Abstract
Background
Pseudomonas aeruginosa is one of the most relevant human opportunistic bacterial pathogens. Two strains (PAO1 and PA14) have been mainly used as models for studying virulence of P. aeruginosa. The strain PA14 is more virulent than PAO1 in a wide range of hosts including insects, nematodes and plants. Whereas some of the differences might be attributable to concerted action of determinants encoded in pathogenicity islands present in the genome of PA14, a global analysis of the differential host responses to these P. aeruginosa strains has not been addressed. Little is known about the host response to infection with P. aeruginosa and whether or not the global host transcription is being affected as a defense mechanism or altered in the benefit of the pathogen. Since the social amoeba Dictyostelium discoideum is a suitable host to study virulence of P. aeruginosa and other pathogens, we used available genomic tools in this model system to study the transcriptional host response to P. aeruginosa infection.
Results
We have compared the virulence of the P. aeruginosa PAO1 and PA14 using D. discoideum and studied the transcriptional response of the amoeba upon infection. Our results showed that PA14 is more virulent in Dictyostelium than PA01using different plating assays. For studying the differential response of the host to infection by these model strains, D. discoideum cells were exposed to either P. aeruginosa PAO1 or P. aeruginosa PA14 (mixed with an excess of the non-pathogenic bacterium Klebsiella aerogenes as food supply) and after 4 hours, cellular RNA extracted. A three-way comparison was made using whole-genome D. discoideum microarrays between RNA samples from cells treated with the two different strains and control cells exposed only to K. aerogenes. The transcriptomic analyses have shown the existence of common and specific responses to infection. The expression of 364 genes changed in a similar way upon infection with one or another strain, whereas 169 genes were differentially regulated depending on whether the infecting strain was either P. aeruginosa PAO1 or PA14. Effects on metabolism, signalling, stress response and cell cycle can be inferred from the genes affected.
Conclusion
Our results show that pathogenic Pseudomonas strains invoke both a common transcriptional response from Dictyostelium and a strain specific one, indicating that the infective process of bacterial pathogens can be strain-specific and is more complex than previously thought.
Background
Nosocomial infections caused by opportunistic pathogens are one of the most important health problems in developed countries. Depending on the geographic location, P. aeruginosa is the first or second causative agent of nosocomial infections [1,2]. P. aeruginosa infects patients suffering from AIDS, people at intensive care units, and burned people among others, and is the major cause of morbidity and mortality in patients with cystic fibrosis, the most prevalent hereditary disease in Caucasian populations [3]. A successful infection by this type of pathogens depends on the interplay of multiple factors including the susceptibility of the host, the virulence of the strain and its resistance to antibiotics [4]. Previous work has shown that the physiological fitness and the virulence of P. aeruginosa and other opportunists are affected by the expression of antibiotic resistance mechanisms such as MDR-pumping systems [5-8].
The pathogenicity of Pseudomonas aeruginosa involves various components operating at different levels. The flagella and pili facilitate contact with the bacterium's cell target and play a role in its adhesion, which is a critical step in the infection [9,10]. After contact, the type III secretion system is able to inject into the cytoplasm of the target cell a series of cytotoxic molecules that act at various levels. The mechanism of action involves, in many cases, the presence of host cofactors still unidentified [11]. Other virulence factors involve products secreted into the extracellular medium by systems I and II such as elastase, alkaline phosphatase and exotoxin A among others. The expression of many of these virulence factors is regulated by a mechanism of bacteria-to-bacteria cell signalling known as quorum-sensing [12]. Despite the functional and genomic similarity among different P. aeruginosa strains [13,14], some differences in their pathogenicity have been observed [15]. For example, the clinical isolate PA14 is more virulent than PAO1 in a wide range of hosts [15-17]. It has been shown that the genome of PA14 contains two pathogenicity islands that are not present in PAO1 and it has been proposed that the virulence in this organism (and the difference between PA14 and PAO1) is the result of a pool of pathogenicity genes interacting in various combinations in different genetic backgrounds [15]. In spite of these suggestions, the cause of the different virulence behavior of PAO1 and PA14 is not yet fully understood.
Although most of the work on pathogenesis has been focused on understanding the bacterial factors that render a virulence phenotype, increasing attention is being paid to the host and those aspects connected to the susceptibility or resistance to infection. Understanding the host-pathogen relationship, at both the cellular and molecular level, is essential to identify new targets and develop new strategies to fight infection. Molecular analysis of host-pathogen interactions would benefit from the use of model systems allowing a systematic study of the factors involved. In this regard the social amoeba D. discoideum has proven particularly useful for its ease of handling, genetic tractability [18-22] and fully sequenced genome [23].
D. discoideum is a soil microorganism that feeds on bacteria by phagocytosis. The interaction between bacteria and their natural predators (Dictyostelium, other protists and worms) is believed to have shaped both predators [24] and bacterial evolution. As a consequence, some of the mechanisms developed by bacteria to avoid the activity of their natural predators in the environment might have been adapted later in evolution to allow the infection of higher organisms such as humans [25]. Specifically, it was found that the quorum-sensing mechanisms and type III secretion, which are essential factors in the infectivity to humans are also responsible for the infectivity of P. aeruginosa in D. discoideum [18,20,21].
Our previous studies have shown the utility of this model system of infection to analyze the virulence of other opportunistic pathogens like Stenotrophomonas maltophilia [7]. It has been also demonstrated the validity of D. discoideum as a model of infection by intracellular pathogens such as Legionella, Cryptococcus and Mycobacterium [19,22]. Consequently, the conservation of the mechanisms of infection needed to infect mammals and D. discoideum in a wide variety of pathogens reinforces the use of this system as a valid model to study host-pathogen relations. We have used whole-genome D. discoideum microarrays to study global host transcription upon infection with Pseudomonas aeruginosa PAO1 and PA14 to determine whether or not transcription is being affected as a defense mechanism or altered in the benefit of the pathogen.
Results
Pseudomonas aeruginosa strains PAO1 and PA14 show a different virulence behavior in D. discoideum
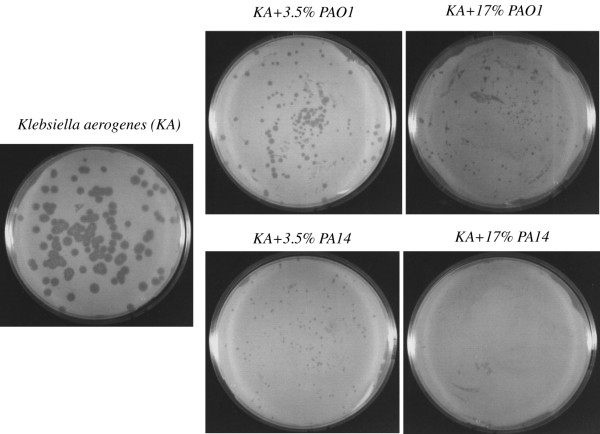
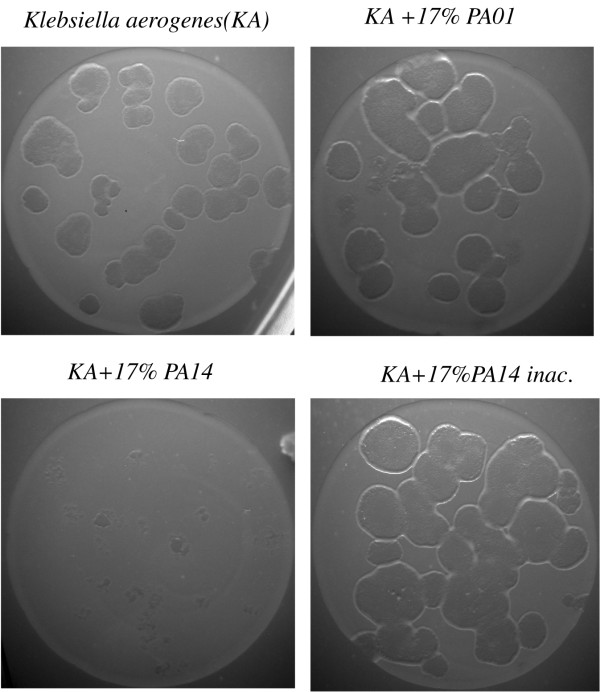
PAO1 and PA14 are two clinical isolates of P. aeruginosa frequently used as model strains to analyze the virulence of this bacterial pathogen. Since they behave differently in some aspects dealing with the expression of virulence determinants, we wanted to compare the differential response of the host to these strains. For this purpose, we made use of D. discoideum as a model for virulence. As a first step a plating assay of virulence was set up. Figure 1 shows a representative experiment of three independent assays in which D. discoideum cells were grown in association with bacteria on nutrient SM plates. Klebsiella aerogenes, a non -pathogenic bacteria, was used as an appropriate food supply and P. aeruginosa mixed at the indicated proportions. An effect in the size of the clearing plaques could already be seen when only 3.5% of P. aeruginosa cells were mixed with 96.5% of K. aerogenes cells and this effect was even clearer using 17% of P. aeruginosa cells. When the behavior of the strains was analyzed in more detail, it was found that PAO1 is reproducibly more permissive than PA14 as observed by the higher growth of D. discoideum on PAO1. The differences in the area of the cleared bacterial lawn between PAO1 and PA14 were measured for the condition corresponding to the 3.5 % mixture. The average area and the standard deviation were 1.65 ± 1.2 mm2 for PAO1 and 0.11 ± 0.07 mm2 for PA14 (the number of clear plaques measured in each condition was 50). The significance of differences between groups as determined by Student's t-test was p < 10-8. To further confirm these results a different plating assay was performed on non-nutrient agar. PAO1, PA14 and K. aerogenes were previously grown in LB overnight, washed out of the media by centrifugation and deposited with D. discoideum cells in agar plates at the indicated proportions. Under these conditions the difference in the virulence between PAO1 and PA14 was even more evident as shown in a representative experiment in Figure 2. Interestingly PAO1 is permissive to D. discoideum growth under these non-nutrient conditions. However, PA14 still shows a strong virulence against D. discoideum. All together these results suggest that PA14 is more virulent than PAO1 in the D. discoideum model of virulence.
Figure 1.
PA14 is more virulent than PAO1 in SM-plating assay. Approximately 100 D. discoideum cells were cultivated in SM-plates with the indicated proportion of Klebsiella and Pseudomonas strains (PAO1 or PA14) previously grown and adjusted to the same optical density. Plates were maintained at 22°C for 5 days. Growth of D. discoideum is severely affected by the presence of Pseudomonas but the inhibition is stronger when PA14 is used.
Figure 2.
PA14 is more virulent than PAO1 in PDF-agar plating assay. D. discoideum cells were cultivated in non-nutrient agar on a lawn of Klebsiella and Pseudomonas (PAO1 and PA14) at the indicated proportion. Under these conditions PA14 maintain a high virulence as seen by the strong inhibition of D. discoideum growth. Heat inactivated PA14 is used as a control.
Pseudomonas aeruginosa induces a specific gene expression response in Dictyostelium
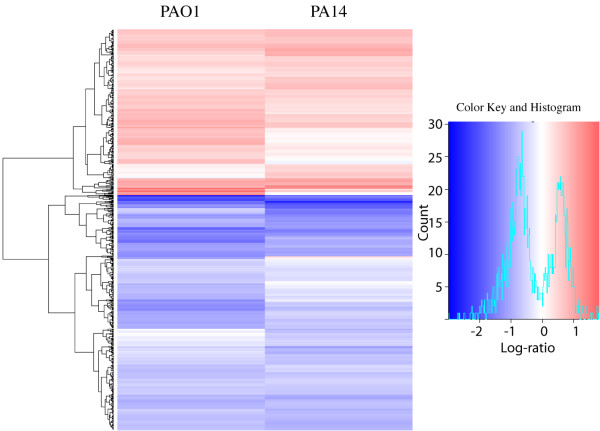
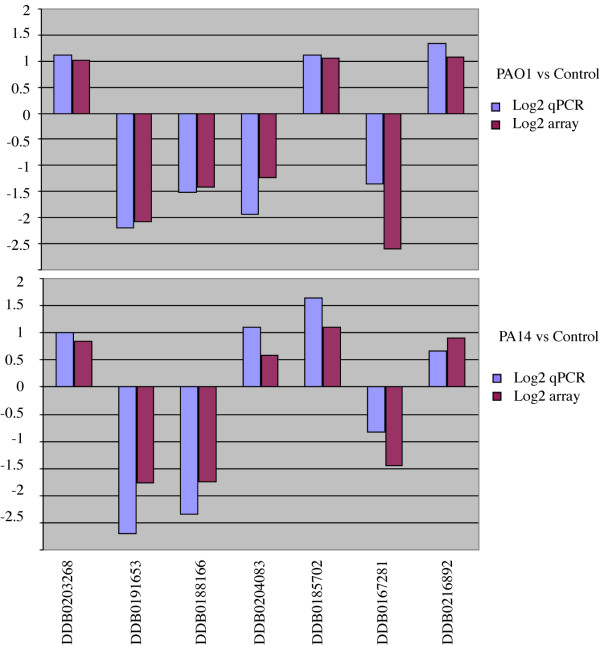
Little is known about the interplay between the host and the pathogen in terms of gene expression responses. We wanted to determine if there is a specific gene expression response of D. discoideum to their interaction with P. aeruginosa. D. discoideum cells were exposed to P. aeruginosa strains PAO1 and PA14 mixed with an excess of K. aerogenes in HL5 for 4 hours. K. aerogenes alone was used as a control to which the gene expression levels were compared. RNA was extracted from D. discoideum and used to study the global pattern of gene expression using whole-genome D. discoideum microarrays (see Additional file 1 for the complete data). Using a P < 0.05 cutoff, there were 752 genes whose expression was significantly different between the PAO1-treated cells and the controls and 624 genes between PA14-treated cells and controls (Table 1 summarizes the results at different P values and log-ratios). The heat map shown in Figure 3 indicates that the responses were broadly comparable between the two strains with very few genes oppositely altered in the two conditions. The differences in the gene expression are approximately in the range between four-fold repression and three-fold induction (log-ratios between of -2 to +1.5 as shown in the histogram of Figure 3). These results were validated by real time PCR of the same samples used for the transcriptomic assays, measuring the expression of 7 representative genes that were up-regulated or down-regulated in the different conditions. Figure 4 shows a good correlation between the data obtained from the microarray transcriptomic experiment as compared with that obtained by quantitative RT-PCR. Although the log-ratio changes in the gene expression showed some differences the overall trend were consistent, supporting the reliability of our data.
Table 1.
Differential genes at p < 0.05 and different Log 2 ratios
| PAO1 vs Control | PA14 vs Control | PAO1+PA14 vs Control | PAO1 vs PA14 | |
| Log 2 ratio | 752 | 623 | 364 (Table 2) | 169 (Table 3) |
| (>+0.5 or <-0.5) | 461 down 291 up | 396 down 227 up | 258 down 106 up | 60 down 109 up |
| Log 2 ratio | 150 | 125 | 70 | 35 |
| (>+1 or <-1) | 126 down 24 up | 105 down 20 up | 66 down 4 up | 14 down 21 up |
Figure 3.
Pseudomonas aeruginosa induces gene expression changes in Dictyostelium. Heat map comparing the genes significantly altered (p < 0.05) between PAO1-treated cells versus control (975) and PA14-treated cells versus control (838). Each row of the plot is a gene and was colored according to the log2ratio of expression with red meaning up-regulation in relation to the controls and blue downregulation. The histogram shows the range of changes in a log2 scale. The data presented are for the three independent experiments combined. The heatmap was generated using the heatmap.2 function of the gplots package in R [47]. The dendrogram was generated using Euclidean distance and the "complete" agglomeration method.
Figure 4.
Correlation of microaray and real-time PCR. Real-time PCR measurements of the mRNA levels for seven representative genes whose expression were affected in the array. Upper panel shows a direct comparison of the changes in a log2 scale for PAO1 versus control and the lower panel shows the same genes for PA14 versus control. Blue bars corresponded to quantitative real time PCR and the purple bars to the array data. The array data and the real time PCR displayed are the combination of three independent biological experiments. The correlation coefficients were: R2 = 0.87 for PAO1 and R2 = 0.91 for PA14.
Common and specific responses of D. discoideum to the infection with PAO1 and PA14 strains
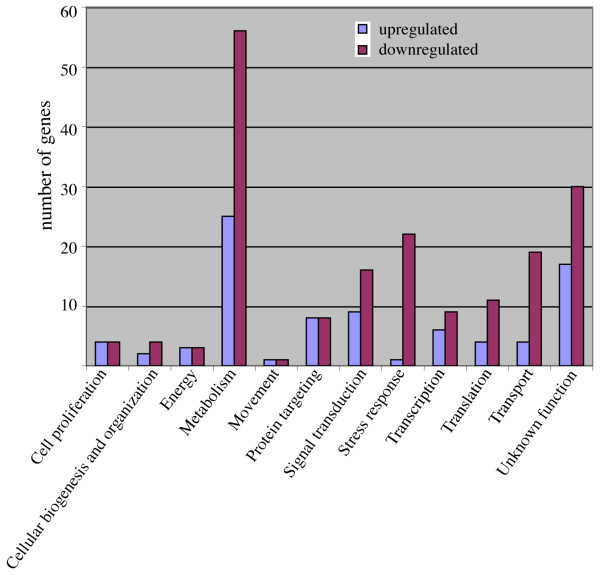
As shown in Table 1 there were 364 genes that showed similar differential regulation with both bacterial strains compared with the controls (labeled as PAO1+PA14 vs control). We have considered in the analysis those genes showing differences in log-ratios that are higher than +0.5 or lower than -0.5. Interestingly the expression of another group of 169 genes (labeled as PAO1 vs PA14) was different depending on whether the infecting strain was PAO1 or PA14. We have studied in detail both groups by manual annotation and categorizing using the extended categorization for D. discoideum previously described [26]. Genes of unknown function and those showing weak homologies were not included in the list. Table 2 contains the genes that were similarly regulated upon infection with any of both strains, and in Figure 5 the genes are categorized by function (see Additional file 2 for the complete data). The first interesting conclusion from this experiment is the existence of a common transcriptional response that affects many different genes that are involved in a wide range of functions. The proportion of the genes that were downregulated by the treatment with both strains of P. aeruginosa is higher (258 genes) compared to those upregulated (106 genes). This difference is more evident in categories such as stress response and transport (Figure 5).
Table 2.
Genes differentially expressed upon infection with PAO1 and PA14 versus Klebsiella
| Gene ID | Gene function and name | Log 2 ratio: PAO1-C | PA14-C |
| Cell proliferation | |||
| DDB0216882 | Cyclin-dependent kinase regulatory | -0,89 | -0,97 |
| DDB0188449 | cdc40, conserved splicing factor | 0,68 | 0,78 |
| DDB0205486 | CDK family protein kinase | 0,81 | 0,89 |
| DDB0168249 | cdk1, "cyclin-dependent_kinase, p34-cdc2_protein" | -0,78 | -0,83 |
| DDB0216532 | cdk10;"putative_CDK_family_protein_kinase | 0,91 | 1,08 |
| DDB0185341 | PP-loop family | -0,60 | -0,70 |
| DDB0205486 | putative protein serine/threonine kinase, CDK family protein kinase | 0,68 | 0,64 |
| DDB0218360 | PhoPQ-activated pathogenicity-related protein | -0,59 | -0,57 |
| Cellular biogenesis and organization | |||
| DDB0189693 | copA, coatomer protein complex alpha subunit | 0,52 | 0,68 |
| DDB0216892 | lvsB;"BEACH_domain-containing_protein" | 1,05 | 0,91 |
| DDB0202609 | Transport protein particle (TRAPP) | -0,77 | -0,61 |
| DDB0187558 | putative mitochondrial import inner membrane translocase | -0,65 | -0,65 |
| DDB0186481 | atg9, APG9, "autophagy_protein_9" | -0,88 | -0,77 |
| DDB0217942 | Putative Mpv17/PMP22 family | -0,66 | -0,93 |
| Energy | |||
| DDB0217090 | Isocitrate lyase family | -0,80 | -0,84 |
| DDB0192001 | ppkA, "poly_P_kinase, polyphosphate_kinase" | -0,65 | -0,51 |
| DDB0190821 | sdhB, complex II, iron-sulfur protein (IP) subunit | 0,74 | 0,53 |
| DDB0190821 | sdhB;"complex_II,(ubiquinone), succinic_dehydrogenase" | 0,59 | 0,61 |
| DDB0167662 | similar to Coenzyme Q9 | -0,71 | -0,72 |
| DDB0204006 | AMPK beta-2 chain | 0,52 | 0,73 |
| Metabolism | |||
| DDB0187528 | cysteine dioxygenase | 0,69 | 0,66 |
| DDB0185702 | hgd;"homogentisate_1,2-dioxygenase" | 1,05 | 1,10 |
| DDB0184361 | Thiamine pyrophosphate enzyme | -0,57 | -0,88 |
| DDB0186120 | gshA, "gamma_glutamylcysteine_synthetase, glutamate-cysteine_ligase" | -1,72 | -0,94 |
| DDB0167249 | Aldehyde dehydrogenase | -0,77 | -0,94 |
| DDB0192169 | alrA;"aldehyde_reductase, aldo-keto_reductase" | -0,54 | -0,88 |
| DDB0218652 | alrB;"aldo-keto_reductase" | -0,60 | -0,69 |
| DDB0189745 | alrC;"aldo-keto_reductase" | -0,84 | -0,72 |
| DDB0186332 | alrE;"aldo-keto_reductase" | -1,23 | -0,99 |
| DDB0204015 | D-Lactate dehydrogenase | 0,92 | 0,65 |
| DDB0187572 | Endoglucanase_E_like | 0,64 | 0,78 |
| DDB0203268 | glgB;"1,4-alpha-glucan_branching_enzyme, branching_enzyme" | 1,02 | 0,85 |
| DDB0187562 | glk;glucokinase | 0,69 | 0,56 |
| DDB0217973 | Gluconolactonase | 0,85 | 0,82 |
| DDB0202855 | Glycoside hydrolase | -1,63 | -1,45 |
| DDB0202233 | Glycosyl hydrolase family 7 | -1,26 | -1,38 |
| DDB0167594 | Glycosyl hydrolases family | 0,60 | 0,56 |
| DDB0204016 | gpt10;"putative_glycophosphotransferase" | -0,69 | -1,11 |
| DDB0204037 | Legume lectins beta-chain signature | -0,86 | -1,16 |
| DDB0206405 | Mannosyl oligosaccharide glucosidase | -0,80 | -0,68 |
| DDB0205896 | NAD-dependent epimerase/dehydratase family protein | -2,12 | -1,73 |
| DDB0204752 | Phosphoglycerate mutase family | 0,64 | 0,61 |
| DDB0190464 | Predicted kinase related to galactokinase and mevalonate kinase | 0,52 | 1,02 |
| DDB0186919 | zinc-containing alcohol dehydrogenase | -2,09 | -1,99 |
| DDB0168737 | zinc-containing alcohol dehydrogenase (ADH) | -1,35 | -1,03 |
| DDB0169356 | carboxylic ester hydrolase | -1,03 | -0,72 |
| DDB0190523 | CAS1;"cycloartenol_synthase" | 0,85 | 0,88 |
| DDB0185601 | cutA;"fatty_acid_elongase_3-ketoacyl-CoA_synthase, long_chain_fatty_acid_elongase" | -0,93 | -0,62 |
| DDB0188166 | delta-24-sterol methyltransferase | -1,42 | -1,74 |
| DDB0186908 | eapA;"alkyl-dihydroxyacetonephosphate_synthase" | 0,72 | 0,81 |
| DDB0187604 | enoyl-CoA hydratase/isomerase domain-containing protei | -0,65 | -0,63 |
| DDB0205302 | Enoyl-CoA hydratase/isomerase family | -0,64 | -0,68 |
| DDB0205157 | fadB, des5-2, "delta_5_fatty_acid_desaturase" | 0,89 | 1,11 |
| DDB0190288 | fcsB, fatty acyl-CoA synthetase, long-chain-fatty-acid-CoA ligase | -1,45 | -1,57 |
| DDB0191679 | GNS1/SUR4 family | -1,03 | -0,59 |
| DDB0205505 | mfeB;"hypothetical_peroxisomal_multifunctional_enzyme_2" | -1,06 | -0,65 |
| DDB0191653 | patatin family protein | -2,07 | -1,77 |
| DDB0218187 | Perilipin family | -1,35 | -1,04 |
| DDB0184443 | Saposin (B) Domains | 0,59 | 0,59 |
| DDB0190553 | Similar to sterol-C4-methyl oxidase-like | -0,71 | -0,91 |
| DDB0217332 | stearoyl-CoA desaturase | 0,90 | 1,02 |
| DDB0206478 | allC;"allantoate_amidinohydrolase, allantoicase" | -1,12 | -1,25 |
| DDB0205700 | cysteine desulfurase | -1,31 | -1,26 |
| DDB0169540 | MOSC domain | -1,00 | -0,67 |
| DDB0187599 | 5 prime nucleotidase family | -0,90 | -0,70 |
| DDB0187063 | 3-methyl-2-oxobutanoate hydroxymethyltransferase | -1,83 | -1,41 |
| DDB0190860 | adenine phosphoribosyltransferase | -0,98 | -0,75 |
| DDB0203073 | adenosine deaminase-related growth factor | -1,35 | -0,74 |
| DDB0215237 | ATP:D-ribose_5-phosphotransferase, ribokinase | -0,56 | -0,51 |
| DDB0206047 | CTP synthase | -1,01 | -1,20 |
| DDB0187738 | Cytidine and deoxycytidylate deaminase zinc-binding region | -0,76 | -0,73 |
| DDB0191911 | putative RNA methylase | -1,01 | -1,21 |
| DDB0185785 | Putative RNA methylase family | -0,59 | -0,63 |
| DDB0219236 | pyrK, cytidylate kinase | -0,98 | -0,71 |
| DDB0217423 | rnrB_2, ribonucleotide reductase small subunit | -0,79 | -0,84 |
| DDB0215284 | tRNA/rRNA methyltransferase SpoU family protein | -0,77 | -0,93 |
| DDB0186269 | Thioredoxin family | -1,41 | -1,91 |
| DDB0206431 | FAD binding domain | -1,63 | -1,13 |
| DDB0187958 | gchA, "GTP_cyclohydrolase_I" | -0,91 | -0,89 |
| DDB0185963 | Oxysterol-binding protein | -0,55 | -0,75 |
| DDB0205608 | pks18, putative polyketide synthase | 0,54 | 0,60 |
| DDB0168380 | pks5, putative polyketide synthase | 0,57 | 0,99 |
| DDB0219613 | stlB, putative polyketide synthase | 0,68 | 1,05 |
| DDB0186173 | Histidine acid phosphatase | -0,79 | -0,80 |
| DDB0184156 | Acetyltransferase (GNAT) family | 0,81 | 0,78 |
| DDB0205937 | Aldehyde dehydrogenase family | 1,02 | 0,81 |
| DDB0183800 | dihydrolipoamide_dehydrogenase | -0,60 | -0,57 |
| DDB0188526 | FAD binding domain | -1,14 | -0,93 |
| DDB0169374 | haloacid dehalogenase-like hydrolase | -1,26 | -1,45 |
| DDB0204714 | hemA, ALAS, "5-aminolevulinate_synthase, ALA_synthase" | 1,00 | 0,99 |
| DDB0187575 | monooxygenase, FAD-binding | -1,30 | -1,14 |
| DDB0203608 | NADH:flavin oxidoreductase/NADH oxidase domain-containing protein | -2,99 | -2,49 |
| DDB0186877 | Predicted hydrolases or acyltransferases | -0,70 | -0,83 |
| DDB0203708 | Putative dehydrogenase domain | 0,60 | 0,66 |
| DDB0186921 | Putative quinone oxidoreductase | -0,68 | -0,80 |
| DDB0218378 | selD, selenophosphate synthase | 0,83 | 0,85 |
| DDB0219578 | short chain dehydrogenase | -0,56 | -0,76 |
| DDB0168766 | short chain dehydrogenase | 0,75 | 0,90 |
| DDB0201995 | Short-chain alcohol dehydrogenase | -0,64 | -0,69 |
| DDB0191047 | Sucrolytic enzyme/ferredoxin homolog protei | -0,76 | -0,55 |
| DDB0205223 | Ubiquinone biosynthesis protein | 1,02 | 0,99 |
| Movement | |||
| DDB0190345 | Actin | -0,62 | -0,64 |
| DDB0216677 | tubB;"beta_tubulin" | 0,78 | 0,66 |
| Protein targeting | |||
| DDB0189280 | CLN3 protein;Major Facilitator Superfamily | -1,38 | -0,81 |
| DDB0186130 | pigF, phosphatidylinositol glycan, class Fphosphoethanolamine N- methyltransferase family | -1,13 | -1,33 |
| DDB0187271 | Protein prenyltransferase, alpha subunit | -0,69 | -0,66 |
| DDB0187195 | Ubiquitin family protein | -0,68 | -0,56 |
| DDB0217546 | vps13B, vacuolar protein sorting-associated protein | 0,80 | 0,57 |
| DDB0205767 | Importin-beta N-terminal domain | -1,39 | -0,91 |
| DDB0219696 | CSN3, COP9 signalosome complex subunit 3 | 0,86 | 0,56 |
| DDB0188097 | mppB;"mitochondrial_processing_peptidase_beta_subunit" | 0,85 | 0,93 |
| DDB0188792 | npl4, nuclear protein localization 4 | -1,46 | -1,26 |
| DDB0189322 | Peptidase family M41 | 0,66 | 0,82 |
| DDB0204548 | putative E3 ubiquitin ligase | 0,56 | 0,88 |
| DDB0203213 | Putative serine protease | 0,57 | 0,64 |
| DDB0190305 | RING-finger-containing ubiquitin ligase | -1,71 | -2,23 |
| DDB0191910 | sigB, GP63, orfGP63", "leishmanolysin_family_protein, peptidase | -1,44 | -1,19 |
| DDB0219558 | usp12, putative ubiquitin carboxyl-terminal hydrolase (UCH) | 0,81 | 0,50 |
| DDB0188490 | usp40, putative ubiquitin carboxyl-terminal hydrolase (UCH) | 0,83 | 0,60 |
| Signal transduction | |||
| DDB0167328 | ArfGAP, Arf GTPase activating protein | 0,58 | 0,67 |
| DDB0187828 | gacX, RacGAP | 0,62 | 0,52 |
| DDB0217797 | gpaG, "G-protein_subunit_alpha_7" | -0,93 | -0,77 |
| DDB0205484 | GTPase-activator protein for Ras-like | 0,58 | 0,69 |
| DDB0202545 | rabX;"Rab_GTPase" | -0,59 | -0,76 |
| DDB0186244 | abkD, AdckB2, "putative_ABC1_family_protein_kinase" | 0,80 | 0,88 |
| DDB0217600 | nek3, putative protein serine/threonine kinase | 0,55 | 0,56 |
| DDB0203684 | tyrosine kinase-like | 0,53 | 0,56 |
| DDB0189806 | vwkA;"protein_serine/threonine_kinase" | -0,77 | -0,67 |
| DDB0205355 | Calcineurin-like phosphoesterase | -1,27 | -0,86 |
| DDB0218779 | pdsA, "PDE, pde1, pdeA", "cAMP_phosphodiesterase" | 0,79 | 1,40 |
| DDB0204820 | rabS;"Rab_GTPase" | -0,80 | -0,87 |
| DDB0190872 | NLI interacting factor-like phosphatase | -0,55 | -0,60 |
| DDB0185382 | Protein phosphatase 5, catalytic subunit | -0,68 | -0,62 |
| DDB0186390 | protein tyrosine phosphatase | -0,85 | -0,74 |
| DDB0218065 | ptpB, DdPTPa, "phosphotyrosine_phosphatase_ptp2 | -1,20 | -1,01 |
| DDB0189698 | Tyrosine specific protein phosphatases family | -0,76 | -0,89 |
| DDB0203756 | G-protein-coupled receptor (GPCR) family protein | 0,69 | 0,99 |
| DDB0189216 | gacD, RacGAP | -0,63 | -0,58 |
| DDB0217433 | Regulator of G protein signaling | 0,90 | 0,72 |
| DDB0169375 | cGMP-specific phosphodiesterase | -0,83 | -0,79 |
| DDB0167494 | plbF, PLB, "phospholipase_B-like" | -1,17 | -1,01 |
| DDB0168860 | sgkA, "SK, SPHK", "sphingosine_kinase" | -0,97 | -0,79 |
| DDB0167227 | Cytochrome b5-like Heme/Steroid binding domain | -0,71 | -1,57 |
| DDB0216720 | Tetraspanin family | -0,73 | -0,68 |
| Stress response | |||
| DDB0202483 | AhpC/TSA family | -0,51 | -0,68 |
| DDB0203727 | AhpC/TSA family protein. Thioredoxin-like | -1,25 | -1,89 |
| DDB0203727 | AhpC/TSA family protein. Thioredoxin-like | -1,11 | -2,00 |
| DDB0205904 | AhpC/TSA family protein. Thioredoxin-like | -1,10 | -1,63 |
| DDB0168230 | Cytochrome P450 | 0,61 | 0,67 |
| DDB0217979 | cytochrome P450 family protein | -1,11 | -1,24 |
| DDB0187276 | cytochrome P450 family protein | -0,73 | -0,88 |
| DDB0186118 | cytochrome P450 family protein | -0,54 | -0,51 |
| DDB0167587 | Glutathione S-transferase | -2,24 | -2,02 |
| DDB0218804 | Glutathione S-transferase | -1,07 | -0,86 |
| DDB0185602 | putative FMN-dependent NAD(P)H:quinone reductase | -0,66 | -0,93 |
| DDB0168563 | putative glutathione S-transferase | -0,64 | -1,23 |
| DDB0201962 | Ku70-binding family protein | -0,60 | -0,51 |
| DDB0191833 | TFIIH4, "TFIIH_subunit, general_transcription_factor_IIH,_polypeptide_4" | -0,81 | -0,90 |
| DDB0204089 | NUDIX hydrolase family | -0,89 | -1,14 |
| DDB0185428 | Strictosidine synthase | -1,74 | -1,01 |
| DDB0169033 | trap1, Dd-trap1, "TNF_receptor-associated_protein", member of the HSP90 fam | -1,93 | -1,56 |
| DDB0188234 | Chaperone clpB | -1,00 | -1,25 |
| DDB0192088 | heat shock cognate protein | -0,82 | -0,99 |
| DDB0192086 | heat shock protein, 70 kDa heat shock protein | -0,94 | -1,18 |
| DDB0169051 | Hsp20/alpha crystallin family | -1,13 | -1,30 |
| DDB0169044 | Hsp20/alpha crystallin family | -1,07 | -1,12 |
| DDB0169207 | hspG12, heat shock protein Hsp20 domain-containing protein | -0,72 | -0,86 |
| Transcription | |||
| DDB0204405 | CRTF;"transcription_factor" | 0,97 | 1,15 |
| DDB0167879 | IWS1 C-terminus | 0,70 | 0,69 |
| DDB0205969 | snd1, tudor domain-containing protein | 0,66 | 0,79 |
| DDB0188840 | TFIIAL, "transcription_factor_IIA" | -0,79 | -0,55 |
| DDB0167865 | ddx52, DEAD/DEAH box helicase | -0,55 | -0,55 |
| DDB0184074 | ddx6, DEAD/DEAH box helicase | 0,62 | 0,66 |
| DDB0184228 | DEAD/DEAH box helicase | -0,63 | -0,60 |
| DDB0206136 | myb domain-containing protein | 0,59 | 0,58 |
| DDB0189583 | rpmA;"DNA-dependent_RNA_polymerase" | -1,33 | -1,17 |
| DDB0186101 | pwp2, ortholog of H. sapiens and S. cerevisiae PWP2 | -0,80 | -0,88 |
| DDB0192008 | rpa2, RNA polymerase I, second largest subunit | -0,98 | -1,13 |
| DDB0218008 | rpc4;"putative_RNA_polymerase_III_subunit" | -0,58 | -0,55 |
| DDB0216877 | tRNA pseudouridine synthase | -0,56 | -0,65 |
| DDB0204724 | DNA helicase TIP49, TBP-interacting protei | 0,63 | 0,64 |
| DDB0219410 | pirin-like protein | -0,99 | -0,81 |
| Translation | |||
| DDB0167043 | Ribosomal protein L10 | -0,86 | -1,37 |
| DDB0190639 | Fibrillarin | -0,65 | -0,84 |
| DDB0184302 | Mitochondrial small ribosomal subunit Rsm22 | -0,70 | -0,72 |
| DDB0205674 | MPP10, U3 small nucleolar ribonucleoprotein | -0,77 | -0,66 |
| DDB0201601 | mrpl11, S60 ribosomal protein L11, mitochondrial | -0,91 | -1,26 |
| DDB0204554 | Ribosomal protein L28 | -0,50 | -0,69 |
| DDB0188692 | Ribosomal protein S8e | -0,94 | -0,79 |
| DDB0183814 | Ribosomal RNA processing protein 4 | 0,58 | 0,56 |
| DDB0188661 | rps9, "rp1024, v12", "40S_ribosomal_protein_S9 | 0,60 | 0,70 |
| DDB0219852 | u3 small nucleolar RNA interacting protein 2, putative | -0,75 | -0,63 |
| DDB0191852 | eukaryotic translation initiation factor 3 subunit 5 | 0,59 | 0,54 |
| DDB0203843 | Eukaryotic translation initiation factor 6 (EIF-6)-like protein | -0,96 | -0,64 |
| DDB0189529 | gfm2, mitochondrial translation elongation factor G | -0,59 | -0,50 |
| DDB0202851 | NMD3 family | -0,95 | -0,97 |
| DDB0168814 | aspartyl-tRNA_synthetase | 0,70 | 0,98 |
| Transport | |||
| DDB0217304 | ABC transporter AbcG17 | -1,58 | -1,44 |
| DDB0167281 | ABC transporter mdrA2 | -2,65 | -1,44 |
| DDB0167281 | ABC transporter mdrA2 | -2,26 | -1,45 |
| DDB0191940 | abcB2;"ABC_transporter_B_family_protein" | 0,87 | 0,93 |
| DDB0188931 | abcE1;"RNaseL_inhibitor-like_protein, non-transporter_ABC_protein" | 0,61 | 0,74 |
| DDB0189332 | amino acid permease family protein | -0,72 | -0,87 |
| DDB0168564 | Amino acid/polyamine transporter | -1,28 | -1,06 |
| DDB0190286 | mcfF, Mitochondrial carrier protein | -0,99 | -0,86 |
| DDB0216936 | mftA;"carrier_protein_RIM" | -0,91 | -1,05 |
| DDB0188529 | nucleoporin family protein | -0,94 | -0,72 |
| DDB0189222 | ccsA, copper chaperone for superoxide dismutase | -0,69 | -0,62 |
| DDB0205129 | Co/Zn/Cd efflux system component | -0,56 | -0,71 |
| DDB0202441 | nhe1, DdNHE1, "Na-H_exchanger, sodium/hydrogen_exchanger" | -0,77 | -0,59 |
| DDB0168533 | porA;porin | 0,68 | 0,59 |
| DDB0218156 | P-type cation-transporting ATPase | -0,69 | -0,57 |
| DDB0189480 | mcfT, mitochondrial substrate carrier family protein | -0,74 | -0,86 |
| DDB0202337 | Nodulin, Major Facilitator Superfamily | -0,60 | -0,68 |
| DDB0185520 | Nucleoside transporter | -0,53 | -0,56 |
| DDB0203447 | Sugar (and other) transporter | 1,01 | 0,71 |
| DDB0168979 | Sugar transport proteins signature 1 | -0,78 | -1,28 |
| DDB0189650 | sodium/potassium-transporting ATPase alpha chain 2 | -1,48 | -0,81 |
| DDB0190036 | Major Facilitator Superfamily | -1,13 | -1,05 |
| DDB0205693 | Major Facilitator Superfamily | -1,33 | -1,08 |
Figure 5.
Functional categories of the genes affected similarly by the exposure with PAO1 and PA14. The genes whose expression was altered by PAO1 and PA14 were manually annotated (see Table 2) and grouped in functional categories. The size of the blue bars indicates the number of genes upregulated in each category related to the control and purple bars the number of genes downregulated.
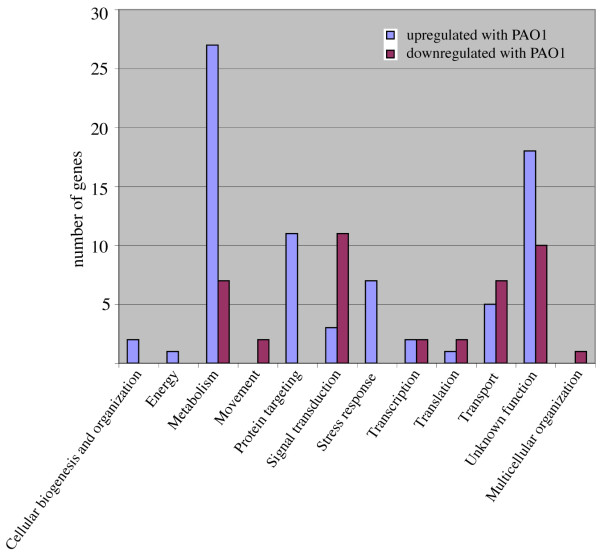
Table 3 displays the D. discoideum genes whose expression changed differentially between PAO1 and PA14 infection and Figure 6 shows the number of genes in each category (see Additional file 3 for complete data). In general a higher proportion of the genes showed a higher level of expression by PAO1 infection (109 genes) when compared with the levels observed by PA14. On the other hand, 60 genes behaved oppositely showing lower levels of expression upon exposure to PAO1 compared to those levels obtained after PA14 infection. Interestingly, all the genes represented in the categories stress-response and protein targeting had a higher level of expression in the cells exposed to PAO1 compared to PA14. The behavior of these genes in comparison with the control is also displayed in Table 3.
Table 3.
Genes differentially expressed upon infection with PAO1 versus PA14
| Gene ID | Gene function and name | Log2 ratio: PAO1-14 | PAO1-C | PA14-C |
| Cellular Biogenesis and organization | ||||
| DDB0187116 | vps13A, vacuolar protein sorting-associated protein | 0,81 | 0,67 | -0,13 |
| DDB0189855 | vps46, Vacuolar Protein Sorting | 0,83 | 0,59 | -0,24 |
| Energy | ||||
| DDB0204335 | cxgE, "cox7E, coxVIIe", "cytochrome_c_oxidase_subunit_VII_E" | 1, 20 | 0,80 | -0,40 |
| Metabolism | ||||
| DDB0190752 | diaminopimelate epimerase | 1,08 | 0,02 | -1,06 |
| DDB0204319 | Hydroxymethyltransferase | 0,63 | 0,43 | -0,20 |
| DDB0168738 | putative arginine deiminase | -0,62 | -0,78 | -0,16 |
| DDB0205389 | acly, ATP citrate lyase | 0,89 | 0,75 | -0,15 |
| DDB0169357 | methylenetetrahydrofolate dehydrogenase | 0,78 | 0,21 | -0,57 |
| DDB0187393 | NAD-dependent epimerase/dehydratase family protein | -0,91 | -1,18 | -0,28 |
| DDB0205386 | putative ATP citrate synthase | 0,96 | 0,79 | -0,17 |
| DDB0205339 | rpe;"ribulose_phosphate_3-epimerase" | 0,78 | 0,43 | -0,35 |
| DDB0187942 | Short-chain alcohol dehydrogenase of unknown specificit | 0,70 | 0,43 | -0,27 |
| DDB0187544 | smlA | 1,43 | 1,00 | -0,43 |
| DDB0217455 | zinc-containing alcohol dehydrogenase (ADH) | -1,21 | -1,16 | 0,06 |
| DDB0217374 | zinc-containing alcohol dehydrogenase (ADH) | -1,02 | -1,37 | -0,35 |
| DDB0190948 | acid ceramidase-like protein | -0,87 | -0,31 | 0,56 |
| DDB0188248 | Acyltransferase | 0,64 | 0,16 | -0,48 |
| DDB0219652 | cinB, "esterase/lipase/thioesterase_domain-containing_protein | 0,92 | 1,28 | 0,36 |
| DDB0189754 | esterase/lipase/thioesterase domain-containing protein | 1,07 | 1,07 | 0,00 |
| DDB0185740 | esterase/lipase/thioesterase domain-containing protein | 1,08 | 1,27 | 0,20 |
| DDB0184141 | Phosphate acyltransferases | 0,55 | -0,26 | -0,82 |
| DDB0167446 | pks16, putative fatty acid synthase | 1,19 | 1,81 | 0,63 |
| DDB0189182 | Putative esterase/lipase/thioesterase | 0,99 | 1,43 | 0,44 |
| DDB0191907 | dUTP diphosphatase | 0,61 | 0,14 | -0,47 |
| DDB0217901 | purH, AICAR transformylase/IMP cyclohydrolase | 0,89 | 0,34 | -0,55 |
| DDB0167009 | pyr4;"dihydroorotate_dehydrogenase, dihydroorotate_oxidase" | 0,68 | 0,18 | -0,50 |
| DDB0217842 | rpiA;"ribose-5-phosphate_isomerase" | 0,63 | 0,42 | -0,21 |
| DDB0189571 | Sulfite reductase, alpha subunit (flavoprotein) | -0,89 | -0,09 | 0,79 |
| DDB0202318 | Cyclopropyl sterol isomerase | 0,73 | 0,31 | -0,42 |
| DDB0167227 | Cytochrome b5-like Heme/Steroid binding domain | 0,86 | -0,71 | -1,57 |
| DDB0168923 | dihydropteridine reductase | 0,60 | 0,14 | -0,47 |
| DDB0185998 | ERG24, Ergosterol biosynthesis | 0,59 | -0,02 | -0,61 |
| DDB0219255 | Fe(II) oxygenase superfamily | 1,14 | 0,91 | -0,23 |
| DDB0217308 | Predicted iron-dependent peroxidase | -0,55 | -0,12 | 0,43 |
| DDB0192180 | putative O-methyltransferase | 0,56 | 0,71 | 0,15 |
| DDB0202301 | putative SAM dependent methyltransferase | 0,88 | 0,94 | 0,05 |
| DDB0167345 | short-chain dehydrogenase/reductase (SDR) family protein | 0,73 | 0,56 | -0,17 |
| Movement | ||||
| DDB0188280 | myoB, "DMIB, abmB", "myosin_IB" | -0,85 | -0,30 | 0,55 |
| DDB0167337 | myoD, DMID, "myosin_ID_heavy_chain" | -0,81 | -0,76 | 0,05 |
| Multicellular organization | ||||
| DDB0216906 | comC;"FIBROSURFIN_PRECURSOR" | -0,65 | -0,39 | 0,26 |
| Protein destination | ||||
| DDB0189735 | homolog to co-chaperone p23 | 0,56 | 0,45 | -0,11 |
| DDB0187409 | Acetyltransferase (GNAT) family | 0,57 | 0,33 | -0,24 |
| DDB0189799 | SET domain-containing protein | 0,52 | 0,33 | -0,19 |
| DDB0202482 | 26S proteasome non-ATPase regulatory subunit 9 | 0,78 | 1,25 | 0,47 |
| DDB0219654 | Cysteine proteinase 1 precursor | 0,70 | 0,61 | -0,09 |
| DDB0167298 | Dipeptidyl aminopeptidase | 0,51 | -0,07 | -0,59 |
| DDB0190542 | Probable proteasome subunit beta type 3 | 0,57 | -0,06 | -0,63 |
| DDB0216902 | prtA, M3L, "proteosomal_alpha-subunit_M3" | 0,93 | 0,94 | 0,01 |
| DDB0216901 | prtB, M3R, "proteosomal_alpha-subunit_7-1" | 1,37 | 1,30 | -0,06 |
| DDB0186869 | small ubiquitin-like protein | 0,61 | 0,33 | -0,28 |
| DDB0217344 | ubiquitin-like domain containing CTD phosphatase | 0,52 | 0,29 | -0,23 |
| Signal transduction | ||||
| DDB0169410 | gpaB, "Ga2, Galpha2, gpa2", "G-protein_subunit_alpha_2" | -0,82 | -0,55 | 0,27 |
| DDB0190318 | rab1C;"Rab_GTPase" | 0,87 | 0,76 | -0,10 |
| DDB0202066 | pakC, STE20 family protein kinase | -0,61 | -0,18 | 0,43 |
| DDB0169250 | putative protein serine/threonine kinase | -0,87 | -0,58 | 0,29 |
| DDB0205782 | roco6;"putative_protein_serine/threonine_kinase | -0,61 | -0,48 | 0,14 |
| DDB0167076 | sepA, putative protein serine/threonine kinase | -0,57 | -0,31 | 0,26 |
| DDB0217465 | fslH, G-protein-coupled receptor (GPCR) family protein | 0,71 | 0,24 | -0,46 |
| DDB0205174 | grlF, GABA-B receptor-like protein | -1,41 | -1,54 | -0,12 |
| DDB0168770 | grlJ, GABA-B receptor-like protein | -0,63 | -0,96 | -0,32 |
| DDB0204083 | grlL, GABA-B receptor-like protein | -1,84 | -1,25 | 0,60 |
| DDB0229801 | grlQ, G-protein-coupled receptor (GPCR) family protein | 0,77 | 0,98 | 0,21 |
| DDB0167432 | gacFF, RacGAP | -0,57 | 0,08 | 0,66 |
| DDB0167384 | putative guanine nucleotide exchange factor (GEF) | -0,70 | -0,01 | 0,70 |
| DDB0167541 | dpoA;"prolyl_oligopeptidase" | -0,66 | -0,11 | 0,55 |
| Stress response and cell rescue | ||||
| DDB0218719 | AhpC/TSA family protein | 0,82 | 0,76 | -0,05 |
| DDB0205882 | AhpC/TSA family protein. Thioredoxin-like | 0,75 | 0,57 | -0,18 |
| DDB0217383 | Cytochrome P450 | 1,16 | 0,42 | -0,74 |
| DDB0168563 | putative glutathione S-transferase | 0,59 | -0,64 | -1,23 |
| DDB0167491 | alyA;lysozyme | 0,63 | 0,73 | 0,10 |
| DDB0167489 | alyB;lysozyme | 0,69 | 0,75 | 0,07 |
| DDB0167490 | alyC;lysozyme | 0,71 | 0,87 | 0,16 |
| Transcription | ||||
| DDB0206051 | member of NOD protein family | 1,05 | 0,90 | -0,15 |
| DDB0202276 | srfC;"putative_MADS-box_transcription_factor" | -0,90 | -0,30 | 0,60 |
| DDB0217613 | wrky1;"putative_WRKY_transcription_factor" | 1,04 | 1,11 | 0,07 |
| DDB0167422 | putative histone acetyltransferase | -0,64 | -0,55 | 0,09 |
| Translation | ||||
| DDB0201621 | mrps2, ribosomal protein S2, mitochondrial | 0,75 | 0,49 | -0,27 |
| DDB0218535 | Eukaryotic elongation factor 1 (EF1) alpha subfamily | -0,83 | -0,18 | 0,65 |
| DDB0167339 | tRNA-ribosyltransferase | -0,52 | -0,56 | -0,04 |
| Transport facilitation | ||||
| DDB0187089 | abcC5;"ABC_transporter_C_family_protein" | -1,12 | -0,68 | 0,44 |
| DDB0218568 | Copper-transporting P-type ATPase | -1,14 | -0,60 | 0,54 |
| DDB0204460 | patA, PAT1, "Ca2+-ATPase, P-type_ATPase" | -0,93 | -0,15 | 0,78 |
| DDB0205031 | potassium channel tetramerization domain-containing protein | 0,92 | 0,84 | -0,08 |
| DDB0217251 | P-type cation-transporting ATPase | -2,09 | -1,46 | 0,63 |
| DDB0186223 | P-type cation-transporting ATPase | -0,69 | -0,40 | 0,29 |
| DDB0167361 | vatE;"vacuolar_H+-ATPase_E_subunit" | 0,54 | 0,47 | -0,08 |
| DDB0217699 | phospholipid-translocating P-type ATPase family protein | -0,67 | -0,63 | 0,04 |
| DDB0187945 | Sugar (and other) transporter | -0,90 | -0,78 | 0,11 |
| DDB0206579 | mcfQ, mitochondrial substrate carrier family protein | 0,65 | 0,78 | 0,13 |
| DDB0192172 | mcfZ, mitochondrial substrate carrier family protein | 0,73 | 0,92 | 0,18 |
| DDB0183815 | Mitochondrial carrier protein | 0,55 | 0,81 | 0,26 |
Figure 6.
Functional categories of the genes differentially affected by the exposure to PAO1 and PA14. The genes whose expression was differentially altered in PAO1 versus PA14 were manually annotated (see Table 3) and grouped in functional categories. The size of the blue bars indicates the number of genes upregulated in PAO1 versus PA14 in each category and purple bars the number of genes downregulated in PAO1 versus PA14.
Discussion
P. aeruginosa is able to infect D. discoideum cells using several virulence traits that are similar to those used to infect mammalian cells and other hosts [18]. The clinical P. aeruginosa isolates PA14 and PAO1 have been used independently to study the infection of Dictyostelium by Pseudomonas in two different laboratories [20,21]. However, no direct comparison had been reported so far between these strains in this pathogenicity model. We now report that PA14 is indeed more virulent in D. discoideum using different plating assays. Since P. aeruginosa is phagocytosed at much lower rate than the non-pathogenic K. aerogenes (commonly used to grow Dictyostelium) [20], the assays were designed to provide sufficient food to D. discoideum to avoid cell starvation. Thus, K. aerogenes was always used in excess together with the pathogenic strains. In the first assay (Figure 1) a nutrient plate was used to allow the growth of bacteria and D. discoideum simultaneously. Under these conditions the presence of PA14 inhibits D. discoideum growth to a greater extent than PAO1. To avoid differences in the growth rates between bacteria that might alter their final proportions, a non-nutrient assay was performed (Figure 2). In these experiments, D. discoideum feed on bacteria that have been previously grown and deposited at different proportions in non-nutrient agar. Interestingly, PAO1 is not virulent in this condition suggesting that bacterial growth is necessary for the expression of the virulence in this strain. However, even in these conditions PA14 is capable of inhibiting D. discoideum growth. Some studies have suggested that PA14 pathogenicity is multifactorial and required the action of multiple virulence mechanisms [15,27]. These differences between strains prompted us to study the transcriptional profile of D. discoideum upon infection with PAO1 and PA14 to gain insights not only into the possible common transcriptional response but also into any specific response that could explain the observed differences in their virulence. Pilot experiments showed that 4 hours of exposure of D. discoideum cells to either Pseudomonas aeruginosa strains did not result in any apparent cell death or change in cell morphology (data not shown). Since we wanted to study the early transcriptional response we chose this short time of exposure to avoid changes due to cell death. The existence of a rapid gene expression response between 1–6 hours upon exposure of D. discoideum cells to Legionella, an intracellular pathogen, has also been described [28].
Our results show the existence of a common transcriptional response to the infection with P. aeruginoa PAO1 or PA14 that affects 364 genes grouped in many different cellular functions. The complexity of the observed transcriptional changes could be the result of the induction of D. discoideum defensive responses or triggered by P. aeruginosa to make a less hostile cell environment that would support a better survival of the pathogen. In this scenario downregulation of genes involved in stress response might be beneficial for a successful infection. Interestingly, we have observed a clear down-regulation of genes dedicated to stress in the common response to PAO1 and PA14 but also in the specific response to the more virulent strain PA14. For example the gene coding for Strictosidine synthase (DDB0185428) which is involved in the synthesis of alkaloids related to defense mechanisms in plants [29], Trap1 (DDB0169033) that plays a central role in cell cycle regulation and differentiation [30] or the genes coding for lysozymes involved in bacterial degradation (DDB0167491) [31], to mention just a few.
Besides stress response other categories are affected by an overall downregulation such as metabolism, translation and transport facilitation. A subcategorization of the genes included in metabolism (see supplementary table 2) showed that all the genes coding for proteins involved in nucleotide metabolism were downregulated in the common response suggesting an effect on cell proliferation. Moreover, a number of other downregulated genes are directly involved in cell growth as demonstrated functionally in previous studies. This is the case for example of DDB0192001 (ppkA, polyphosphate kinase), whose disruption leads to reduced growth on bacteria [32,33], DDB0186120 (gcsA, glutamylcysteine synthetase), which is essential for cell growth as mutants in the gene are not viable in the absence of glutathione [34]. DDB0168860 (sgkA, sphingosine kinase) that is involved in cell proliferation [35], among others that have been annotated in Supplementary Table 2, 3.
Two different expression microarray analysis in mammals upon infection with Pseudomonas aeruginosa have been reported. In the first report epithelial cells were exposed to the pathogen for 3 hours, a short exposure similar to our experimental design. Unfortunately the number of genes represented in the array was very limited (1500 cDNAs) [36]. Only 22 genes were differentially regulated and we have not found any homologous gene in common. The other work reported the analysis of Pseudomonas aeruginosa corneal infection using an oligonucleotide microarray [37]. This experiment is not directly comparable to ours since a long exposure to the pathogen (1 day) was performed to assure an infection process. As a consequence most of the regulated genes were associated with the immune response and apoptosis, aspects that are not present in Dictyostelium.
D. discoideum is also susceptible to the infection by Legionella pneumophila, a facultative intracellular parasite, which uses different infective mechanisms from P. aeruginosa. It is important to note here that the transcriptional response of D. discoideum upon infection with Legionella [28] was essentially different to the one we report for P. aeruginosa. Only 8 genes were found to be altered in both experiments (DDB0186332, DDB0219578, DDB0167879, DDB0205386, DDB0185740, DDB0167345, DDB0201617, DDB0202615). This indicates that the host response is rather specific of the type of infection and the bacterial pathogen involved. Nevertheless, some responses can be also common. For instance, DDB0202615 (nramp1, natural resistance-associated macrophage protein) whose expression is downregulated in PAO1 and PA14, plays an important role in Legionella infection since the null mutant has increased sensitivity to the infection [38]. Nramp1 transports metal cations out of the phagolysosome in an ATP-dependent process. This activity is believed to be necessary to avoid the growth of intracellular pathogens and might also contribute to the efficient killing of other bacterial pathogens.
The variety of genes whose expression is altered by P. aeruginosa infection suggests a complex scenario in which a combined downregulation of the expression of some of the mentioned genes might affect D. discoideum fitness thus favoring the infection. The precise role of these genes in the pathogenesis and the mechanisms that regulates their expression will promote further investigation.
Conclusion
Our results showed that P. aeruginosa PA14 is more virulent than PAO1 in the D. discoideum model using different plating assays. The transcriptional responses of D. discoideum infected by either P. aeruginosa PAO1 or PA14 were analyzed by whole-genome microarrays and the expression of 364 genes changed similarly upon infection with any of both strains as compared with the control. Effects on metabolism, signaling, stress response and cell cycle can be inferred from the genes affected. Interestingly there were 169 genes differentially regulated between PAO1 and PA14, and this differential response might contribute to the different virulence behavior displayed by these two model strains. This is a starting point to begin to understand the complex relationships between environmental opportunistic pathogens and their natural hosts. Besides, our data support the idea that the host responses to different isolates of the same bacterial pathogen are largely different, thus indicating that the crosstalk between the pathogen and its host is more specific and more complex than previously thought.
Methods
D. discoideum growth and plating assays
Dictyostelium AX4 cells were grown axenically in HL5 medium (14 g/l tryptone, 7 g/l yeast extract, 0.35 g/l Na2HPO4, 1.2 g/l KH2PO4, 14 g/l glucose, pH 6.5) or in association with Klebsiella aerogenes on SM plates (10 g/l glucose, 1 g/l yeast extract, 10 g/l peptone, 1 g/l MgSO4·6H20, 1.9 g/l KH2PO4, 0.6 g/l K2HPO4, 20 g/l agar, pH 6.5) [39].
For the nutrient SM-plating assay Pseudomonas aeruginosa (PAO1 and PA14) and Klebsiella aerogenes (KA) were grown overnight in LB. After washing, bacteria were resuspended with PDF (20 mM KCl; 9 mM K2HPO4, 13 mM KH2PO4, 1 mM CaCl2; 1 mM MgSO4; pH: 6.4) and the optical density (OD) determined at 600 nm. After adjusting to 0.5 OD units, 300 μl of Klebsiella and Pseudomonas at the indicated proportions were plated in SM-agar plates with approximately 100 D. discoideum cells.
For non-nutrient plating assay bacteria were grown as before, washed and resuspended in PDF (at an OD of 15 units at 600 nm. 100 μl of bacteria at the indicated proportions were mixed with D. discoideum cells and deposited in a drop over a PDF-based non-nutrient agar plates.
Microarrays
Dictyostelium cells (5 × 107 cells) were deposited in 10 ml of HL5 (without antibiotics) in shaking culture and exposed during 4 hours to 1.0 OD (approximate multiplicity of infection: 1000) of Klebsiella aerogenes as a control or to a mixture of Pseudomonas aeruginosa (PAO1 and PA14) and Klebsiella (used in excess to provide similar conditions of food supply). The proportion of Klebsiella to Pseudomonas (either PAO1 or PA14) was 7:3. Previous experiments showed that these proportions are adequate for a clear inhibition of Dictyostelium growth in plating assays (see the results section). Dictyostelium cells were separated from the bacteria by gentle centrifugation (twice at 1000 rpm, 5 minutes) and RNA isolated by Trizol (Life Technologies) according to manufacturer's instructions. Three independent biological experiments were performed making a total of three independent treatments for each condition (Klebsiella control, PAO1, and PA14). RNA from the three treatments were paired in all three combinations (Control/PAO1; Control/PA14 and PAO1/PA14) and hybridized to three different arrays. The same was performed for the other two biological replicates making a total of 9 microarrays hybridized. One of the three biological replicates was hybridized in the opposite dye orientation to the other two. The arrays, and protocols for labelling, hybridisation and scanning were as previously described [40]. Background fluorescence was subtracted [41], linear models were fitted and the significance of apparent changes in expression was assessed using limma [42,43]. The data were normalised within each array with the printtip loess algorithm to counteract scanning and spatial biases, and further between each array to normalise mean absolute deviations using the 'scale' algorithm [44].
Preliminary analysis using ANOVA methods indicated that many genes had significant differences in expression between treatments, so we proceeded to examine each pairwise contrast in turn. We filtered out less reliable data by selecting genes with p-values adjusted for multiple testing [45] less than 0.05, making use of the moderated t statistics calculated by the eBayes function of limma. Since many genes passing this cutoff showed small changes in expression, we filtered further by absolute log2ratio. The commonly-used cutoff of greater than 2-fold change would have excluded a large number of genes that appeared to change in expression quite consistently, so we used the less stringent criterion of absolute log2ratio >0.5. A lower log2ratio would have included genes with differences in expression too small to be corroborated by other methods. The array design is available from ArrayExpress [46] under the accession A-SGRP-3. The array experiment was deposited in the ArrayExpress database under the accession E-TABM-464.
Quantitative PCR
The same RNA samples subjected to microarray study were used as templates for retrotranscription with High Capacity cDNA Reverse transcription kit (Applied Biosystems) using 250 ng of each RNA in a final volume of 20 μl. For each sample, a triplicated blank was used. This cDNAs served as template in the PCR reaction carried out in 7900 HT Fast Real- Time PCR System using Power Sybrgreen PCR Master Mix 2× with 300 nM oligonucleotides concentration in a final volume of 10 μl. The results were acquired with SDS 2.3 software by Applied Biosystems and handled with EXCEL software by Microsoft. A total of seven genes were studied for each sample and their amount were related to one control gene, DDB0217951, whose expression is not affected by the treatments with the different strains.
Authors' contributions
SC and JC performed the biological experiments and analyzed the data. GB, JS and AI designed and constructed the microarray, and provided informatics and analysis tools; GB contributed to the array experimental design, carried out the array experiments, and contributed to the analysis of results; RK was array project manager. JM contributed to the experimental design of the array and biological experiments. RE designed, coordinated the experiments and drafted the manuscript. All the authors read and approved the final manuscript.
Supplementary Material
Microsoft excel document containing all the results and genes contained in the microarray.
Microsoft excel document containing the array data for the genes that showed differences in both PAO1 and PA14. The genes were manually annotated and filtered for p < 0.05 and log2 ratio >0.5 or <-0.5. Table 2 was obtained from these data.
Microsoft excel document containing the array data and manual annotation of the genes that showed differential regulation between PAO1 and PA14. The genes were filtered for p < 0.05 and log2 ratio >0.5 or <-0.5. Table 3 was obtained from these data.
Acknowledgments
Acknowledgements
This work was supported by grants BMC2006-00394 from the Spanish Ministerio de Educación y Ciencia (R.E.), CCG07-CSIC/SAL-1959 from Comunidad de Madrid/CSIC (R.E.) and BIO2005-04278, LSHM-CT-2005-518152 and LSHM-CT-2005-018705 to J.M. We would like to thank Theresa Feltwell for technical assistance with the microarray experiment. GB, JS and AI were supported by the Wellcome Trust. JC is recipient of a pre-doctoral fellowship from Spanish Ministerio de Educación y Ciencia.
Contributor Information
Sergio Carilla-Latorre, Email: scarilla@iib.uam.es.
Javier Calvo-Garrido, Email: jcalvo@iib.uam.es.
Gareth Bloomfield, Email: garethb@mrc-lmb.cam.ac.uk.
Jason Skelton, Email: jps@sanger.ac.uk.
Robert R Kay, Email: rrk@mrc-lmb.cam.ac.uk.
Alasdair Ivens, Email: alicat@sanger.ac.uk.
José L Martinez, Email: jlmtnez@cnb.csic.es.
Ricardo Escalante, Email: rescalante@iib.uam.es.
References
- Fluit AC, Verhoef J, Schmitz FJ. Frequency of isolation and antimicrobial resistance of gram-negative and gram-positive bacteria from patients in intensive care units of 25 European university hospitals participating in the European arm of the SENTRY Antimicrobial Surveillance Program 1997–1998. Eur J Clin Microbiol Infect Dis. 2001;20:617–625. doi: 10.1007/s100960100564. [DOI] [PubMed] [Google Scholar]
- Fluit AC, Verhoef J, Schmitz FJ. Antimicrobial resistance among isolates cultured from patients hospitalized with lower respiratory tract infection in Europe. Int J Infect Dis. 2002;6:144–146. doi: 10.1016/s1201-9712(02)90077-9. [DOI] [PubMed] [Google Scholar]
- Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. American journal of respiratory and critical care medicine. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Baquero F. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev. 2002;15:647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Diez B, Sanchez P, Baquero F, Martinez JL, Navas A. Differential interactions within the Caenorhabditis elegans-Pseudomonas aeruginosa pathogenesis model. Journal of theoretical biology. 2003;225:469–476. doi: 10.1016/s0022-5193(03)00288-1. [DOI] [PubMed] [Google Scholar]
- Sanchez P, Linares JF, Ruiz-Diez B, Campanario E, Navas A, Baquero F, Martinez JL. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. The Journal of antimicrobial chemotherapy. 2002;50:657–664. doi: 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- Alonso A, Morales G, Escalante R, Campanario E, Sastre L, Martinez JL. Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. The Journal of antimicrobial chemotherapy. 2004;53:432–434. doi: 10.1093/jac/dkh074. [DOI] [PubMed] [Google Scholar]
- Linares JF, Lopez JA, Camafeita E, Albar JP, Rojo F, Martinez JL. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. Journal of bacteriology. 2005;187:1384–1391. doi: 10.1128/JB.187.4.1384-1391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infection and immunity. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo R, Sokol S, Soong G, Gomez MI, Prince A. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialoGM1 and toll-like receptor 2 as well as toll-like receptor 5. American journal of respiratory cell and molecular biology. 2004;30:627–634. doi: 10.1165/rcmb.2003-0260OC. [DOI] [PubMed] [Google Scholar]
- Kipnis E, Sawa T, Wiener-Kronish J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Medecine et maladies infectieuses. 2006;36:78–91. doi: 10.1016/j.medmal.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerging infectious diseases. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A, Rojo F, Martinez JL. Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin. Environmental microbiology. 1999;1:421–430. doi: 10.1046/j.1462-2920.1999.00052.x. [DOI] [PubMed] [Google Scholar]
- Morales G, Wiehlmann L, Gudowius P, van Delden C, Tummler B, Martinez JL, Rojo F. Structure of Pseudomonas aeruginosa populations analyzed by single nucleotide polymorphism and pulsed-field gel electrophoresis genotyping. Journal of bacteriology. 2004;186:4228–4237. doi: 10.1128/JB.186.13.4228-4237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibaud L, Kohler T, Coudray A, Prigent-Combaret C, Bergeret E, Perrin J, Benghezal M, Reimmann C, Gauthier Y, van Delden C, et al. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cellular microbiology. 2008;10:729–740. doi: 10.1111/j.1462-5822.2007.01080.x. [DOI] [PubMed] [Google Scholar]
- Kurz CL, Ewbank JJ. Infection in a dish: high-throughput analyses of bacterial pathogenesis. Current opinion in microbiology. 2007;10:10–16. doi: 10.1016/j.mib.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Pukatzki S, Kessin RH, Mekalanos JJ. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc Natl Acad Sci USA. 2002;99:3159–3164. doi: 10.1073/pnas.052704399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, van Delden C, Curty LK, Kohler T. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bact. 2002;184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal C, Steinert M. Dictyostelium discoideum as a model to study host-pathogen interactions. Methods in molecular biology (Clifton, NJ) 2006;346:507–515. doi: 10.1385/1-59745-144-4:507. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas A, Cobas G, Talavera M, Ayala JA, Lopez JA, Martinez JL. Experimental validation of Haldane's hypothesis on the role of infection as an evolutionary force for Metazoans. Proc Natl Acad Sci USA. 2007;104:13728–13731. doi: 10.1073/pnas.0704497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Heuner K. Dictyostelium as host model for pathogenesis. Cellular microbiology. 2005;7:307–314. doi: 10.1111/j.1462-5822.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- Urushihara H, Morio T, Saito T, Kohara Y, Koriki E, Ochiai H, Maeda M, Williams JG, Takeuchi I, Tanaka Y. Analyses of cDNAs from growth and slug stages of Dictyostelium discoideum. Nucl Acids Res. 2004;32:1647–1653. doi: 10.1093/nar/gkh262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Sifri CD, Goumnerov BC, Rahme LG, Ausubel FM, Calderwood SB. Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. Journal of bacteriology. 2002;184:952–961. doi: 10.1128/jb.184.4.952-961.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbrother P, Wagner C, Na J, Tunggal B, Morio T, Urushihara H, Tanaka Y, Schleicher M, Steinert M, Eichinger L. Dictyostelium transcriptional host cell response upon infection with Legionella. Cellular microbiology. 2006;8:438–456. doi: 10.1111/j.1462-5822.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- Sarosh BR, Meijer J. Transcriptional profiling by cDNA-AFLP reveals novel insights during methyl jasmonate, wounding and insect attack in Brassica napus. Plant molecular biology. 2007;64:425–438. doi: 10.1007/s11103-007-9164-9. [DOI] [PubMed] [Google Scholar]
- Morita T, Amagai A, Maeda Y. Translocation of the Dictyostelium TRAP1 homologue to mitochondria induces a novel prestarvation response. J Cell Sci. 2004;117:5759–5770. doi: 10.1242/jcs.01499. [DOI] [PubMed] [Google Scholar]
- Muller I, Subert N, Otto H, Herbst R, Ruhling H, Maniak M, Leippe M. A Dictyostelium mutant with reduced lysozyme levels compensates by increased phagocytic activity. J Biol Chem. 2005;280:10435–10443. doi: 10.1074/jbc.M411445200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gomez-Garcia MR, Shi X, Rao NN, Kornberg A. Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum, with a role in cytokinesis. Proc Natl Acad Sci USA. 2007;104:16486–16491. doi: 10.1073/pnas.0706847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gomez-Garcia MR, Brown MR, Kornberg A. Inorganic polyphosphate in Dictyostelium discoideum: influence on development, sporulation, and predation. Proc Natl Acad Sci USA. 2005;102:2731–2735. doi: 10.1073/pnas.0500023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Choi CH, Lee CH, Jeong SY, Kim JS, Kim BY, Yim HS, Kang SO. Glutathione is required for growth and prespore cell differentiation in Dictyostelium. Dev Biol. 2005;284:387–398. doi: 10.1016/j.ydbio.2005.05.034. [DOI] [PubMed] [Google Scholar]
- Min J, Traynor D, Stegner AL, Zhang L, Hanigan MH, Alexander H, Alexander S. Sphingosine kinase regulates the sensitivity of Dictyostelium discoideum cells to the anticancer drug cisplatin. Eukaryot Cell. 2005;4:178–189. doi: 10.1128/EC.4.1.178-189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa JK, Norris A, Bangera MG, Geiss GK, van 't Wout AB, Bumgarner RE, Lory S. Interaction of pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc Natl Acad Sci USA. 2000;97:9659–9664. doi: 10.1073/pnas.160140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hazlett LD. Analysis of Pseudomonas aeruginosa corneal infection using an oligonucleotide microarray. Invest Ophthalmol Vis Sci. 2003;44:3409–3416. doi: 10.1167/iovs.03-0162. [DOI] [PubMed] [Google Scholar]
- Peracino B, Wagner C, Balest A, Balbo A, Pergolizzi B, Noegel AA, Steinert M, Bozzaro S. Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic. 2006;7:22–38. doi: 10.1111/j.1600-0854.2005.00356.x. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Meth Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Bloomfield G, Tanaka Y, Skelton J, Ivens A, Kay RR. Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum. Genome Biol. 2008;9:R75. doi: 10.1186/gb-2008-9-4-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooperberg C, Fazzio TG, Delrow JJ, Tsukiyama T. Improved background correction for spotted DNA microarrays. J Comput Biol. 2002;9:55–66. doi: 10.1089/10665270252833190. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed TP. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the flase discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- ArrayExpress http://www.ebi.ac.uk/arrayexpress
- Warnes GR. Includes R source code and/or documentation contributed by Ben Bolker and Thomas Lumley. gplots: Various R programming tools for plotting data. R package version 2.3.2. 2007. http://cran.r-project.org/web/packages/gplots/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microsoft excel document containing all the results and genes contained in the microarray.
Microsoft excel document containing the array data for the genes that showed differences in both PAO1 and PA14. The genes were manually annotated and filtered for p < 0.05 and log2 ratio >0.5 or <-0.5. Table 2 was obtained from these data.
Microsoft excel document containing the array data and manual annotation of the genes that showed differential regulation between PAO1 and PA14. The genes were filtered for p < 0.05 and log2 ratio >0.5 or <-0.5. Table 3 was obtained from these data.