Abstract
The ribosome is essential for protein synthesis. The composition and structure of ribosomes from several organisms have been determined, and it is well documented that ribosomal RNAs (rRNAs) and ribosomal proteins (RPs) constitute this important organelle. Many RPs also fill various roles that are independent of protein biosynthesis, called extraribosomal functions. These functions include DNA replication, transcription and repair, RNA splicing and modification, cell growth and proliferation, regulation of apoptosis and development, and cellular transformation. Previous investigations have revealed that RP regulation in colorectal carcinomas (CRC) differs from that found in colorectal adenoma or normal mucosa, with some RPs being up-regulated while others are down-regulated. The expression patterns of RPs are associated with the differentiation, progression or metastasis of CRC. Additionally, the recent literature has shown that the perturbation of specific RPs may promote certain genetic diseases and tumorigenesis. Because of the implications of RPs in disease, especially malignancy, our review sought to address several questions. Why do expression levels or categories of RPs differ in different diseases, most notably in CRC? Is this a cause or consequence of the diseases? What are their possible roles in the diseases? We review the known extraribosomal functions of RPs and associated changes in colorectal cancer and attempt to clarify the possible roles of RPs in colonic malignancy.
Key Words: Ribosomal proteins, extraribosomal functions, colorectal cancer
INTRODUCTION
Ribosome,the essential cellular organelle for protein synthesis in all cells, consists of ribosomal RNAs (rRNAs) and ribosomal proteins (RPs). Human ribosomes are made up of four rRNAs species and about 80 different RPs. The integrated ribosome has two subunits: large (L,60S) and small (S,40S) subunits accordingly that composed of different rRNAs species and RPs. RPs combine with rRNAs, the core bone structures of ribosome directly or bind to each other. Also, the accessory factors called proteins associated with ribosome (PAR) combine with two ribosomal subunits to regulate its synthesis and recycling, such as initiation facters (IFs), elongation facters (EFs). The catalytic activation of decoding mRNAs to proteins depends on the integrated compound of rRNAs, RPs and PARs.
After transcription and modification in nucleolus, the rRNAs combine with different RPs to form two subunits (60S,40S) which are transported to cytoplasm where they assemble to an integrated ribosome to decode mRNAs into proteins and detach after protein synthesis. The whole assembly and detachment process of two subunits are called “Ribosome Recycling” (Fig. 1).
Fig. (1).
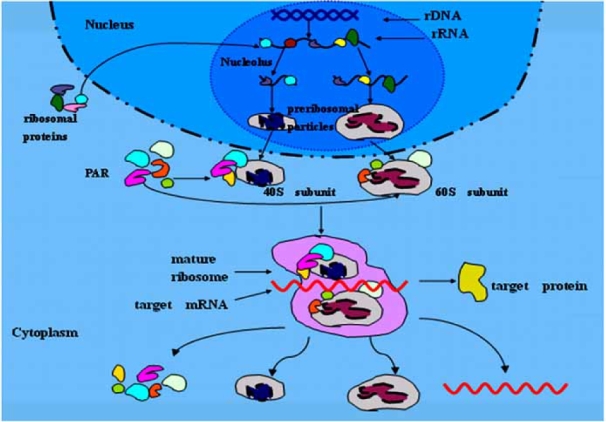
Diagram of the human ribosome synthesis and recycling. rRNAs are transcribed in nucleolus, then combine with different RPs which are synthesized in cytoplasm to form two subunits (40S,60S) called preribosomal particles after modification in the nucleolus. In addition to combination with some PARs, the two subunits are assembled to be a mature ribosome to decode target mRNAs into proteins. After protein synthesis, two subunits detach and PARs, target mRNA are released.
All 80 different RP genes were placed on a cytogenetic map of the human genome in 2001 successfully. The DNA sequences, chromosomal locations of all 4 RNAs and 80 proteins genes are well documented and most of their locations and functions in the integrated ribosome are also clarified. Most of the RPs are significantly conserved during evolution, their amino acid sequences are nearly identical among mammals, which suggests their important roles in organisms although they differ in their specific functions in the mature ribosome. Most studies on RPs focused on their functions in protein synthesis in the past. The accepted conclusion is: RPs stabilize specific rRNA structures in mature ribosomal subunits and promote correct folding of rRNAs during ribosomal assembly. The recent solution of the ribo-some structure by X-ray crystallography and cryo-electron microscopy has improved our understanding of RPs functions during the steps that lead to protein synthesis. Many RPs might function as RNA chaperones not only in the ribo-somal assembly process but also in the stabilization of important domains of the rRNA, such as the peptidyltransferase center of the large subunit. In addition, RPs could coordinate the interaction between the ribosome and mRNA, as well as the initiation and elongation factor [1]. One might predict that genetic defects in any of ribosomal components such as rRNAs or RP genes would cause serious problems with the translational apparatus, hence result in early embryonic death as most researchers assumed that protein synthesis is RP’s only function [2]. However, RP genes were always identified in screening the differentiated expressed genes of human diseases especially in genetic diseases and cancers. The disturbances of their structures or expression levels were associated with various diseases. Many RPs also fill various roles that are independent of protein biosynthesis, called extrari-bosomal functions. Some researches revealed that most of RPs just enhanced but were not prerequisite for the protein biosynthesis [3]. The Minute drosophila with RP genes mutation of RP49,RPL9 and RPS13, demonstrated strongly that a quantitative deficiency of RPs could yield abnormal but viable phenotypes [4–6]. In 1996, Wool defined extraribosomal functions for 29 RPs of human, E. coli and other species, including DNA replication, transcription, DNA repair, RNA splicing and modification, cell growth, proliferation, apopto-sis and development regulation, cellular transformation and others [7]. All the known extraribosomal functions of RPs are summarized in Table 1.
Table 1.
Extraribosomal Functions of Ribosomal Proteins. All the RPs from the Studies are Classified into Different Categories According to their Specific Extraribosomal Functions. RPs in Bold Letters are from the Studies in Colorectal Cancers
| Extraribosomal functions | Specific ribosomal protein (species) |
|---|---|
| DNA replication | RPS1, RPL14 (E. coli) |
| transcription regulation | RPS10 (E. coli) |
| RPS20 (Yeast) | |
| self-translation regulation | RPS4, RPS7, RPS8, RPL4, RPL1, RPL10 (E. coli) |
| RPS30 (yeast) | |
| RPS14, RPS26, RPL7, RPL13A, RPL26 (Human) | |
| RNA splicing and modification | RPS12 (E. coli) |
| RPS12, RPL26 (Human) | |
| DNA topoisomerase activation | RPL41 (Human) |
| DNA repair | RPS9 (E. coli) |
| RPS3 (Drosophila melanogaster) | |
| RPS3, RPLP0 (Human) | |
| developement regulation | RPS2, RPS6, RPL19 (Drosophila melanogaster) |
| RPS15A (Strongylocentrotus purpuratus) | |
| RPS18 (Arabidopsis thaliana) | |
| RPS19 (Ascaris lumbricoides) | |
| RPS4X, RPS4Y1 (Human) | |
| cell growth or proliferation regulation | RPS2, RPS6, RPS13, RPL7, RPL18, RPS27A, RPL31 (Human) |
| cell apoptosis regulation | RPS3, RPS3A, RPS20, RPL35A, RPL7, RPL7A, RPL13A (Human) |
| tumor suppressor gene regulation | RPS29, RPL5, RPL11, RPL23 (Human) |
| proto-oncogene regulation | RPL7A (Human) |
| cell malignant transformation | RPL5, RPS27A, RPL31 (Human) |
| tumor progression, invasion,metastasis, differentiation | RPS19, RPLP0, RPL31 (Human) |
| miscellaneous | RPS15A (X. laevis) |
| RPL32 (Mouse) | |
| RPLP2, RPL5 (Rat) | |
| RPL22 (Human) |
RIBOSOMAL PROTEINS AND COLORECTAL CARCINOMA
RP gene mutations or disturbance in their expression levels were found in many inherited genetic diseases such as Diamond-Blackfan anaemia syndrome, Tuner syndrome, Noonan syndrome, Camurati-Engelmann disease, Bardet-Biedl syndrome 4 [8]. The similar results appeared in carcinoma of breast [9], prostate [3], uterine cervix [10], esophagus [11], liver [12] and also in the glioblastoma and multiforme brain tumors [13].
From year 1988, RPs began to be screened out from cDNA libraries of CRC. The first one was isolated representing mRNA associated with tumor progression and metastasis in 1988 and was identified as RPP2 in 1990 [14,15]. RPL31 cDNA was selected from a normal colon cDNA library on the basis of overexpression in familial adenomatous poly-posis (FAP). The mRNA was found overexpressed in all 23 CRC by slot-blot hybridization. In situ hybridization results illustrated that RPL31 was more abundant in CRC than normal colonic epithelium or stromal tissues suggesting its role in proliferation and neoplasia [16]. In 1991, Northern blot analysis showed RPS3, S6, S8, S12, L5, and P0 increased in CRC and adenomatous polyps except RPL26 and RPL35. Increased mRNA levels of these RPs existed in one tumor sample, indicating those RPs were coordinately controlled. These phenomena cannot be simply due to the presence of a higher percentage of dividing cells in tumor since the proliferative indices of CRC was not significantly different from that of normal colonic mucosa in every case, however, these results suggested that increased synthesis of ribosomes was an early event in colon neoplasia [17]. RPS19 cDNA was isolated from a colon tumor-enriched subtraction library in 1992. It was highly expressed in primary CRC tissue than the paired normal colon tissue in 5 of 6 cases by Northern blotting. It increased concomitantly with tumor progression in two pairs of cell lines derived from the same patients (SW480 and SW620; COLO201 and COLO205) and decreased after butyrate-treatment in HT29 cell line. Therefore, high expression of RPS19 was correlated with higher malignant potential of colon carcinoma cells because there was no association between the high levels and the cell growth states [18]. Ubiquitin PRS27a was much more in tumor tissues than in adjacent normal mucosa especially in advanced CRC by Northern blot. It was expressed in early stage of cell growth without effecting other protein synthesis, which was almost identical to that of proto-oncogene c-jun or c-fos, the known early growth response genes.Thus, overexpression of ubiquitin PRS27a was correlated with CRC and was an early event of carcinogenesis [19]. The study from Graham et al. showed RPP0 mRNA level were higher in primary CRC than in paired adjacent normal colonic epithelium in 36 of 38 cases, increasing with the Dukes’ stage. Its level was also higher in metastatic CRC of liver than primary hepatocellu-lar carcinoma. However, this change can’t be confirmed in gastric cancers [20]. Barnard group also found RPS6, L18, LP0, L37 overexpressed in CRC cases but their expression levels had no correlation with the Dukes’ stage. To their surprise, only RPLP0 was significantly overexpressed in hepa-tocellular carcinoma and none was found in gastric cancers. These findings support the notion that RPs were selectively expressed in different tissues and tumors, even though in the tumors of endodermal origin [21]. Kumar group discoveried that RPL18 was overexpressed in CRC tissue by interaction with double-stranded RNA (dsRNA)-activated protein kinase (PKR) and inhibited dsRNA binding to PKR competitively. Thus, overexpression of RPL18 may promote protein synthesis and cell growth in certain cancerous tissue especially in CRC through inhibition of PKR activity [22]. Frigerio’s group cloned 8 RPs that strongly expressed in one CRC case (L5, L21, L27a, L28,S5,S9, S10, S29). The results was confirmed in 6 CRC cases by Northern blot and RPL5 still showed variant splicing in the tumor. However, no association was observed between the expression levels and the severity of the disease [23]. In 1996,we cloned several differentially expressed RPs in three cDNA libraries by suppression subtractive hybridization (SSH) including RPS2, S12, S27a, L5, L7a, L10a in colorectal adenoma to normal mocusa library; RPS12 in colorectal adenocarcinoma to normal mucose library and RPS15, S25, S11, L18a in colo-rectal adenocarcinoma to colorectal adenoma library [24].
By the high throughput methods, more differentially expressed RPs were screen out easily. Zhang et al. identified 48 RPs highly expressed in CRC by serial analysis of gene expression (SAGE) and ascribed the result to uncontrolled growth of tumor cells [25]. Kitahara et al. found 9 RPs (RPL8,L18,L18a,L29,L6,L3,S19,L7,S5) upregulated in 8 CRC compared to normal epithelia by cDNA microarrays after Laser-capture Microdissection [26]. Notterman et al. found mRNA level of RPS29 was highest in adenomas and RPL3 was highest in CRC by clustering analysis detecting18 colon adenocarcinomas, 4 adenomas and paired normal tissue using oligonucleotide arrays. They assumed the overex-pression of RPs was the result of the high growth speed of tumor cells but not the cause of tumorigenesis [27]. Some similar results were achieved in a study by detecting 11 colon adenocarcinomas, 9 adenomas and their paired normal tissue using oligonucleotide arrays. Ten RP genes (RPS3, S4X, S27a, S3, L6, L9, S3A, S2, L3) were commonly over-expressed in adenoma and adenocarcinomas. They also ascribe it to the high proliferative speed of tumor cells [28].
However, some reverse results appeared in Bertucci’s study. They found several Rps (RPL5,L6,L15,L29,L31,L39) were down-regulated in metastatic CRC by oligonucleotide arrays detecting 50 colon adenocarcinomas and their paired normal mucosa [29]. The similar result appeared firstly in Cao’s study in 1997 [30]. In their study, RPLP2 was down-regulated in CRC.
In 2003, Kasai et al. demonstrated the different protein expressed profiles of 12 RPs (RPSa,S8,S11,S12,S18,S24,L7, L13a,L18,L28,L32,L35a) in normal colorectal mucosa and CRC for the first time. The immunohistochemistry (IHC) results of 18 CRC and paired normal mucosa showed RPS11 and RPL7 were highly expressed in CRC (especially in immature mucosal cells located in the crypt base) but can be detected hardly in the normal mucosa. RPL7 may fill a role in neuroendocrine function of the enterochromaffin cells since its protein was frequently found in enterochromaffin cells expressing chromogranin A. According to their own results and the pervious results which indicated RPS11 was down-regulated in apoptotic ovarian carcinoma cells, they assumed it may inhibit cell apoptosis. The other 10 RPs exhibited a similar staining pattern that were more abundant in normal mucosa than CRC tissue, especially in columnar and goblet cells. Interestingly, these RPs were expressed more abundantly in mature epithelial cells of the upper crypts rather than the immature epithelial cells of the lower crypts, suggesting that the biosynthesis of these ribosomal proteins is significantly enhanced in association with maturation of the mucosal epithelia [31]. We constructed digital gene expression profiles for normal mucosa, colorectal adenoma and CRC by a bioinformatics method based on expressed sequence tag (EST) database (dbEST) of National Center for Biotechnology Information (NCBI) in 2005 [32]. The profiles contain 43 RPs for normal mucosa, 29 for adenoma and 77 for carcinoma. The primary analysis showed that RPs are the highly expressed genes in each profile especially in adenoma, although the expression levels or categories of RPs differed in the three profiles (unpublished data). Table 2 shows all the RPs genes screened out from CRC.
Table 2.
All the RPs from Studies are Classified into Two Groups According to their Extraribosomal Functions. Some have the Known Extraribosomal Functions and the others don’t. RPs in Bold Letters are form the Research Data of CRC
| Classification | RPs |
|---|---|
| extraribosomal functions known | RPS2, RPS3, RPS3A, RPS4X, RPS6, RPS8, RPS9, RPS10, RPS11, RPS12, RPS13, RPS18, RPS19, RPS27A, RPS29, RPSA, RPL3, RPL4, RPL5, RPL7, RPL11, RPL13A, RPL18, RPL23, RPL26, RPL31, RPL32, RPL35A, RPLP0, RPLP2 |
| extraribosomal functions unknown | RPS5, RPS15, RPS23, RPS24, RPS25, RPS28, RPL6, RPL8, RPL9, RPL10A, RPL15, RPL18A, RPL21, RPL23A, RPL27A, RPL28, RPL29, RPL35, RPL37, RPL39, RPL7A |
Generally, the results abovementioned support the notion that different expression patterns of RPs existed in CRC. Some RPs were more abundant in CRC at their mRNA or protein level, but some were on the contrary and even more, some of them were remarkably more abundant in adenoma than in CRC tissue and normal mucosa. The mRNA levels for some specific RPs were associated with the Dukes’ stage, the invasion or metastasis state of the tumor. Most researchers ascribed the overexpression to the hyperproliferation of tumor cells and the down-regulated expression to cell mature and differentiation. The complex expression pattern of RPs indicated their complex roles in CRC and the complexity of the perturbations associated with tumor formation in colon. It was suggested that a single clinical entity included tumors with different molecular mechanisms in fact. Most of the studies only focused on the different expression profiles of RPs superficially which can hardly illustrate their exact roles in carcinogenesis, however, much left to do more detail in their function to reveal the real mechanism.
POSSIBLE ROLES OF RIBOSOMAL PROTEINS IN COLORECTAL CARCINOGENESIS
1. Different Expression Patterns of RPs
The expression patterns of RPs were tissue and disease specific both in normal and tumor tissues, [23] even though in the tumors of endodermal origin. For example, the specific RP is expressed unbalancedly in gastric cancer, hepa- tocellular carcinoma and colorectal carcinoma [20,21]. Moreover, RPs are also expressed unbalancedly in tumors with identical morphological type, histological type or differentiation state. The greatest confusion is that the expression levels even varied in different stages of the same tumor [23]. All of these indicate the complexity of mechanisms in cancer development and progression as well as the relationship between RPs and CRC. Different RPs may fill different roles in the same phase and the same RP may also play different roles in different phases in tumorigenesis. Generally, we may draw two different conclusions for the possible roles of RPs in tumorigenesis: (1) perturbation of RPs disturbs their functions in protein biosynthesis, which is the consequence or the important process of tumorigenesis; (2) RPs directly participate the tumorigenesis by their extraribosomal functions, which means the perturbative expression of RPs induces the tumorigenesis.
2. Protein Biosynthesis Function of RPs in Cancer
The intact ribosomes are composed of rRNAs, RPs and PAR. Their exact function depends on the precise combination of those three components. Ribosome biogenesis and translation are regulated at multiple levels including transcriptional, translational and post-translational levels and associated with accurate cell growth and proliferation. Some tumor suppressors and oncogenes were found to modulate the RPs biosynthesis or ribosome translation initiation or both directly [1]. MYC, a proto-oncogene product, regulated the mature ribosome biogenesis by modifying the genes of necessary factors in ribosomal assembly. Its overexpression in tumor cells increased the expression and activity of ribo-somal components. Therefore, regulation of protein synthesis could be an important mechanism by which MYC regulates cell growth and initiates tumorigenesis. PTEN, a tumor suppressor, regulated the mature ribosome formation expecially the RPS6 through suppressing S6K activity in the PI3K pathway [35–38]. Therefore, the disturbance of genes upstream results in dysregulation of RP expression quantitatively or/and qualitatively so the ribosome can hardly function exactly or produce normal proteins. The perturbation of RPs may reduce the levels of “survival/protective” factors, leading to cancer development and progression [37]. Tumor suppressors and oncogenes may promote cellular transformation by altering ribosomal components. This means RP perturbation is the consequence or the important process but not the direct cause of tumorigenesis. All these studies suggest that the perturbation of ribosome components or their function in protein synthesis may promote cellular transformation, such as RPS19, which causes Diamond-Blackfan anaemia syndrome as well as a high susceptibility to cancer when it mutated.
3. Extraribosomal Functions of RPs in Cancer
Undoubtedly, the increased overall ribosome biogenesis is a common feature of active proliferation and the proliferation rate of tumor cells is always higher than normal ones. Meanwhile, tumor suppressors and oncogenes can modulate the biosynthesis of RPs directly according to the hypothesis above. However, independent, noncoordinate changes in expression of an individual ribosomal protein gene, or of a subset of ribosomal protein genes, can occur under various cellular conditions, and have no direct association or correlation with proliferative and/or protein synthetic activities per se. Therefore, it will be arbitrary and unwise to take the increased RPs mRNA levels simply as a consequence of the high proliferation rate of tumor cells. The reasons are: Firstly, the proliferation levels of CRC is not always significantly different from that of normal colonic mucosa [17]. Secondly, RPs showed different expression patterns: not the all RPs increased in the same tumor or tissue, and the same RP expressed differentially in different tumors or different stages of diseases. Thirdly, some RPs only increased in senescent cells, [38] some in quiescent cells and some in tumor cells well differentiated [31] but some decreased in metas-tatic CRC [29]. Fourthly, p53 mutation, a common event in CRC, which is considered the key factor in colon carcino-genesis can upregulate only several RPs but not all of them; [39] Fifthly, overexpression of specific RPs did not increase the synthesis of proteins [40]. RPs could still been produced when rRNA synthesis was blocked which means RPs synthesis can not be consistent with rRNA synthesis. Under this condition, RPs seemed to have no necessary relationship with ribosome biosynthesis. The Minute drosophila, the viable but abnormal phenotypes models of RP genes mutation, strongly demonstrated the extraribosomal functions of RP49, RPL9 and RPS13. Another powerful illustration is RPS3 which has endonuclease activity and could be used for DNA repair, [41] and also activate caspase 8 and caspase 3 to induce cell apoptosis. The dual-function was performed by its different domains [42]. RPS6, a regulator of cell growth, [3]was regulated precisely by extracellular signal which was always disordered in tumor [43–45]. RPS6 kinase significantly upregulated in tumors of PTEN mutation, and inhibition of their activity would slowdown the growth of tumors by anti-cancer medicine [46–47]. These studies support an idea: RPs participate the tumorigenesis by their extrariboso-mal functions directly. Firstly, RPs directly regulate the expression of oncogene and tumor suppressors on DNA replication, transcription and translation. The recombinantion of human trk proto-oncogene with RPL7a activated its oncogenic function [48]. RPS29, whose mRNAs was much higher in quiescent cells than that in growth phase of endo-thelial cells, had tumor suppressor activity for ras transformed NIH3T3 cells [49]. Secondly, RPs can combine “survival/protective” factors to cause cancer or suppress car-cinogenesis. For example, the enhanced RPL5, RPL11 and RPL23 proteins may form a complex with MDM2 and consequently inhibited the combination MDM2 with p53, resulting in reduced MDM2-mediated p53 ubiquitination and also induced p53 activity and G1 arrest [50–53]. The overexpres-sion of RPS3a gene caused cell malignant transformation and tumorigenesis of nude mouse and was assumed to promote cell transformation by suppressing cell apoptosis since it induced synthesis of anti-apoptosis proteins [54]. Lastly, RPs may play critical roles in DNA repair and cell apoptosis leading to tumourgenesis, such as RPS3.
Indeed, nobody can divide the dual-function of RPs of protein biosynthesis and extraribosomal fuctions arbitrarily in colon carcinogenesis. The two relative distinct fuctions must co-function in tumourgenesis. Some RPs involved in cell mature and differentiation are important in tumor progressing, invasion, metastasis, but possiblely they effect proteins synthesis that are critical in cell mature and differentiation. Undoubtedly, RPs can participate to this process through their extraribosomal functions, no matter what the mechanisms of pathogenesis are.
CONCLUSION
The extraribosomal functions of RPs highlight the studies in pathogenesis of diseases. It challenges our pervious assumption that protein biosynthesis is the only function of RPs. Actually, no one knows whether RPs were designed specifically for the ribosome or whether they were co-opted from among a set of pre-existent proteins that already had defined functions. The two possibilities are by no means exclusive, nor it is likely that all the proteins are added at one time. Therefore the extraribosomal functions of RPs could be a reasonable explanation for their unbalancedly expression patterns in different diseases or tissues although their exact functions are still unknown definitely [7]. It remains to be determined whether the different expression profiles are a cause or a consequence of tumor formation. Independent of this, the two relative distinct functions of RPs must co-function in the tumorigenesis. Although, RPS3 was identified as an endonuclease participating into DNA repair and may be involved in carcinogenesis, but unluckily no direct evidence supported this assumption. Because of no experiment ascertaining the relation between the overexpres-sion of RPs and high proliferating speed, it will be a great challenge to reveal the real roles of RPs in diseases especially when multiple RPs were abnormal in one disease [54]. Moreover, the tissue and disease-specific expression patterns of RPs also prevent us to answer the question effortlessly. Although lots have done revealing the different expression profiles of RPs, much left to do more detail to illustrate their exact roles in carcinogenesis.
Recently, the new term “Ribosomics” emerged which refers to the analysis of ribosomal components at the ge-nomic/proteomic level, for example using cryo-electron microscopy and X-ray crystallography [55]. The genes and proteins sequences of rRNA, RPs as well as the interactions of them have provided abundant data for us to reveal the exact roles of RPs [56]. Taking all the studies in CRC together, we can see that single or minority RPs will be not enough for explaining their functions in the cancers. Therefore, “Ribosomics” may be a powerful tool for research on CRC mechanisms and, for us, to best understand CRC pathogenesis.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant: 30371605). We thank Dr. Cheng Liang from Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis 46202, USA for his expert comments on the manuscript.
ABBREVIATIONS
- rRNA
Ribosomal RNA
- RP
Ribosomal protein
- CRC
Colorectal carcinoma
- IF
Initiation factors
- EF
Elongation factors
- PAR
Proteins associated with ribosome
- L,60S
Large subunits
- S,40S
Small subunits
- FAP
Familial adenomatous polyposis
- RPL
Ribosomal protein large subunit
- RPS
Ribosomal protein small subunit
- dsRNA
Double-stranded RNA
- PKR
dsRNA-activated protein kinase
- SSH
Suppression subtractive hybridization
- IHC
Immunohistochemistry
- SAGE
Serial analysis of gene expression
- EST
Expressed sequence tag
- dbEST
Expressed sequence tag database
- NCBI
National Center for Biotechnology Information
REFERENCES
- 1.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 2.Uechi T, Tanaka T, Kenmochi N. A complete map of the human ribosomal protein genes: assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics. 2001;72:223–230. doi: 10.1006/geno.2000.6470. [DOI] [PubMed] [Google Scholar]
- 3.Vaarala MH, Porvari KS, Kyllonen AP, Mustonen MV, Lukkarinen O, Vihko PT. Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int J Cancer. 1998;78:27–32. doi: 10.1002/(sici)1097-0215(19980925)78:1<27::aid-ijc6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 4.Kongsuwan K, Yu Q, Vincent A, Frisardi MC, Rosbash M, Lengyel JA, Merriam J. A Drosophila Minute gene encodes a ribosomal protein. Nature. 1985;317:555–558. doi: 10.1038/317555a0. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt A, Hollmann M, Schafer U. A newly identified Minute locus, M(2)32D, encodes the ribosomal protein L9 in Drosophila melanogaster. Mol Gen Genet. 1996;251(3):381–387. doi: 10.1007/BF02172530. [DOI] [PubMed] [Google Scholar]
- 6.Saeboe-Larssen S, Lambertsson A. A novel Drosophila Minute locus encodes ribosomal protein S13. Genetics. 1996;143(2):877–885. doi: 10.1093/genetics/143.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 8.Yang F, Liu WP. The progress of ribosomal protein genes and human diseases. J Clin Exp Pathol. 2005;20:354–356. [Google Scholar]
- 9.Henry JL, Coggin DL, King CR. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 1993;15:1403–1408. [PubMed] [Google Scholar]
- 10.Cheng Q, Lau WM, Chew SH, Ho TH, Tay SK, Hui KM. Identification of molecular markers for the early detection of human squamous cell carcinoma of the uterine cervix. Br J Cancer. 2002;86:274–281. doi: 10.1038/sj.bjc.6600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Yang C, Zhou J, Wang X, Wu M, Liu Z. Cloning and characterization of full-length human ribosomal protein L15 cDNA which was overexpressed in esophageal cancer. Gene. 2001;263:205–209. doi: 10.1016/s0378-1119(00)00570-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39:129–138. doi: 10.1002/hep.20017. [DOI] [PubMed] [Google Scholar]
- 13.Lopez CD, Martinovsky G, Naumovski L. Inhibition of cell death by ribosomal protein L35a. Cancer Lett. 2002;180:195–202. doi: 10.1016/s0304-3835(02)00024-1. [DOI] [PubMed] [Google Scholar]
- 14.Elvin P, Kerr IB, McArdle CS, Birnie GD. Isolation and preliminary characterisation of cDNA clones representing mRNAs associated with tumour progression and metastasis in colorectal cancer. Br J Cancer. 1988;57:36–42. doi: 10.1038/bjc.1988.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharp MG, Adams SM, Elvin P, Walker RA, Brammar WJ, Varley JM. A sequence previously identified as metastasis-related encodes an acidic ribosomal phosphoprotein, P2. Br J Cancer. 1990;61:83–88. doi: 10.1038/bjc.1990.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chester KA, Robson L, Begent RH, Talbot IC, Pringle JH, Primrose L, Macpherson AJ, Boxer G, Southall P, Malcolm AD. Identification of a human ribosomal protein mRNA with increased expression in colorectal tumours. Biochim Biophys Acta. 1989;1009:297–300. doi: 10.1016/0167-4781(89)90119-x. [DOI] [PubMed] [Google Scholar]
- 17.Pogue-Geile K, Geiser JR, Shu M, Miller C, Wool IG, Meisler AI, Pipas JM. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11:3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondoh N, Schweinfest CW, Henderson KW, Papas TS. Differential expression of S19 ribosomal protein, laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res. 1992;52:791–796. [PubMed] [Google Scholar]
- 19.Wong JM, Mafune K, Yow H, Rivers EN, Ravikumar TS, Steele GD, Jr, Chen LB. Ubiquitin-ribosomal protein S27a gene overexpressed in human colorectal carcinoma is an early growth response gene. Cancer Res. 1993;53:1916–1920. [PubMed] [Google Scholar]
- 20.Barnard GF, Staniunas RJ, Bao S, Mafune K, Steele GD, Jr, Gollan JL, Chen LB. Increased expression of human ribosomal phosphoprotein P0 messenger RNA in hepatocellular carcinoma and colon carcinoma. Cancer Res. 1992;52:3067–3072. [PubMed] [Google Scholar]
- 21.Barnard GF, Staniunas RJ, Mori M, Puder M, Jessup MJ, Steele GD, Jr, Chen LB. Gastric and hepatocellular carcinomas do not overexpress the same ribosomal protein messenger RNAs as colonic carcinoma. Cancer Res. 1993;53:4048–4052. [PubMed] [Google Scholar]
- 22.Kumar KU, Srivastava SP, Kaufman RJ. Double-stranded RNA-activated protein kinase (PKR) is negatively regulated by 60S ribosomal subunit protein L18. Mol Cell Biol. 1999;19:1116–1125. doi: 10.1128/mcb.19.2.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frigerio JM, Dagorn JC, Iovanna JL. Cloning, sequencing and expression of the L5, L21, L27a, L28, S5, S9, S10 and S29 human ribosomal protein mRNAs. Biochim Biophys Acta. 1995;1262:64–68. doi: 10.1016/0167-4781(95)00045-i. [DOI] [PubMed] [Google Scholar]
- 24.Luo MJ, Lai MD. Identification of differentially expressed genes in normal mucosa, adenoma and adenocarcinoma of colon by SSH. (2001) World J Gastroenterol. 7:726–731. doi: 10.3748/wjg.v7.i5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 26.Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544–3549. [PubMed] [Google Scholar]
- 27.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 28.Lin YM, Furukawa Y, Tsunoda T, Yue CT, Yang KC, Nakamura Y. Molecular diagnosis of colorectal tumors by expression profiles of 50 genes expressed differentially in adenomas and carcinomas. Oncogene. 2002;21:4120–4128. doi: 10.1038/sj.onc.1205518. [DOI] [PubMed] [Google Scholar]
- 29.Bertucci F, Salas S, Eysteries S, Nasser V, Finetti P, Ginestier C, Charafe-Jauffret E, Loriod B, Bachelart L, Montfort J, Victorero G, Viret F, Ollendorff V, Fert V, Giovaninni M, Delpero JR, Nguyen C, Viens P, Monges G, Birnbaum D, Houlgatte R. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- 30.Cao J, Cai X, Zheng L, Geng L, Shi Z, Pao CC, Zheng S. Characterization of colorectal-cancer-related cDNA clones obtained by subtractive hybridization screening. J Cancer Res Clin Oncol. 1997;123:447–451. doi: 10.1007/BF01372549. [DOI] [PubMed] [Google Scholar]
- 31.Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA, Nakayama J. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem. 2003;51:567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Lv BJ, Zhang H, Chen J, Zhu YM, Gu XM, Lai MD. Construction and utilization of analysis platform for digital expression profile. J Zhejiang University (Eng Sci) 2006;40:186–191. [Google Scholar]
- 33.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 34.Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boon K, Caron HN, van, Asperen R, Valentijn L, Hermus MC, vanSluis P, Roobeek I, Weis I, Voute PA, Schwab M, Versteeg R. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steller H, Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 38.Seshadri T, Uzman JA, Oshima J, Campisi J. Identification of a transcript that is down -regulated in senescent human fibroblasts. Cloning, sequence analysis, and regulation of the human L7 ribo-somal protein gene. J Biol Chem. 1993;268:18474–18480. [PubMed] [Google Scholar]
- 39.Loging WT, Reisman D. Elevated expression of ribosomal protein genes L37, RPP-1, and S2 in the presence of mutant p53. Cancer Epidemiol Biomarkers Prev. 1999;8:1011–1016. [PubMed] [Google Scholar]
- 40.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Chubatsu LS, Admon A, Stahl J, Fellous R, Linn S. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J Biol Chem. 1995;270:13620–13629. doi: 10.1074/jbc.270.23.13620. [DOI] [PubMed] [Google Scholar]
- 42.Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560:81–85. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 43.Terada N, Patel HR, Takase K, Kohno K, Nairn AC, Gelfand EW. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the "polypyrimidine tract" mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5’TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kawasome H, Papst P, Webb S, Keller GM, Johnson GL, Gelfand EW, Terada N. Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity. Proc Natl Acad Sci USA. 1998;95:5033–5038. doi: 10.1073/pnas.95.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziemiecki A, Muller RG, Fu XC, Hynes NE, Kozma S. Oncogenic activation of the human trk proto-oncogene by recombination with the ribosomal large subunit protein L7a. EMBO J. 1990;9:191–6. doi: 10.1002/j.1460-2075.1990.tb08095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coppock D, Kopman C, Gudas J, Cina-Poppe DA. Regulation of the quiescence-induced genes: quiescin Q6, decorin, and ribosomal protein S29. Biochem Biophys Res Commun. 2000;269:604–610. doi: 10.1006/bbrc.2000.2324. [DOI] [PubMed] [Google Scholar]
- 50.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 51.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cel Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frank J. Cryo-electron microscopy as an investigative tool: the ribosome as an example. Bioessays. 2001;23:725–732. doi: 10.1002/bies.1102. [DOI] [PubMed] [Google Scholar]
- 56.Nadano D, Ishihara G, Aoki C, Yoshinaka T, Irie S, Sato TA. Preparation and characterization of antibodies against human ribosomal proteins: heterogeneous expression of S11 and S30 in a panel of human cancer cell lines. Jpn J Cancer Res. 2000;91:802–810. doi: 10.1111/j.1349-7006.2000.tb01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


