Abstract
Nipah virus (NiV) and Hendra virus are the type species of the highly pathogenic paramyxovirus genus Henipavirus, which can cause severe respiratory disease and fatal encephalitis infections in humans, with case fatality rates approaching 75%. NiV contains two envelope glycoproteins, the receptor-binding G glycoprotein (NiV-G) that facilitates attachment to host cells and the fusion (F) glycoprotein that mediates membrane merger. The henipavirus G glycoproteins lack both hemagglutinating and neuraminidase activities and, instead, engage the highly conserved ephrin-B2 and ephrin-B3 cell surface proteins as their entry receptors. Here, we report the crystal structures of the NiV-G both in its receptor-unbound state and in complex with ephrin-B3, providing, to our knowledge, the first view of a paramyxovirus attachment complex in which a cellular protein is used as the virus receptor. Complex formation generates an extensive protein–protein interface around a protruding ephrin loop, which is inserted in the central cavity of the NiV-G β-propeller. Analysis of the structural data reveals the molecular basis for the highly specific interactions of the henipavirus G glycoproteins with only two members (ephrin-B2 and ephrin-B3) of the very large ephrin family and suggests how they mediate in a unique fashion both cell attachment and the initiation of membrane fusion during the virus infection processes. The structures further suggest that the NiV-G/ephrin interactions can be effectively targeted to disrupt viral entry and provide the foundation for structure-based antiviral drug design.
Keywords: crystallography, viral attachment
The recently emerged Nipah virus (NiV) is an enveloped, negative-sense single-stranded RNA paramyxovirus that, along with the closely related Hendra virus (HeV), is the type species of the genus Henipavirus. Both NiV and HeV have an unusual broad species tropism, are highly pathogenic in a variety of vertebrate animals including humans, and have been given biosecurity level 4 status (1). Since their initial discovery in Australia and Malaysia (2, 3), sporadic HeV spillover events have been reported from 1995 to 2007 (4); however, NiV outbreaks have occurred on a regular basis in Bangladesh and India, with human case fatality rates approaching 75% (5–7). Both serologic and virologic studies have demonstrated that the natural reservoirs for HeV and NiV are several species of large fruit bats commonly referred to as flying foxes in the genus Pteropus (8).
NiV contains two membrane anchored glycoproteins within their envelope, the receptor-binding G glycoprotein (G) and the fusion (F) glycoprotein. The G glycoprotein is a type II membrane protein containing 602-aa residues and, in contrast to most other well characterized paramyxoviruses, lacks hemagglutinating and neuraminidase activities and does not bind to carbohydrate moieties (2, 3). The main role of NiV-G is to recognize and attach the virus to receptors within the host cell membrane, but it also facilitates the F-mediated membrane fusion process via an as yet undefined mechanism that is initiated through binding to its cognate receptor. Ephrin-B2 and ephrin-B3, which were recently identified as the cellular receptors for both NiV and HeV (9–12), are members of a large family of cell surface proteins that bind to the Eph receptors, the largest subgroup of receptor tyrosine kinase (13, 14) and, along with their ephrin partners, mediate bidirectional signaling during a variety of cell–cell interactions (15, 16). The identification of these broadly expressed and highly conserved molecules as the major receptors for the henipaviruses has aided in the appreciation and explanation of their broad species and tissue tropisms, as well as the resultant pathogenic processes seen in both humans and animal hosts (17). The Ephs and the ephrins are divided into two subclasses, A and B, based on their affinities for each other and on sequence conservation (18). All ephrins contain a 20-kDa conserved extracellular Eph-binding domain, which is also recognized by the henipavirus G glycoproteins.
Results and Discussion
Structure of NiV-G.
NiV-G contains a C-terminal globular head that extends from the viral membrane on a stalk (19). The recombinant NiV-G head region was functionally active in receptor binding, as judged by in vitro binding assays (Methods) and was monomeric both in solution and in the crystals. NiV-G (Fig. 1) had an overall disk-like shape, with dimensions of ≈55 × 55 × 45 Å. The fold was that of a β-propeller with six blades surrounding a central cavity, overall similar to that of structurally characterized hemagglutinin–neuraminidase (HN) viral attachment glycoproteins (19). Each of the six blade modules (B1 to B6) contained a four-stranded (strands S1 to S4) antiparallel β-sheet. Like most of the other known propeller structures, NiV-G uses a “Velcro” system to close the circle between the first and the last blades (20, 21). The NiV-G Velcro was further enhanced by a disulfide bond (C181–C601) connecting the N and C termini of the β-propeller. An unusually large number of disulfide bonds stabilized the structure with one within each of the six blades and one connecting blades B3 and B4. For comparison, the other paramyxovirus subfamilies contain two to five disulfide bonds in the head region of their attachment proteins (22).
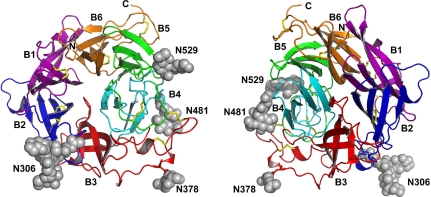
Fig. 1.
Crystal structure of the globular head region of NiV-G. (Left) View of the top (ephrin-binding) face of the molecule. (Right) View of the bottom face. The individual β-propellers are shown in different colors and are labeled. The carbohydrate modifications are in gray and the disulfide bridges are in yellow.
N-linked glycosylation is known to facilitate viral G protein folding and stabilization and NiV-G contains five potential N-linked glycosylation sites. Of these, we observed glycan electron density for N306, N378, N481, and N529 (N417 is not glycosylated as predicted). All carbohydrate modifications were extending in the solvent and did not interact with the protein. This was in contrast to the structure of the measles virus (MeV) attachment protein (H), where the N-linked glycans interact with the top face of the protein β-propeller and are suggested to function in blocking the sialic-acid binding site (22–24).
A comparison of the NiV-G structure with the contents of the FSSP database (25) revealed that the closest structural homologs of NiV-G were the HN glycoproteins from human parainfluenza virus type III (hPIV3) (26), parainfluenza virus 5 (SV5) (27), and Newcastle disease virus (NDV) (28). The β-propeller domains of these proteins could be superimposed on NiV-G, with rmsd between α carbon positions of ≈2.2 Å (for selected ≈370 of the 425 β-propeller residues, sharing ≈20% sequence identity). Despite this overall structural homology, NiV-G significantly diverges from the attachment glycoproteins of other paramyxoviruses and possesses neither hemagglutinating nor neuraminidase activities (2, 3). Indeed the henipaviruses are the only known Paramyxovirinae subfamily members that have no carbohydrate-binding activity and have instead developed the ability to bind host cell protein receptors (ephrin-B2 and B3).
Overall Structure of the NiV-G/Ephrin-B3 Complex.
Gel filtration and analytical ultracentrifugation experiments indicated that the interacting regions of NiV-G and ephrin-B3, in the absence of the NiV-G stalk, bind each other with a 1:1 stoichiometry. Indeed, the crystal structure of their complex revealed a heterodimeric G protein/receptor assembly (Fig. 2A). Ephrin-B3 bound at the center of the top face of the NiV-G β-propeller, interacting with several of the loops connecting the propeller β-strands. Consequently, the NiV-G/ephrin-B3 complex had an elongated shape, with overall dimensions of 75 × 55 × 55 Å. The C terminus of ephrin-B3, which points toward the cellular membrane of the host cell, and the N terminus of NiV-G, which connects to the stalk region, were located on the opposite sides of the complex. There were two copies of the complex in the asymmetric unit of the crystal, which had an rmsd between equivalent α carbon positions of 0.5 Å and only differed in the conformation of a flexible surface NiV-G loop (B1H1–B1S1).
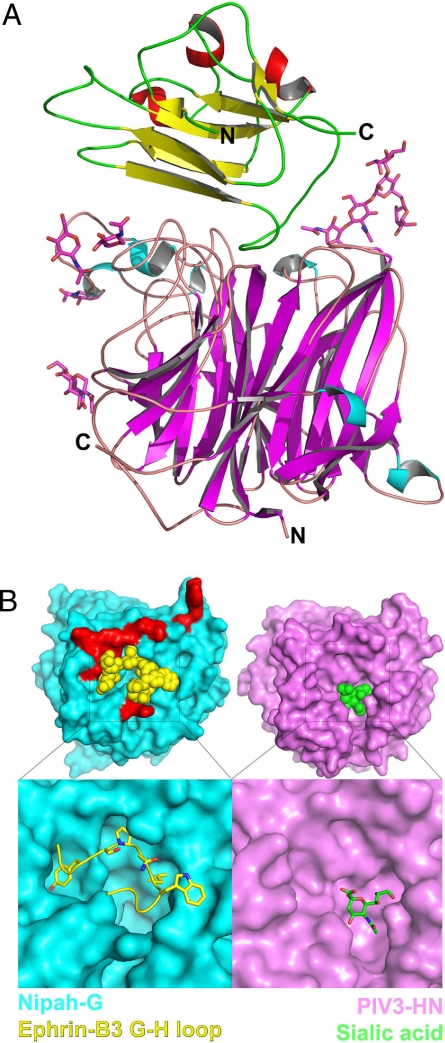
Fig. 2.
Crystal structure of the NiV-G/ephrin-B3 complex. (A) Side view of the NiV-G/ephrin-B3 complex. The β-strands of NiV-G are colored in magenta, and the α-helices are in cyan. The β-strands of ephrin-B3 are colored in yellow and the α-helices are in red. The carbohydrate moieties, shown as stick models, do not interact with ephrin-B3 but extend in the solvent. The N and C termini of the molecules are labeled. (B) The molecular surfaces of the henipavirus (cyan) and the parainfluenza virus (magenta) attachment proteins along the top (or receptor-binding) face of the molecules. The lower images are close-up views of the receptor-binding pockets with the bound receptor (ephrin-B3 G–H loop in yellow, sialic acid in green). Only the G–H loop of ephrin-B3 is shown. In red are shown the NiV-G residues that interact with ephrin-B3 residues outside of the G–H loop, highlighting the polar region of the NiV-G/ephrin interface.
Structure of NiV-G in the Complex.
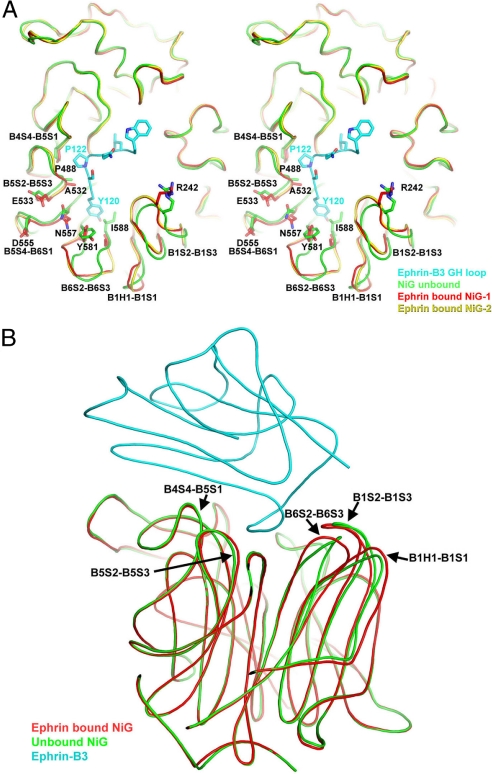
The overall structure of the ephrin-binding region of NiV-G in the complex (Fig. 3A) was very similar to that of the unbound protein (Fig. 3B). The significant conformational changes in the attachment protein involved loops at the protein–protein interface. Most notable were the reorganizations of the B6S2–B6S3, B1H1–B1S1, and B1S2–B1S3 loops, which moved by 3 Å or more, as well as the more subtle adjustments in the B4S4–B5S1, B5S2–B5-S3, and B5S4–B6S1 loops (see Fig. 3 and discussion further below).
Fig. 3.
Structural rearrangements in NiV-G upon ephrin binding. (A) A view of the top face of NiV-G. The ephrin G–H loop is in cyan, the unbound NiV-G is in green, and the two-asymmetric-unit copies of the ephrin-bound NiV-G are in red and yellow. The regions that are structurally different in the bound and free proteins are labeled. (B) A side view of the NiV-G/ephrin complex with superimposed structure of the unbound NiV-G in green. The NiV-G loops, which are structurally different in the bound and free molecules, are labeled.
Structure of Ephrin-B3.
The NiV-G/ephrin-B3 complex described here also provides, to our knowledge, the first structure of ephrin-B3. Structures of several other ephrin family members, including ephrin-B1, -B2, and -A5, have been reported previously, both alone and in complexes with their corresponding Eph receptors (15). Ephrin-B3 shares significant sequence homology (≈40% amino acid identity) with ephrin-B1 and -B2, and supporting information (SI) Figs. S1B and S2 show the sequence alignment and the organization of secondary structure elements of the three B class ephrins. As anticipated, the overall structure of ephrin-B3 was very similar to those of ephrin-B1 and -B2 (29), and these could be superimposed with rmsd values between equivalent α carbon positions of ≈1.5 Å (Fig. 4B).
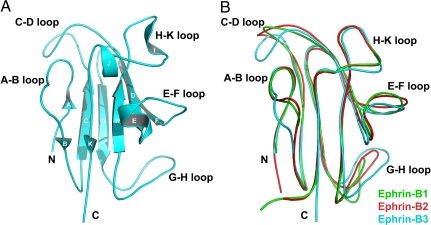
Fig. 4.
Structure of the henipavirus cellular receptor, ephrin-B3. (A) Structure of ephrin-B3. Secondary structure elements, as well as the N and C termini of the molecule, are labeled. (B) Superimposition of the structures of ephrin-B1 (green), ephrin-B2 (red), and ephrin-B3 (cyan). The structurally variable loops are labeled.
Notably, the most structurally distinct region of the ephrins is their Eph-binding (G–H) loop (reviewed in ref. 18), which is also used by ephrin-B3 to bind the NiV-G glycoprotein. In most ephrin structures, it has an extended architecture, but in some, such as ephrin-B1, it is more flexible (29) (Fig. 4B). The different conformations of the G–H loop suggest that it could undergo structural readjustments upon binding to any respective protein partner. Interestingly, ephrin-B2 and -B3, which serve as the henipavirus cellular receptors, seem to have more rigid G–H loop conformations (30), whereas ephrin-B1, which does not bind the henipavirus G glycoproteins, has a more flexible loop conformation.
Interestingly, whereas all other structurally characterized ephrins contain N-linked glycosylation, we could not detect any glycosylation in ephrin-B3. It has been suggested that glycosylation might enhance the Eph/ephrin interactions and the fact that ephrin-B3 lacks such modification could explain its lower binding affinity to most B class Ephs, including EphB1, EphB2, and EphB4 (13).
Ligand–Receptor Interface.
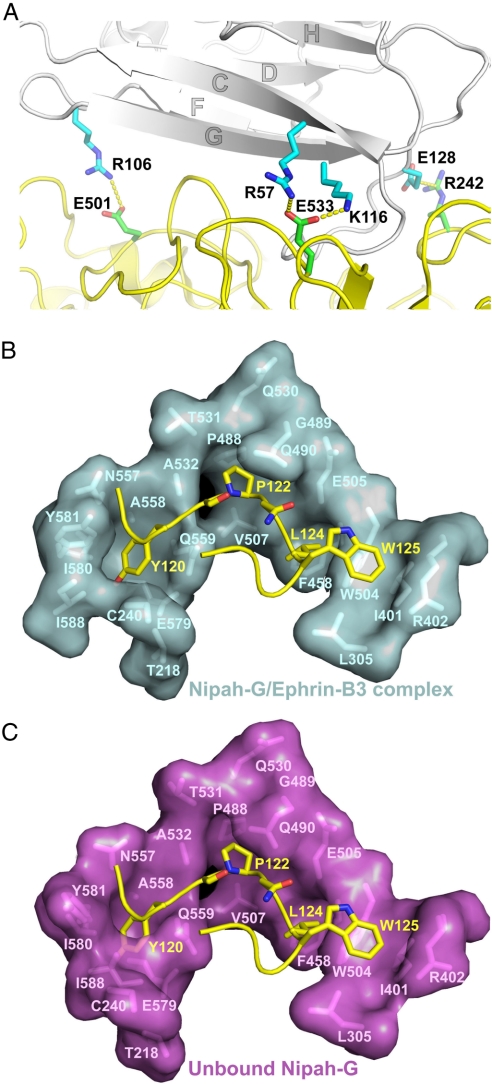
Ephrin-B3 bound along the upper face of the NiV-G β-propeller. The attachment interface was large and continuous, burying ≈2,600 Å2 of molecular surface. The interface could be divided into two regions. The first region encompassed the ephrin-docking site toward the outer rim of the β-propeller (red in Fig. 2B) and was mostly polar, relying on hydrogen-bond networks at solvent-excluded regions for binding and recognition. The interacting residues at the surface of the ephrin-B3 β sandwich came from three β-strands, C, F, and G, and the connecting F–G and G–H loops (Fig. 5B and Fig. S1). They interacted with several of the extended outer NiV-G loops, including B1S2–B1S3, B3H2–B3H3, B4S4–B5S1, B5S2–B5S3, and B5S4–B6S1. This area contained four salt bridges, including ephrin R57, R106, and E128, interacting with NiV-G E533, E501, and R242, respectively, as well as ephrin K116, interacting with both E533 and D555 of NiV-G (shown in Fig. 5A). In addition to the salt bridges, an intricate hydrogen bond network further stabilized the NiV-G/ephrin complex, including both main-chain/side-chain (L101–S491, P107–Y398, R112–Q530, A532–S118, E119–Y581, and T114–Q530) and side-chain/side-chain (D108–Y389, D108–Q388, and Q118–Y581) bonds.
Fig. 5.
Structure of the NiV-G/ephrin interface. Interacting residues are labeled. (A) Salt bridges at the polar (peripheral) region of the NiV-G/ephrin interface. NiV-G is in yellow and ephrin-B3 in gray. (B) The G–H ephrin-B3 loop bound to the NiV-G surface channel. (C) The same surface in the unbound NiV-G molecule. The position of the G–H ephrin-B3 loop is still shown to illustrate that the binding pockets for ephrin residues P122, L124, and W125 are already fully formed in the unbound attachment protein and undergo little or no conformational rearrangements upon ephrin binding. On the other hand, the Y120 binding pocket is only formed upon ephrin binding.
The second region of the interface centered around the G–H loop of ephrin-B3, which was inserted in a channel on the surface of NiV-G (Figs. 2B and 5B). The sides of the channel were defined by the B3H3–B3S3, B4S2–B4S3, B4S4–B5S1, and B6S3–B6S4 NiV-G loops, as well as the B6S1 strand. The binding here was dominated by van der Waals contacts between two predominantly hydrophobic surfaces because ephrin buries Y120, P122, L124, and W125 (Fig. 5B). Each of these four hydrophobic residues bound in its own hydrophobic pocket on the surface of NiV-G that is part of the continuous channel. The pocket for Y120 was formed by I588, I580, Y581 A558, Q559, and the C216–C240 disulfide bridge. P122 sat on top of P488 and its pocket was defined by V507, A532, T531, G489, and Q490. L124 sat on top of P485 and was surrounded by E505, G506, and W504. Finally, W125 rested on W504, and its pocket was formed by I401, F485, and L305. In this part of the interface, there were also several intermolecular hydrogen bonds involving main-chain atoms, including S121–Q559 and P122–G506. The location of the ephrin-binding channel on the top face of NiV-G was similar to the location of the sialic acid binding site in the parainfluenza virus HN (Fig. 2B), although, of course, the latter is much smaller in size. It is interesting that this location was quite different from the proposed location of the receptor-binding sites of the MeV H glycoprotein, which is the only other paramyxovirus with a solved structure that utilizes proteins as host cell receptors. Although there are as yet no reported structures of a MeV H/receptor complex, mutagenesis experiments suggest that the receptors might bind along the sides of the MeV H β-barrel and not on the top face, as observed here for NiV-G (22–24).
Ephrin Recognition by NiV-G.
Interestingly, the same ephrin surface elements were involved in both NiV-G and Eph binding. As in the NiV-G/ephrin complex, the Eph/ephrin high-affinity interface (31) contains two regions: one involving the hydrophobic ephrin G–H loop, which is inserted in a hydrophobic channel, and one involving ephrin strands C, F, and G, which forms an intricate network of hydrogen bonds and salt bridges with residues surrounding the Eph channel. The total surface area of the Eph/ephrin and NiV-G/ephrin interfaces was also similar: 2,400 and 2,600 Å2, respectively. Nevertheless ephrin-B2 and -B3 bind NiV-G with higher affinity (32–34). One possible explanation of this observation is that the Eph/ephrin recognition proceeds via an induced-fit mechanism, whereas the NiV-G/ephrin recognition seems to proceed via a lock-and-key type binding. Indeed, the Eph loops forming the side of the ephrin-binding channel are mostly unstructured in the unbound receptor and fold upon ligand binding, thus requiring energy to generate the extensive interaction surface that was complementary to the ephrin G–H loop. On the other hand, during henipavirus attachment to the ephrins, two relatively rigid molecular surfaces, which were already complementary to each other both in shape and in chemical nature, interacted with no need for significant conformational changes in either molecule. Fig. 5B compares the NiV-G molecular surface at the ephrin-binding channel in the free and ephrin-bound structures, documenting that three of the four hydrophobic pockets (those accepting P122, L124, and W125) did not undergo any significant rearrangements and that only the Y120 binding pockets was altered to accommodate the incoming ephrin. Indeed, of the three surface loops that differed significantly in the bound and unbound NiV-G structures (Fig. 3A), two (B6S2–B6S3 and B1S1–B1S3) were part of the Y120 binding pocket, whereas the third one (B1H1–B1S1) was intrinsically very flexible and had distinct conformations even in the two copies of the molecule in the asymmetric unit of the crystal (Fig. 3A, red and yellow).
A likely biological rationale for the higher affinity of the henipavirus G/ephrin interactions is that they mediate essentially irreversible viral attachment and, therefore, the tighter that the binding is, the more efficient the virus infection process would be. On the other hand, the Eph/ephrin interactions are signaling events that might need to be regulated, so that the specific interaction affinities of the individual Eph and ephrin members, together with their local concentration at the interaction sites within the membrane, determine the exact signaling effects in the interacting cells. In addition, the henipaviruses might need to compete with the endogenous Eph receptors for ephrin binding and have, therefore, evolved a higher-affinity attachment interface.
The Ephrins As Cell Receptors for the Henipaviruses.
The henipaviruses use only two cellular receptors (ephrin-B2 and -B3) among the nine ephrin family members. Notably, even the closely related ephrin-B1 is not recognized and the NiV-G/ephrin-B3 structure now provides the molecular bases for this specificity: a close inspection of the binding interface revealed that the ephrin binding pocket (Fig. 5B) will not readily accommodate the ephrin-B1 G–H loop because the L124→Y and W125→M substitutions will result in steric clashes. Indeed, this observation is validated by previous mutagenesis results documenting that alterations of these particular ephrin residues abolish attachment protein binding and that substitution of the ephrin-B1 residues (Y and M) with the ephrin-B2/-B3 residues (L and W) confers viral binding (12).
In addition to the identity of the individual ephrin G–H loop residues, the overall conformation and flexibility of this loop might also play a role in the receptor selectivity of the henipavirus attachment proteins. Indeed, although both ephrin-B2 and -B3 seem to have G–H loops with extended and relatively rigid conformations, ephrin-B1 has a more flexible G–H loop (29), which may not be compatible with the lock-and-key ephrin/G protein binding mechanism.
Regarding the HeV G glycoprotein and ephrin binding, in light of the high degree of sequence homology between NiV-G and HeV-G, along with their identical receptor recognition pattern and host cell tropisms, we would expect that the overall structures of the NiV and HeV G and their cognate receptor complexes would be similar. Indeed, only three of the 22 NiV-G ephrin contact residues differed in HeV-G (Fig. S1A), and these were all conservative substitutions, unlikely to significantly affect the G protein/ephrin complex formation.
Implications for Viral Membrane Fusion.
The first step during paramyxovirus infection of a permissive host cell is the recognition and binding of the viral attachment protein to a suitable cellular receptor, which are located on the juxtaposed membrane surfaces. Although the precise number of viral glycoproteins and cellular receptor molecules required for the assembly of a functional entry complex is unknown, it is expected that the complex is oligomeric and large. Indeed, the F glycoprotein is a trimer (35, 36) and the G glycoprotein is a tetramer (dimer of dimers) (37), whereas the ephrins themselves are often clustered (18). The G and F glycoproteins of paramyxoviruses work in concert to facilitate the membrane fusion process, and the preponderance of data to date suggest that they associate with each other within the viral membrane. It has been suggested that the G glycoprotein stalk region is primarily involved in these interactions with their partner F glycoproteins (reviewed in ref. 22).
The membrane fusion process in paramyxoviruses appears to be directly triggered by receptor binding to the viral attachment glycoprotein. Although the precise molecular details of this process have yet to be clarified, there are two potential mechanisms that could be envisioned. A currently favored model that receptor binding triggers significant conformational changes in the attachment protein, which, in turn, serve as the trigger for the membrane fusion activity of the associated F glycoprotein (38). The majority of studies aimed at addressing this process have been carried out with HN glycoproteins that employ sialic acid receptors. Also supporting such a model are the observations that most high-affinity viral attachment protein interactions with cellular receptors induce significant structural rearrangements within the viral attachment protein(s), such as those associated with CD4 and coreceptor engagement of the envelope glycoprotein of HIV-1 that eventually lead to gp41 six-helix bundle formation concomitant with membrane merger. On the other hand, lower affinity interactions between virus attachment proteins and cellular receptors often lead to entry processes involving endocytosis, which is followed by low-pH triggering of the viral fusion process (39).
Interestingly, even though the NiV-G/ephrin interactions are specific and of very high affinity, the NiV-G rearrangements are relatively small and strictly localized to the interaction interface. Our data, therefore, support an alternative model that the henipavirus membrane fusion process is likely triggered not via major conformational changes in the attachment protein but perhaps by more subtle rearrangements and repositioning relative to each other of the G and F glycoproteins that might result, for example, from the displacement of F interaction epitopes from the top face of the G glycoprotein β-propeller by the incoming ephrin receptor molecules. However, we cannot exclude the possibility that receptor engagement might induce more significant conformational changes between the oligomers of G themselves (the dimer of dimers).
It has also been proposed that the fusion process in the sialic acid receptor-using paramyxoviruses follows a different molecular mechanism than the fusion process in the protein receptor-using paramyxoviruses (which include the henipaviruses and morbilliviruses) (22). This proposition is based in two premises: first, that the location of the receptor binding site in the sialic acid-binding viruses (at the top face of the β-propeller) is very different from the proposed receptor binding site locations in the protein receptor-binding viruses (the sides of the β barrel for MeV H); and second, that whereas the extent of fusion in the sialic acid-binding viruses is proportional to the strength of the attachment/fusion protein association, in the protein receptor-binding MeV, these features are inversely related. Although there are currently no reported structures of a MeV H/receptor complex, the NiV-G/ephrin structure documents that the protein receptor, at least in the henipavirus case, binds at approximately the same region within the G glycoprotein as the sialic acid receptors, suggesting that these two fusion events may, in fact, be more similar than previously appreciated. Nevertheless, as discussed above, our structures would be consistent with the hypothesis that the henipavirus fusion event is regulated by the dissociation of a preexisting G–F complex by competing ephrin binding. This would also correlate with the likely requirement that the F glycoprotein be unassociated with its attachment glycoprotein partner in order for F to proceed with the conformational alterations leading to six-helix bundle formation and membrane fusion (40).
Structural Insights Into Antiviral Drug Design.
Because the majority of the NiV-G/ephrin-B3 interactions involve the extended G–H ephrin loop, the structure suggests that ephrin-based peptides could potentially be designed to serve as antiviral agents, competing with the cellular receptor for NiV-G binding. It is interesting to note that phage display has been used to identify Eph-binding peptides, which turned out to contain G–H-loop-like sequences and which target the surface region used by the Eph receptors for ephrin binding (41, 42). The NiV-G structures suggest that on one hand, it would be easier to engineer ephrin-based NiV-G binding peptides, than ephrin-based Eph binding peptides, because the ephrin-binding channel in NiV-G is already formed in the unbound molecule, whereas in the Eph receptors, it forms only subsequent to ligand binding. However, on the other hand, because the NiV-G/ephrin interaction has a lower Kd value than the Eph/ephrin binding and involves a slightly larger contact area outside of the G–H ephrin loop/channel interface (see Fig. 2), it would be more difficult to identify small peptides that effectively compete with ephrin for NiV-G binding. Indeed, we tested several G–H-loop-derived peptides immediately available in our laboratory (Methods), but, unfortunately, although they bound to NiV-G with various affinities, they were unable to compete with the full-length ephrin (data not shown).
The same considerations are also valid for potential small-molecule inhibitors of the NiV-G/ephrin interactions. On the one hand, the fact that the NiV-G ephrin-binding channel does not significantly change upon ephrin binding provides the rationale for an in silico screen using the NiV-G structures as a starting point, whereas on the other hand, the very high affinity of the NiV-G/ephrin binding and the large interface area will undoubtedly make such a worthy task more challenging. In addition to computational structure-based screens, simple high-throughput screens of compound libraries could also potentially provide small-molecule leads that bind NiV and block ephrin binding.
Finally, based on the structures reported here, we propose that a viable therapeutic approach might be to directly use modified ephrins as a treatment modality in patients infected with NiV or HeV. Indeed, our data indicate that one could use structure-based protein engineering approaches to generate mutations in ephrin-B3 or -B2 that still retain their subnanomolar affinity for the viral attachment proteins but reduce their affinity for their functional Eph receptors. Moreover, because monomeric ephrins do not normally induce a cellular response in Eph-expressing cells, the use of monomeric ephrins for the treatment of viral infections may not elicit significant side effects.
Methods
Construct Design, Expression, and Crystallization of Nipah-G and Ephrin-B3.
All constructs of NiV-G and ephrin-B3 were cloned into a modified pAcGP67 baculovirus expression vector, expressed in insect cells, and crystallized as described in SI Materials and Methods.
Data Collection and Structure Determination.
The crystallographic data were collected and processed as described in SI Materials and Methods. Crystallographic data statistics are listed in Table S1. The structure of unbound NiV-G was determined by using the SAD technique with the anomalous signal from protein-bound iodine as described in detail in SI Materials and Methods. The structure of the NiV-G/ephrin-B3 complex was determined by using molecular replacement techniques as described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Alexander Antipenko (Memorial Sloan-Kettering Cancer Center, New York, NY) for the custom-made baculovirus expression vector and Dr. Yehuda Goldgur for help with data collection. This work was supported by National Institutes of Health Grant NS38486 (to D.B.N.) and, in part, by National Institutes of Health Grants AI057168 and AI054715 (to C.C.B.). The Northeastern Collaborative Access Team beamlines of the Advanced Photon Source are supported by National Center for Research Resources, National Institutes of Health Award RR-15301. Use of the Advanced Photon Source is supported by the U.S. Department of Energy under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3D11 for NiV-G and 3D12 for NiV-G/ephrin-B3).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804797105/DCSupplemental.
References
- 1.Eaton BT, Broder CC, Middleton D, Wang LF. Hendra and Nipah viruses: Different and dangerous. Nat Rev Microbiol. 2006;4:23–35. doi: 10.1038/nrmicro1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chua KB, et al. Nipah virus: A recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 3.Murray K, et al. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94–97. doi: 10.1126/science.7701348. [DOI] [PubMed] [Google Scholar]
- 4.Field HE, et al. Epidemiological perspectives on Hendra virus infection in horses and flying foxes. Aust Vet J. 2007;85:268–270. doi: 10.1111/j.1751-0813.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- 5.Chadha MS, et al. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg Infect Dis. 2006;12:235–240. doi: 10.3201/eid1202.051247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurley ES, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13:1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu VP, et al. Nipah virus encephalitis reemergence, Bangladesh. Emerg Infect Dis. 2004;10:2082–2087. doi: 10.3201/eid1012.040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field HE, Mackenzie JS, Daszak P. Henipaviruses: Emerging paramyxoviruses associated with fruit bats. Curr Top Microbiol Immunol. 2007;315:133–159. doi: 10.1007/978-3-540-70962-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop KA, et al. Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J Virol. 2007;81:5893–5901. doi: 10.1128/JVI.02022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonaparte MI, et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc Natl Acad Sci USA. 2005;102:10652–10657. doi: 10.1073/pnas.0504887102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negrete OA, et al. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature. 2005;436:401–405. doi: 10.1038/nature03838. [DOI] [PubMed] [Google Scholar]
- 12.Negrete OA, et al. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006;2:e7. doi: 10.1371/journal.ppat.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 14.Kullander K, Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol. 2002;3:475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 15.Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasquale EB. Eph receptor signalling casts a wide net on cell behaviour. Nat Rev Mol Cell Biol. 2005;6:462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 17.Eaton BT, Broder CC, Wang LF. Hendra and Nipah viruses: Pathogenesis and therapeutics. Curr Mol Med. 2005;5:805–816. doi: 10.2174/156652405774962308. [DOI] [PubMed] [Google Scholar]
- 18.Himanen JP, Nikolov DB. Eph signaling: A structural view. Trends Neurosci. 2003;26:46–51. doi: 10.1016/s0166-2236(02)00005-x. [DOI] [PubMed] [Google Scholar]
- 19.Lamb RA, Parks GD. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 20.Fulop V, Jones DT. Beta propellers: Structural rigidity and functional diversity. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 21.Paoli M. Protein folds propelled by diversity. Prog Biophys Mol Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 22.Iorio RM, Mahon PJ. Paramyxoviruses: Different receptors - different mechanisms of fusion. Trends Microbiol. 2008;16:135–137. doi: 10.1016/j.tim.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 24.Hashiguchi T, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 2007;6:135–137. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MC, et al. Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J Mol Biol. 2004;335:1343–1357. doi: 10.1016/j.jmb.2003.11.032. [DOI] [PubMed] [Google Scholar]
- 27.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 28.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 29.Nikolov DB, Li C, Barton WA, Himanen JP. Crystal structure of the ephrin-B1 ectodomain: Implications for receptor recognition and signaling. Biochemistry. 2005;44:10947–10953. doi: 10.1021/bi050789w. [DOI] [PubMed] [Google Scholar]
- 30.Toth J, et al. Crystal structure of an ephrin ectodomain. Dev Cell. 2001;1:83–92. doi: 10.1016/s1534-5807(01)00002-8. [DOI] [PubMed] [Google Scholar]
- 31.Himanen JP, et al. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414:933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- 32.Negrete OA, Chu D, Aguilar HC, Lee B. Single amino acid changes in the Nipah and Hendra virus attachment glycoproteins distinguish ephrinB2 from ephrinB3 usage. J Virol. 2007;81:10804–10814. doi: 10.1128/JVI.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Himanen JP, et al. Repelling class discrimination: Ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci. 2004;7:501–509. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- 34.Brambilla R, et al. Similarities and differences in the way transmembrane-type ligands interact with the Elk subclass of Eph receptors. Mol Cell Neurosci. 1996;8:199–209. [PubMed] [Google Scholar]
- 35.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamb RA, Paterson RG, Jardetzky TS. Paramyxovirus membrane fusion: Lessons from the F and HN atomic structures. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bossart KN, et al. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J Virol. 2005;79:6690–6702. doi: 10.1128/JVI.79.11.6690-6702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bossart KN, Broder CC. Paramyxovirus entry. In: Pöhlmann S, Simmons G, editors. Viral Entry into Host Cells. Georgetown, TX: Landes Bioscience; 2007. [Google Scholar]
- 39.Dimitrov DS. Virus entry: Molecular mechanisms and biomedical applications. Nat Rev Microbiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chrencik JE, et al. Structure and thermodynamic characterization of the EphB4/Ephrin-B2 antagonist peptide complex reveals the determinants for receptor specificity. Structure. 2006;14:321–330. doi: 10.1016/j.str.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 42.Koolpe M, Burgess R, Dail M, Pasquale EB. EphB receptor-binding peptides identified by phage display enable design of an antagonist with ephrin-like affinity. J Biol Chem. 2005;280:17301–17311. doi: 10.1074/jbc.M500363200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.