Abstract
Cardiac ion channels are surprisingly dynamic in nature, and are continuously formed, trafficked to specific subregions of plasma membrane, inserted in the plasma membrane, and removed to be degraded or recycled. Due to these movements which affect channel availability, ion channel function is dependent on not just channel biophysical properties but on channel trafficking as well. The development of molecular techniques to tag proteins of interest with fluorescent and other genetically encoded proteins, and of advanced imaging modalities such as total internal reflection microscopy (TIRF) have created new opportunities to understand the intracellular movement of proteins near the plasma membrane, and their dynamics therein. In this paper we present approaches for ion channel biologists to the use of fluorescent and non-fluorescent fusion proteins, techniques for cloning and expression of fusion proteins in mammalian cells, and imaging techniques for live-cell high resolution microscopy. For illustration, original data are presented on creation of a stable cell lane capable of inducible expression of connexin43 tagged to green fluorescent protein and its distribution viewed with both widefield epifluorescence and TIRFm. With revolutionary advances in fluorescence microscopy, ion channel biologists are now entering a new realm of studying channel function, which is to understand the mechanisms of channel trafficking.
Keywords: ion channel, trafficking, plasma membrane, fusion proteins, fluorescence microscopy, TIRF, gap junctions, connexin43
Introduction
Acting as the primary gatekeepers of cellular charge and osmolarity, ion channels are one of the most important classes of transmembrane proteins expressed in biological cells. They play a central role in cell survival and orchestrate key physiological functions including the propagation of action potentials through the nervous system and myocardium [1]. The expression and localization of specific ion channels, not only in certain cell types, but also in precise domains within the plasma membrane itself, is essential for the normal function of many tissues [2–4]. Their disruption, either through genetic mutation or acute pathological states such as ischemia, often leads to serious diseases primarily of the nervous system and heart [5, 6]. While the existence of membranous ion channels was first proposed over fifty years ago [7], substantial gaps remain in our understanding of ion channel basic biology. The patch-clamp technique has dominated the field of ion channel research since its introduction by Neher and Sakmann in 1976 [8, 9]. While this technique is the gold standard for biophysical properties of channels and groups of channels, it is more limited in the information it can provide regarding the dynamic nature of intracellular ion channel movements within and on the surface of cells. The development of novel therapeutics aimed at treatment of disorders associated with altered ion channel expression and localization will depend upon further elucidation of ion channel trafficking.
Gap junctions are a class of intercellular channels which form low resistance conduits between the cytoplasms of adjacent cells via serial coupling of connexin hemichannels [10–12]. While gap junctions are relatively non-specific in distinguishing which molecules they allow to pass, their location at the intercalated discs of the myocardium permits rapid transmission of excitation between adjacent cardiomyocytes in an organized and polarized fashion. Action potentials are propagated throughout the working myocardium in this manner, facilitating well-orchestrated and efficient contractile function [12]. Altered regulation and localization of gap junctions are an intrinsic part of the molecular basis of the arrhythmias of sudden cardiac death [13–15] and have been implicated in dyssynchronous cardiac function during congestive heart failure [16]. However, despite the importance of gap junctions in normal physiology and ischemic heart disease, much is unknown about their regulation. We recently found that gap junctions can be directly and specifically targeted to the cell-cell interface by dynamic microtubules [4]. Through the use of advanced imaging techniques such as total-internal reflection (TIRF) microscopy and fluorescent recovery after photobleaching (FRAP) in combination with fluorescently-tagged fusion proteins, it is possible to study the dynamic behavior of gap junction repopulation in real time [4].
The recent refinement of live-cell imaging techniques, together with the development of spectrally distinct fluorescent protein tags, has opened the door to a new era of basic cell biology research. This paper will discuss those tools and approaches which are currently available and relevant to the study of ion channel dynamics in living cells. Topics covered include: choice of fusion protein tag, cloning approach, expression system, and methods of imaging and acquisition. Supported by complementary assays based on biochemical and genetic techniques, exploitation of these recent advances in live-cell imaging will enhance our understanding of the basic molecular mechanisms underlying ion channel trafficking and dynamics.
Fluorescent fusion protein tags
The cloning of the green fluorescent protein (GFP) gene of the jellyfish Aquoria victoria in 1992 led to its rapid exploitation as a reporter gene and development as a valuable tool for studying protein behavior in living cells when expressed as a genetic fusion product [17, 18]. As a result of cloning additional fluorescent proteins (FPs) from other marine organisms and the genetic modification of these and GFP, an increasingly broad range of spectrally distinct fluorescent proteins are now available. It should be noted, however, that use of these proteins can be complicated by multimeric tendencies, suboptimal brightness, variable photostability, and artifactual changes to the protein of interest. Care should therefore be taken when choosing FPs as fusion tags depending on their intended application [19, 20]. In general, the brightest FPs emit in the green and yellow wavelengths of the spectrum, while those emitting in blue and red display fainter fluorescence. The development of red FP fusion tags was also hindered by their oligomeric nature. A monomeric form of DSRed is available, but the superior photostability of mCherry and the recently described TagRFP render these the two FPs of choice in the visible red spectrum [19, 21]. Based on our own experience, we would recommend the use of the FPs EGFP, EYFP or Venus, and mCherry or TagRFP for the study of ion channels in the green, yellow and red wavelengths respectively. The markedly lower brightness exhibited by blue FPs and those in the far-red spectra such as mPlum make the imaging of single ion channels difficult, although these proteins still have considerable value when encoded as genetic reporters.
An exciting addition to the field of FPs are those whose fluorescent properties can be manipulated in real time by the researcher. Photoactivatable and photoswitchable FPs such as PA-GFP and Kaede permit the study of protein dynamics in a manner comparable to pulse-chase experiments. PA-GFP fluorescence increases 100 times and Kaede experiences an emission shift from green to red upon exposure to 413 and 350 nm excitation respectively [22, 23]. Therefore, the movement of a specific subset of fusion-proteins within the cell can be tracked over time following activation/switching without contaminating background from those fusion proteins not exposed to the lower wavelength excitation.
Non-Fluorescent fusion tags for secondary labeling and fluorescent live cell imaging
Despite the convenience of using genetically encoded FP fusion tags, their physical properties can potentially interfere with the structure, trafficking and function of the protein of interest. Several alternative strategies exist which involve the use of smaller non-fluorescent fusion tags (typically < 30 residues compared to 229 residues for GFP), which then can be fluorescently labeled in a secondary step prior to imaging. The FLAG (DYKDDDDK) and HA (influenza virus hemagglutinin epitope; YPYDVPDYA) affinity tags typically utilized in biochemical assays can be used to introduce foreign epitopes either internally or at a terminus of the protein of interest. Fluorophores are subsequently attached specifically via monoclonal antibodies, or more elegantly, via Fab fragments of monoclonal antibodies.
Enzymes which catalyze postranslational modifications of proteins can also be exploited in the specific fluorescent labeling of ion channels on the cell surface. Two such methods, E. coli BirA biotin ligase and B. subtilis Sfp phosphopantetheinyl transferase, are adapted from bacterial systems and provide efficient, high-affinity labeling of target proteins with minimal background. BirA biotinylates a lysine within a 15 residue acceptor peptide permitting subsequent labeling with streptavidin conjugated fluorophores [24, 25]. One drawback of biotinylation is that the avidin family of proteins exist as multivalant tetramers which could lead to aggregation of tagged (biotinylated) protein and as such, elegant steps have been taken to overcome this caveat [26]. A recent method, termed phosphopantetheinylation, exploits Sfp phosphopantetheinyl transferase in the covalent attachment of coenzymeA-linked fluorophores to a serine within an 11 residue motif termed the ybbR tag. This procedure, which involves fewer steps than that involving BirA, does not lead to aggregation of target ion channels, and the covalent bond provides a high level of stability in comparison to affinity tag-antibody binding [27].
A popular tag used for indirect fluorescent labeling is the tetracysteine motif (CCPGCC) which, unlike the techniques mentioned above, can also be used to label intracellular proteins. Biarsenical dyes, available in both red and green wavelengths, will only fluoresce upon binding to the small tetracysteine motif, providing a method to time protein localization without attaching large GFP-type fluorophores to the proteins [28, 29]. For dynamic studies, the tetracysteine motif method may not have the quantum efficiency for very rapid live cell imaging of protein movements.
Considerations for fusion protein cloning and expression
The imaging laboratory often requires the generation of a wide range of constructs expressing fluorescent fusion proteins of various colors. Traditional restriction cloning methods can prove time consuming and cumbersome when generating a variety of expression vectors. It can be helpful to utilize site-specific recombination cloning technology such as the Gateway system (Invitrogen) which allows for rapid and straightforward shuttling of genes between differing expression vectors, thus generating a wide variety of genetic fusions to a target protein in a short time with relative ease [30]. In our laboratory, we have used the Gateway system to create a library of “destination vectors” consisting of separate mammalian expression vectors each containing a different genetically encoded fluorophore or affinity tag that is fused to the C-/N-terminus of the protein of interest. In addition, a library of non-expressing “entry clones” containing genes encoding the proteins we study has been generated. A single step involving incubation of an entry clone with a destination vector of choice generates an expression vector encoding a fluorescently tagged fusion protein of the protein of interest without any involvement of PCR-based restriction cloning.
Liposome-mediated transient transfection of recombinant DNA is now a well established technique for overexpression of proteins in cultured mammalian cells. Adult cardiomyocytes however, are not permissive to liposome-mediated transfection techniques. Therefore transduction with appropriate viral vectors will be required in the study of ion channel dynamics within this cell type. Ease of transfection and growth renders many established mammalian cell lines valuable tools as general trafficking models. Selection of the appropriate cell line is user dependent and depends heavily on species of interest, endogenous protein expression profile, and imaging qualities. For instance, in previous studies we have used HeLa cells which are human in origin, express N-cadherin as do cardiomyocytes, express relatively low quantities of Cx43 so overexpression does not compete with endogenous protein, and are comparatively flat so image well with planar imaging.
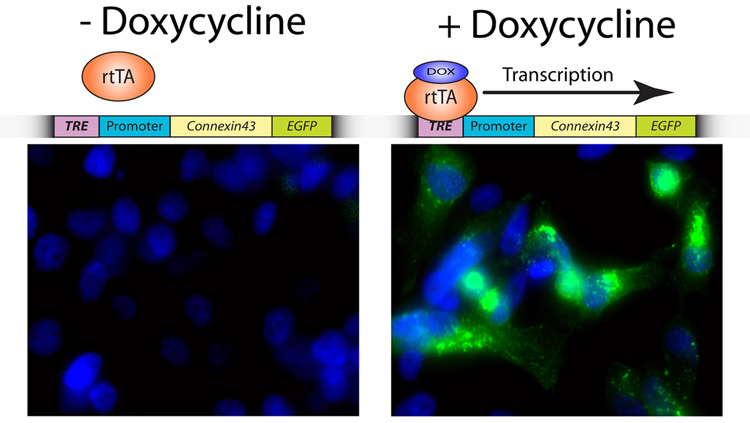
While valuable information can be obtained from transient transfection, variance in cellular levels of protein expression within the same culture dish can result in significant intercellular variability of phenomena studied. To address this concern, it is possible to generate stable clones of cell lines expressing similar levels of protein using antibiotic selection resistance markers which are now included in most commercially available expression vectors. Should chronic overexpression of the fusion protein prove toxic to the cells, it is also possible to generate inducible stable clones using regulatory genetic systems adapted from bacteria. A well accepted system is the tetracycline binary regulatory operon of E. coli which can be used to generate clonal populations of cells which only express the fusion protein in either the presence or absence of tetracycline or its derivative doxycycline [31]. The tight regulation achieved when using this technology not only allows for the expansion of clonal cell lines prior to expression, but also yields consistency in the study of ion channels at the single cell level. Figure one shows a stable HeLa cell line which expresses the gap junction protein Connexin43 with a C-terminal EGFP fusion tag in the presence of doxycycline. We therefore can induce or halt expression of Cx43-GFP in a full population of HeLa cells by adding or removing an otherwise innocuous reagent in the medium.
Figure 1. Doxycycline-induced expression of connexin43-EGFP in a stable cell line.
HeLa cells expressing the rtTA-Advanced transactivator protein (Clontech) were transfected with pTRE-tight tetracycline-responsive vector encoding connexin43-EGFP and a linear selection marker (Clontech). The schematic above image panels illustrates that doxycycline is required for the transactivator protein rtTA to bind to the tetracycline-responsive element (TRE) preceding the promoter and gene of interest, allowing transcription to commence. Cells were incubated with (right panel) or without (left panel) 1.0 µg/ml Doxycycline (Sigma) for 24 h prior to fixation and the addition of nuclear counterstain DAPI (blue). Image acquisition was undertaken using a 60X objective.
Imaging Techniques
The use of widefield epifluorescence microscopy is well established and traditionally used for sensitive image acquisition of fixed cell immunofluorescence in many cell biology laboratories. Recent development of fluorescent tools for tagging specific proteins within live cells furthered interest in the use of more advanced imaging techniques such as confocal, and TIRF microscopy. These techniques have been applied to ion channel research and each has its respective advantages and disadvantages, as discussed below.
Widefield epifluorescence imaging of live cells can provide an excellent overview of protein dynamics within a cell, and images can be acquired rapidly. However, at high objective magnification, there is poor vertical spatial resolution and noise from out of plane signals can significantly confound the image. Post-processing mathematical deconvolution techniques can significantly resolve this problem, but at a cost of increasing imaging time with multiple plane acquisitions. Thus, with some exceptions, deconvolution techniques work best for ‘static’ fixed cell preparations. Another drawback of epifluorescence imaging of live cell preparations is the need for entire specimen exposure to the excitation light. This can result in phototoxic effects on the cell and rapid bleaching of fluorophores. Despite these limitations, epifluorescence imaging remains a useful tool applicable to study ion channel movements within the cell cytoplasm. For studies concentrating on protein dynamics at specific compartments of the cell, such as the plasma membrane, higher vertical spatial resolution is required.
Laser scanning and spinning disc confocal microscopy are widely used techniques in the acquisition of images with higher vertical spatial resolution (typically ~ 500 nm). These techniques exploit pinholes which only accept emission from a specific and precisely controlled plane within the sample, the acquisition of z-stacks can generate impressive 3D images. A disadvantage of confocal microscopy, particularly laser scanning microscopy, is that the entire field of view is exposed to excitation laser light during acquisition. As with epifluorescence imaging yet with a laser source, overexposure can result in phototoxicity of live samples and photobleaching effects on fluorophores. Spinning disc confocal microscopes can acquire confocal images of the entire field of view in a shorter time than their laser scanning counterparts, reducing these unwanted effects. The problem remains, however, that dynamic proteins studied in live cell imaging can move in and out of the focal plane. Furthermore, when studying protein movements that exist within a 5 nm plasma membrane, even confocal imaging will capture mainly cytoplasmic signal. It is for these reasons, that we recommend the use of TIRF microscopy in studying the biology of ion channels at the plasma membrane.
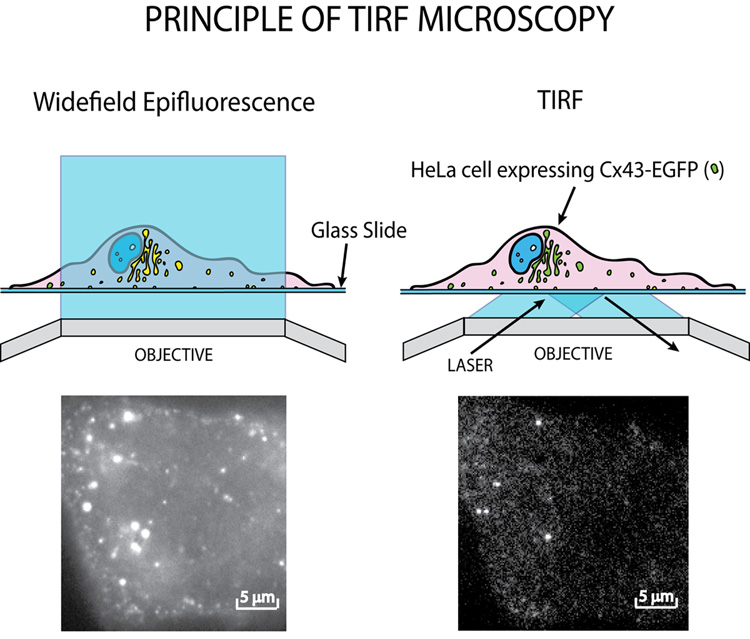
The application of TIRF in the study of cell biology was promoted by Daniel Axelrod in the early 1980s [34, 35]. TIRF microscopy uses specially angled light and exploits the difference in refractive indexes of aqueous medium and a glass coverslip to obtain extremely high vertical spatial resolution. Briefly, an excitation laser beam is directed at the specimen at an angle of incidence greater than or equal to the critical angle of refraction whereby the excitation laser light is reflected. At the point of reflection, an exponentially decaying evanescent wave is generated and penetrates the specimen. Due to exponential decay, only those fluorophores within 10–100 nm of the glass/medium interface are excited, providing high resolution images of the nearest plasma membrane and immediate subcortical regions (Figure 2). Given the limited depth of fluorophore excitation, signal to noise ratios can be high enough to image single ion channels or clusters. With a sensitive CCD camera and rapid data acquisition techniques, TIRF microscopy can be used to visualize ion channel or other membrane protein insertion and lateral diffusion within the plasma membrane at the single molecule level. One limitation of TIRF microscopy is that only the interface of the cell and coverslip may be imaged, the focal plane can not be moved “upwards” as in confocal microscopy. However, the relatively low level of energy from the evanescent wave causes markedly less phototoxicity and photobleaching, compared to the toxic effects of confocal and epifluorescence imaging. Recent improvements in TIRF technology now allow for fully automated multi-wavelength systems which greatly improve consistency and facilitate the use of techniques such as FRAP and fluorescence resonance energy transfer (FRET) in combination with TIRF microscopy. TIRF microscopy is therefore a highly appropriate tool in the study of ion channel trafficking to, and dynamics within, the plasma membrane [4].
Figure 2. Widefield Epifluorescence and TIRF microscopy.
Widefield epifluorescence excites the entire cell being images, activating all fluorophores within the field of view. TIRF microscopy limits excitation depth to 10–100 nm activating only those fluorophores in and just below the plasma membrane. Images are of the same induced conexin43-EGFP expressing cell as viewed with widefield epifluorescence (left) and TIRF (right).
Conclusions
The fluorescent imaging revolution has introduced a host of novel tools for basic cell biologists. Application of these techniques to ion channel biology is beginning to provide novel insight into the biological regulation of channel movements. Fluorescent labeling of proteins is now possible with a variety of colors and types of tags, allowing sensitive detection and tracking of individual ion channels as they are transported to their designated region in the plasma membrane. We expect that imaging will increasingly complement more traditional biochemical and genetic assays, providing directly visualized, real time data. We hope that this brief summary will encourage further development and promote use of this new area of basic ion channel research.
Acknowledgments
Financial Support (RMS): NIH (NHLBI), GlaxoSmithKline Research and Education Foundation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Conflicts: None
References
- 1.Hille B. Ion Channels of Excitable Membranes. Sunderland, Massachusetts, USA: Sinauer; 2001. [Google Scholar]
- 2.Lai HC, Jan LY. The distribution and targeting of neuronal voltage-gated ion channels. Nat Rev Neurosci. 2006;7(7):548–562. doi: 10.1038/nrn1938. [DOI] [PubMed] [Google Scholar]
- 3.Scriven DR, Dan P, Moore ED. Distribution of proteins implicated in excitation-contraction coupling in rat ventricular myocytes. Biophys J. 2000;79(5):2682–2891. doi: 10.1016/S0006-3495(00)76506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw RM, Fay AJ, Puthenveedu MA, et al. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128(3):547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akar JG, Akar FG. Regulation of ion channels and arrhythmias in the ischemic heart. J Electrocardiol. 2007;40(6 Suppl):S37–S41. doi: 10.1016/j.jelectrocard.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Cooper EC, Jan LY. Ion channel genes and human neurological disease: recent progress, prospects, and challenges. Proc Natl Acad Sci U S A. 1999;96(9):4759–4766. doi: 10.1073/pnas.96.9.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornreich BG. The patch clamp technique: principles and technical considerations. J Vet Cardiol. 2007;9(1):25–37. doi: 10.1016/j.jvc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Neher E, Sakmann B. Single-channel currents recorded from membrane of denervated frog muscle fibres. Nature. 1976;260(5554):799–802. doi: 10.1038/260799a0. [DOI] [PubMed] [Google Scholar]
- 10.Revel JP, Yee AG, Hudspeth AJ. Gap junctions between electrotonically coupled cells in tissue culture and in brown fat. Proc Natl Acad Sci U S A. 1971;68(12):2924–2927. doi: 10.1073/pnas.68.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson JD. The Occurrence of a Subunit Pattern in the Unit Membranes of Club Endings in Mauthner Cell Synapses in Goldfish Brains. J Cell Biol. 1963;19:201–221. doi: 10.1083/jcb.19.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gros DB, Jongsma HJ. Connexins in mammalian heart function. Bioessays. 1996;18(9):719–730. doi: 10.1002/bies.950180907. [DOI] [PubMed] [Google Scholar]
- 13.Saffitz JE, Hames KY, Kanno S. Remodeling of gap junctions in ischemic and nonischemic forms of heart disease. J Membr Biol. 2007;218(1–3):65–71. doi: 10.1007/s00232-007-9031-2. [DOI] [PubMed] [Google Scholar]
- 14.Shaw RM, Rudy Y. Ionic mechanisms of propagation in cardiac tissue. Roles of the sodium and L-type calcium currents during reduced excitability and decreased gap junction coupling. Circ Res. 1997;81(5):727–741. doi: 10.1161/01.res.81.5.727. [DOI] [PubMed] [Google Scholar]
- 15.Wit AL, Janse MJ. Reperfusion arrhythmias and sudden cardiac death: a century of progress toward an understanding of the mechanisms. Circ Res. 2001;89(9):741–743. [PubMed] [Google Scholar]
- 16.Spragg DD, Leclercq C, Loghmani M, et al. Regional alterations in protein expression in the dyssynchronous failing heart. Circulation. 2003;108(8):929–932. doi: 10.1161/01.CIR.0000088782.99568.CA. [DOI] [PubMed] [Google Scholar]
- 17.Prasher DC, Eckenrode VK, Ward WW, et al. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111(2):229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 18.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 19.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2(12):905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 20.Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23(12):605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Merzlyak EM, Goedhart J, Shcherbo D, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4(7):555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 22.Ando R, Hama H, Yamamoto-Hino M, et al. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99(20):12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297(5588):1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 24.Chen I, Howarth M, Lin W, et al. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2(2):99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 25.Howarth M, Takao K, Hayashi Y. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc Natl Acad Sci U S A. 2005;102(21):7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howarth M, Chinnapen DJ, Gerrow K, et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat Methods. 2006;3(4):267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin J, Straight PD, McLoughlin SM, et al. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc Natl Acad Sci U S A. 2005;102(44):15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams SR, Campbell RE, Gross LA, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124(21):6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 29.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281(5374):269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 30.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10(11):1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bates IR, Hebert B, Luo Y, et al. Membrane lateral diffusion and capture of CFTR within transient confinement zones. Biophys J. 2006;91(3):1046–1058. doi: 10.1529/biophysj.106.084830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haggie PM, Kim JK, Lukacs GL, et al. Tracking of quantum dot-labeled CFTR shows near immobilization by C-terminal PDZ interactions. Mol Biol Cell. 2006;17(12):4937–4945. doi: 10.1091/mbc.E06-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89(1):141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]