P., Carrera G., Viguerie-Bascands N., Levi-Meyrueis C., Fleury C., Pecqueur C., Nibbelink M., Andre M., Casteilla L., Ricquier D. Kupffer cells are a dominant site of uncoupling protein 2 expression in rat liver. Biochem. Biophys. Res. Commun. 1997;235:760–764. doi: 10.1006/bbrc.1997.6852. [DOI] [PubMed] [Google Scholar]
- 35.Lee F. Y., Li Y., Zhu H., Yang S., Lin H. Z., Trush M., Diehl A. M. Tumor necrosis factor increases mitochondrial oxidant production and induces expression of uncoupling protein-2 in the regenerating rat liver. Hepatology. 1999;29:677–687. doi: 10.1002/hep.510290320. [DOI] [PubMed] [Google Scholar]
- 36.Carretero M. V., Torres L., Latasa U., Garcia-Trevijano E. R., Prieto J., Mato J. M., Avila M. A. Transformed but not normal hepatocytes express UCP2. FEBS Lett. 1998;439:55–58. doi: 10.1016/s0014-5793(98)01335-0. [DOI] [PubMed] [Google Scholar]
- 37.Hodny Z., Kolarova P., Rossmeisl M., Horakova M., Nibbelink M., Penicaud L., Casteilla L., Kopecky J. High expression of uncoupling protein 2 in foetal liver. FEBS Lett. 1998;425:185–190. doi: 10.1016/s0014-5793(98)00230-0. [DOI] [PubMed] [Google Scholar]
- 38.Harper J. A., Stuart J. A., Jekabsons M. B., Roussel D., Brindle K. M., Dickinson K., Jones R. B., Brand M. D. Artifactual uncoupling by uncoupling protein 3 in yeast mitochondria at the concentrations found in mouse and rat skeletal-muscle mitochondria. Biochem. J. 2002;361:49–56. doi: 10.1042/0264-6021:3610049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 40.Haworth R. A., Hunter D. R. Control of the mitochondrial permeability transition pore by high-affinity ADP binding at the ADP/ATP translocase in permeabilized mitochondria. J. Bioenerg. Biomembr. 2000;32:91–96. doi: 10.1023/a:1005568630151. [DOI] [PubMed] [Google Scholar]
- 41.Ullrich O., Grune T., Henke W., Esterbauer H., Siems W. G. Identification of metabolic pathways of the lipid peroxidation product 4-hydroxynonenal by mitochondria isolated from rat kidney cortex. FEBS Lett. 1994;352:84–86. doi: 10.1016/0014-5793(94)00922-8. [DOI] [PubMed] [Google Scholar]
- 42.Aldini G., Dalle-Donne I., Facino R. M., Milzani A., Carini M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Med. Res. Rev. 2007;27:817–868. doi: 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]
- 43.Beyer K., Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry (Moscow) 1985;24:3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- 44.Hoffmann B., Stockl A., Schlame M., Beyer K., Klingenberg M. The reconstituted ADP/ATP carrier activity has an absolute requirement for cardiolipin as shown in cysteine mutants. J. Biol. Chem. 1994;269:1940–1944. [PubMed] [Google Scholar]
- 45.Aquila H., Klingenberg M. The reactivity of -SH groups in the ADP/ATP carrier isolated from beef heart mitochondria. Eur. J. Biochem. 1982;122:141–145. doi: 10.1111/j.1432-1033.1982.tb05859.x. [DOI] [PubMed] [Google Scholar]
- 46.Klingenberg M., Appel M. Is there a binding center in the ADP, ATP carrier for substrate and inhibitors? Amino acid reagents and the mechanism of the ADP, ATP translocator. FEBS Lett. 1980;119:195–199. doi: 10.1016/0014-5793(80)81029-5. [DOI] [PubMed] [Google Scholar]
- 47.Block M. R., Lauquin G. J., Vignais P. V. Chemical modifications of atractyloside and bongkrekic acid binding sites of the mitochondrial adenine nucleotide carrier. Are there distinct binding sites? Biochemistry (Moscow) 1981;20:2692–2699. doi: 10.1021/bi00512a051. [DOI] [PubMed] [Google Scholar]
- 48.Klingenberg M. Molecular aspects of the adenine nucleotide carrier from mitochondria. Arch. Biochem. Biophys. 1989;270:1–14. doi: 10.1016/0003-9861(89)90001-5. [DOI] [PubMed] [Google Scholar]
- 49.Aquila H., Eiermann W., Babel W., Klingenberg M. Isolation of the ADP/ATP translocator from beef heart mitochondria as the bongkrekate-protein complex. Eur. J. Biochem. 1978;85:549–560. doi: 10.1111/j.1432-1033.1978.tb12270.x. [DOI] [PubMed] [Google Scholar]
- 50.Vignais P. V., Vignais P. M. Effect of SH reagents on atractyloside binding to mitochondria and ADP translocation. Potentiation by ADP and its prevention by uncoupler FCCP. FEBS Lett. 1972;26:27–31. doi: 10.1016/0014-5793(72)80534-9. [DOI] [PubMed] [Google Scholar]