Abstract
The characterization of the involvement of different histone post-translational modifications (PTMs) and histone variants in chromatin structure has represented one of the most recurrent topics in molecular biology during the last decade (since 1996). The interest in this topic underscores the critical roles played by chromatin in such important processes as DNA packaging, DNA repair and recombination, and regulation of gene expression. The genomic information currently available has pushed the boundaries of this research a step further, from the study of local domains to the genome-wide characterization of the mechanisms governing chromatin dynamics. How the heterchromatin and euchromatin compartmentalization is established has been the subject of recent extensive research. Many PTMs, as well as histone variants have been identified to play a role, including the replacement of histone H2A by the histone variant H2A.Z. Several studies have provided support to a role for H2A.Z (known as Htz1 in yeast) in transcriptional regulation, chromosome structure, DNA repair and heterochromatin formation. Although the mechanisms by which H2A.Z defines different structural regions in the chromatin have long remained elusive, various reports published last year have shed new insight into this process. The present mini review focuses its attention on the genome-wide distribution of H2A.Z, with special attention to the mechanisms involved in its distribution and exchange as well as on the role of its N-terminal acetylation.
Key Words: H2A.Z, Acetylation, Genome-Wide Distribution, Promoter Regions, Gene Activity, Heterochromatin
INTRODUCTION
Although eukaryotic chromatin shows a heterogeneous organization across the genome, there are common features shared by all of its domains. These include the presence of nucleosomes consisting of canonical and/or variant histones [1], the occurrence of histone PTMs [2] and the association with remodeling complexes responsible for nucleosome mobilization [3]. Thus, histone variants, histone PTMs and ATP-dependent and ATP-independent remodeling complexes can all contribute to the differences in chromatin conformation. While heterochromatin domains are structurally quite well defined, less is known about the genesis of euchromatin, as well as how it is maintained and transmitted. Different chromatin modifications have been identified in recent years that are responsible for promoting the ‘euchromatic’ state by antagonizing silencing. These include the replacement of histone H2A by H2A.Z [4, 5], global acetylation of histones and acetylation of H4K16 in particular [6], and methylation of H3K4 [7, 8] and H3K79 [9, 10].
The sequence of the histone variant H2A.Z substantially departs from that of canonical H2A and is highly conserved in higher eukaryotes, though Saccharomyces cerevisiae (yeast) and Tetrahymena thermophila (ciliate) sequences are notably different. Previous reports have shown that in yeast, the H2A.Z histone (also referred to as Htz1) is involved in antagonizing the spreading of silent heterochromatin [4]. In addition, H2A.Z is also involved in other critical cellular processes such as gene activation [11∗∗-13], chromosome segregation [14], and progression through the cell cycle [15∗]. A key question for understanding of the functional and structural involvement of H2A.Z in all these roles is whether its location through chromatin is random or not, and if not, how are the specific sites of deposition determined and regulated? The purpose of the present work is to provide a comprehensive review of the state-of-the-art knowledge about the genome-wide pattern of H2A.Z distribution in relation to the potential role of this variant in chromatin stability. Special emphasis is placed on the discussion of several papers published during the last two years addressing the mechanisms of H2A.Z recruitment to chromatin and the critical role played by acetylation in this and other processes.
H2A.Z DISTRIBUTION GENOME-WIDE
The genome-wide distribution of the variant H2A.Z still represents a controversial point in the study of chromatin dynamics. Two recent works published simultaneously in Cell during 2005 [16∗∗, 17∗∗] addressed this issue using yeast as a model system. In one of these works Chromatin ImmunoPrecipitation (ChIP) was used to identify H2A.Z-occupied sites across the genome. The resulting localization profile was not only consistent with a non-random localization pattern of H2A, but significantly coincident with the distribution of Swr1, the catalytic subunit of the SWR1 complex, which is thus required for the insertion of H2A.Z into chromatin [18–20]. Notwithstanding, it was found that it was the Yaf9 component of the SWR1 complex that is likely responsible for H2A.Z targeting into chromatin [17∗∗]. H2A.Z is not usually found at defined chromosome elements. However, certain loci such as those consisting of genes tran∗Address correspondence to this author at the Department of Biochemistry and Microbiology, University of Victoria, Victoria, BC, Canada V8W 3P6; E-mail: jeirin@udc.es scribed by RNA polymerase I and III are deficient in this variant, indicating a clear preference for promoter of genes transcribed by RNA polymerase II [17∗∗], Fig. (1a).
A second work examined the deposition profile of H2A.Z across yeast chromosome III using a strain carrying an allele of H2A.Z with an amino-terminal influenza hemagglutinin epitope tag [16∗∗]. ChIP and quantitative real-time PCR experiments revealed a highly uniform and chromosome-wide distribution of H2A.Z, which was positively correlated with its proximity to the nearest 5’ end of genes. By increasing the resolution of these experiments to single inter-genic regions (IGRs) flanked by open reading frames (ORFs) with different orientations a striking enrichment for H2A.Z at 5’ regions was evident, concomitant with a decline in the coding regions. These data were further corroborated by analyses at a single nucleosome resolution, revealing a striking distribution in which H2A.Z seems to appear in two consecutive nucleosomes marking 5’ regions of most euchro-matic genes (whether at active or inactive loci) Fig. (1a). This is in contrast with genes close to telomeres and those from the HMLα silent cassette which lack any detectable H2A.Z [16∗∗].
A clear negative correlation has been observed between Bdf1 (a member of the SWR1 complex) occupancy and the presence of a consensus TATA-box at promoters, raising the possibility that Bdf1 may help recruit SWR1/H2A.Z only to TATA-less promoters. Furthermore, Bdf1 is also positively correlated with H2A.Z occupancy and this histone variant is (as in the case of Bdf1) negatively correlated with the presence of a TATA-box, suggesting that both H2A.Z and Bdf1 occupy overlapping TATA-less promoters, which preferentially utilize TFIID for initiation [17∗∗]. It seems likely that the SWR1 complex is the primary (if not the sole) factor directing specific localized H2A.Z deposition in yeast. However, it still remains to elucidate how SWR1 is recruited to Pol II promoters and what is the nature of the bias for TATA-less promoters.
Nearly all yeast genes contain an ~ 150 bp nucleosome-free region (NFR) which is centered about ~ 200 bp upstream of the initiation codon, containing the initiation site for transcription of their associated genes [21∗]. These regions were shown to be flanked by two nucleosomes containing H2A.Z in most of yeast genes and thus, not only does H2A.Z mark the 5’ ends of genes, but also indicates that the transcription initiation site is typically flanked by two positioned H2A.Z nucleosomes [16∗∗] Fig. (1b).
Relationship Between H2A.Z Distribution and Transcriptional Activity
The organized manner in which H2A.Z is deposited at promoter regions may suggest a implicit relationship with gene transcription. However, no correlation was observed between H2A.Z enrichment at flanking NFRs and transcriptional activity. This observation has very important implications as it suggests that H2A.Z is a hallmark for the identification of the 5’ ends of genes and its deposition does not depend on ongoing transcription [16∗∗]. In a later study, Millar and colleagues [22∗∗] raised antibodies against H2A.Z acetylated at K14 and against a peptide of the unmodified C-terminal domain in order to compare the distribution of bulk and acetylated forms of this variant on a ge-nome-wide scale in yeast chromosomes. Antibodies were used in ChIP and the purified DNA fragments hybridized against whole-genome microarrays, which were spotted with probes corresponding to RNA polymerase II-transcribed ORFs and IGRs. The results showed that H2A.Z is preferentially bound to genes that are repressed, finding an inverse correlation between H2A.Z presence and transcription [22∗∗]. These results are supported by those previously obtained by Zhang and colleagues [17∗∗], but in this case H2A was only weakly, negatively correlated with transcription levels, raising the possibility that H2A.Z may be lost or ejected from promoters during activation to a greater extent than H2A [17∗∗]. Although this would agree with the notion that H2A.Z-containing nucleosomes are less stable than their H2A counterparts in purified yeast chromatin, this is most likely the result of its own and/or the rest of the core histone complement acetylation, which is often present at the promoter regions of actively transcribing genes Fig. (1b). In this regard it is important to note that the stability of chicken H2A.Z-containing nucleosomes has been shown to be dependent on acetylation [23∗].
The deposition profile pattern of H2A.Z has also been recently reported for a vertebrate organism. In this study, a synthetic 16-residue N-terminal peptide from chicken H2A.Z and also its tri-acetyl (K4, K7, K11) counterpart were used to raise antibodies that were subsequently used in ChIP on native nucleosomes from different chicken cell types. The results, thus, obtained showed a consistent enrichment of acetylated H2A.Z at promoter proximal and 5’ end of active housekeeping genes. In contrast, neither acetylated nor bulk H2A.Z was identified at inactive genes. Acetylated H2A.Z, thus, appears to be a feature of active genes, with a prevalent concentration at the 5’ end, but it is not, for example, always present at genes in an unmodified form for subsequent acetylation on gene activation [11∗∗]. Similar results have been obtained in two human housekeeping genes [24], extending the link between gene activity and the presence of acetylated H2A.Z to other vertebrates.
A higher occupancy of H2A.Z at promoters had been previously described [12, 13]. A similar bias toward promoters has also been observed in the case of H2A.Z-K14Ac. However, in the latter instance the association was not as extensive as in the case of bulk H2A.Z, probably due to the preferential loss of nucleosomes from active genes, where K14 is acetylated. Thus, it was concluded that H2A.Z and H2A.Z-K14Ac are both enriched at promoters, but the acetylated version is enriched at the promoters of active genes [22∗∗] Fig. (1b). In this regard, previous results had suggested that H2A.Z facilitates nucleosome loss during gene activity [17∗∗] and hence one possible role for K14 acetylation could be its involvement in the loss of H2A.Z and possibly entire nucleosomes at promoters of active genes. Nevertheless, mutation of acetylatable lysines at H2A.Z N-terminal tails did not alter kinetics of neither H2A.Z nor the H3 loss during gene activation, indicating that the acetylation of H2A.Z alone is not in itself enough to produce such destabilization [22∗∗].
The question of the dynamic redistribution of H2A.Z in response to massive transcriptional changes was examined by Zhang and colleagues [17∗∗] by altering the transcription of hundreds of genes through heat shock treatment and diauxic shift (induction of anaerobic to aerobic respiration by glucose depletion) in yeast. Changes in gene expression were compared with those on H2A.Z occupancy genome-wide. This approach revealed a loss of H2A.Z in activated single promoters and an acquisition of H2A.Z at repressed single promoters upon change in the growth conditions. Supris-ingly, H2A.Z was not required to establish the repressed/ basal state (at least for the heat shock response) instead, this H2A variant appears to poise the promoter to facilitate activation through ejection or loss during later activation program [17∗∗], probably following acetylation. Although at least in one case (the AGA2 gene from yeast) higher H2A.Z levels were observed in the active gene in comparison to when it was inactive, H2A.Z depletion (involving nucleosome loss) was also evidenced in other genes. These, likely represent the two main mechanisms by which the levels of H2A.Z are correlated with transcriptional activity [16∗∗]. Indeed, nucleosome deficiency is characteristic of yeast active promoters [25, 26] and gene activation has been shown to promote H2A.Z loss [17∗∗], as it had been previously reported [12, 13]. Furthermore, H2A.Z chromatin turnover is of greater magnitude than that of canonical H2A. Its highly dynamic status contributes to the full rapid activation of occupied genes, representing a general activator that is deposited during repression (and whose loss promotes activation) [17∗∗].
H2A.Z ACETYLATION
H2A.Z acetylation is a critical modification that is relevant for its deposition into chromatin. Indeed, acetylation by the nucleosome acetyl-transferase of histone H4 complex (NuA4) has been demonstrated to occur both in vitro and in vivo in yeast Htz1 [27∗∗, 28∗] as well as in Drosophila H2AvD (a Drosophila H2A variant hybrid of H2A.X and H2A.Z). In both instances, NuA4 preferentially acetylates H2A over H4 contained in the same nucleosome [29]. The number of acetyl groups added directly to H2A.Z by NuA4 was identified in vitro using ion-trap mass spectrometry, and it showed the presence of a single peak corresponding to a H2A.Z + 42 Da, the molecular weight of a single acetyl group, suggesting that H2A.Z is monoacetylated by NuA4 [27∗∗]. In contrast with this data, immunoprecipitation analysis of flag-tagged H2A.Z in yeast revealed the presence of at least four modified forms in vivo. Individual K-R mutants directed to the potential targets of acetylation at the N-terminal region (K3, K8, K10 and K14) revealed that each of these sites were acetylated in vivo [27∗∗]. The identity of these N-terminal acetylation sites was further corroborated by electrospray ionization mass spectrometry (ESI-MS) carried out on HPLC purified Htz1 isolated from asynchronous cultures. This analysis further revealed that K14 was the predominantly acetylated form [22∗∗].
Several groups have tested yeast strains carrying inactivating mutations of the catalytic subunits of different HAT complexes. For instance, experiments using a severe temperature-sensitive allele of Esa1, the catalytic subunit of NuA4 that is required for viability in yeast, resulted in the elimination of three of the four modified forms, leaving a single NuA4-independent modification [27∗∗]. The study also revealed an additional contribution of the SAGA HAT adding a single acetyl group to H2A.Z. Interestingly, the epistasis analysis performed by these authors suggested that NuA4 acetylation of H2A.Z was a prerequisite for SAGA acetylation [27∗∗]. The contribution of different HATs to H2A.Z acetylation was further studied in genomic loci bearing high levels of H2A.Z-K14Ac. Comparison of the abundance of bulk H2A.Z and that of its K14 acetylated form in Esa1 and Gcn5 (the catalytic subunits of NuA4 and SAGA, respectively) inactivating mutants showed a strong decrease of the latter. A strain consisting of a double mutant exhibited only low background Htz1 acetylation, attesting to the role of these two HAT complexes in most of, if not all, the acetylation of H2A.Z-K14 residues in vivo [22∗∗].
The absence of histone H2A.Z results in alterations of the proper development of yeast [30] and is essential for viability in higher eukaryotes [5]. Unacetylatable H2A.Z mutants also exhibit a decrease in growth rate [27∗∗] and are lethal in Tetrahymena [30]. Since the absence of the acetylation catalytic subunits of NuA4 and SAGA also results in inviability [27∗∗], a possible interpretation of these observations is that acetylation of either H2A.Z or some other NuA4 target, but not both, may be essential for proper cell survival.
Given the reversibility of the histone acetylation process, a screen of several known histone deacetylases (HDACs) was carried out in order to identify the HDACs involved in the H2A.Z deacetylation process. However, no significant alteration on the levels of H2A.Z-K14Ac could be observed with any of these mutants. In addition, H2A.Z-K14Ac levels remained unchanged throughout the cell cycle and in response to methyl methanosulfonate (MMS)-induced DNA damage [28∗]. Therefore, the HDACs involved in this dynamic turnover of H2A.Z-K14Ac remain to be established.
Acetylation and Histone H2A.Z Chromatin Deposition
Recent reports have addressed the acetylation of the his-tone H2A.Z variant by the NuA4 complex in S. cerevisiae [22∗∗, 27∗∗, 28∗]. This complex represents the only essential HAT in yeast, and shares several subunits with the Swr1-ATPase complex, SWR1, which is responsible for the deposition of H2A.Z into chromatin [18–20]. Both complexes share common regulatory targets [14] but how NuA4 and SWR1 are functionally connected remains elusive.
Although incorporation of H2A.Z into chromatin has been shown to be dependent on SWR1, a very recent work by Keogh and colleagues [28∗] have demonstrated that in S. cerevisiae, histone H2A.Z is acetylated by NuA4 at lysine 14 only after it is assembled into chromatin,(Fig. 1)Fig. (1c). Acetylation at this residue is exclusively dependent on NuA4 and unaffected by the mutation of any other of the three lysines at the N-terminal tail of H2A.Z [28∗]. This conclusion was supported by a different approach, in which the evaluation of the modifications present on both the chromatin-associated and soluble fractions of H2A.Z were analyzed [27∗∗]. In this case, soluble H2A.Z was found to be largely unaceylated while the majority of acetylated forms were present in the chromatin-associated fraction. Furthermore, when SWR1 mutants were used, H2A.Z was observed to remain unmodified in both the soluble and chromatin-associated fractions, suggesting that an efficient acetylation of H2A.Z by NuA4 (and by extension, by SAGA) could not occur in the absence of SWR1 [27∗∗].
Fig. (1).
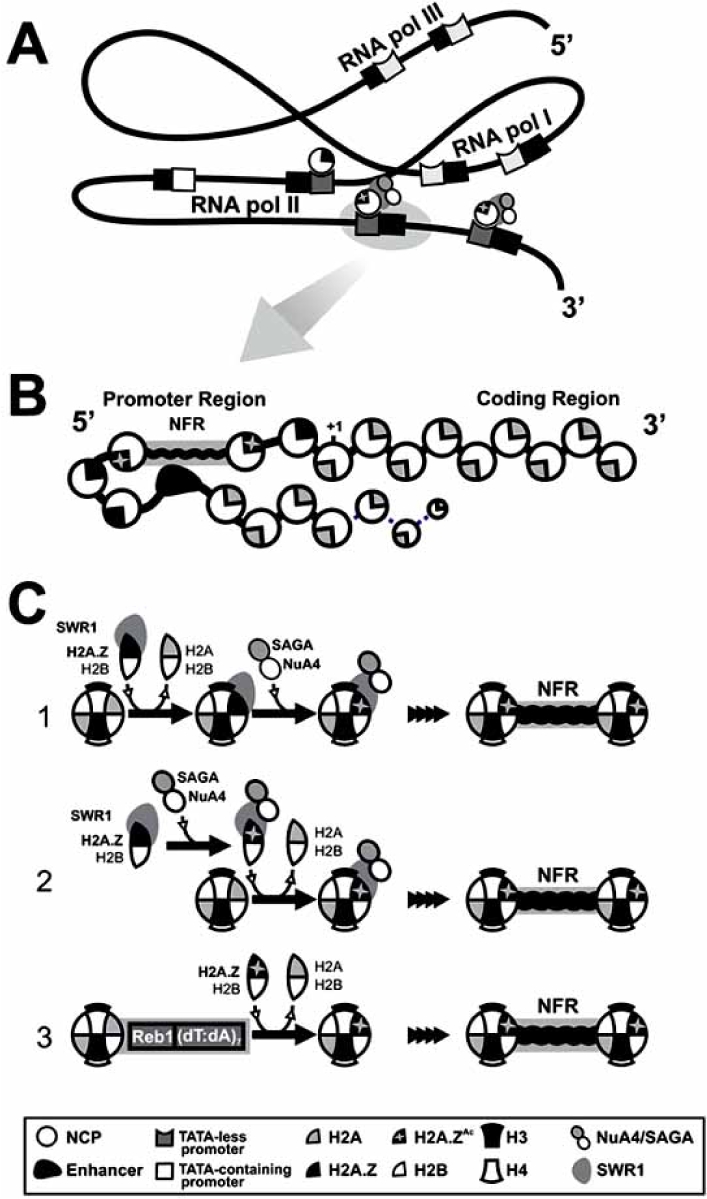
Schematic representation of the H2A.Z genome-wide distribution and the different mechanisms of chromatin recruitment. A) H2A.Z is preferentially enriched at TATA-less promoters of genes transcribed by RNA polymerase II [17∗∗]. Targeting of H2A.Z to these regions is reliant on SWR1, and involves acetylation of N-terminal lysines by NuA4 and SAGA (mostly K14 in the case of yeast [15∗]). B) Nearly all promoters of yeast genes contain an ~ 150 bp nucleosome-free region (NFR) centered about ~ 200 bp upstream of the initiation codon, which contains the transcription initiation site flanked by two H2A.Z-containing nucleosomes. Although H2A.Z occupation at promoters is not correlated with transcriptional activity, H2A.Z-K14Ac is preferentially enriched at promoters of active genes [22∗∗], raising the possibility that acetylated H2A.Z may contribute to nucleosome loss during activation, together with acetylation of other members of the core histone complement. C) H2A.Z is acetylated in vivo by NuA4 and SAGA [22∗∗, 27∗∗-29]. Different studies have reached different conclusions as it pertains to the role of acetylation in H2A.Z deposition in chromatin. One possible mechanism (C.1) involves the acetylation of H2A.Z only after (Legend Fig. 1) contd…. it is recruited into chromatin by SWR1 [28∗]. By contrast, a second model (C.2) has been proposed in which H2A.Z deposition would require its previous acetylation and thus, acetylation itself would regulate H2A.Z recruitment [22∗∗]. In addition, a recent report [16∗∗] has demonstrated that H2A.Z can be recruited to specific chromatin regions regardless of SWR1 and acetylation, through the recognition of specific DNA elements (the Reb1 signal and a dT:dA tract) at promoters of both active and inactive genes (C.3). The recognition of these elements induces the formation of the NFR region flanked by two positioned H2A.Z-containing nucleosomes and it thus, complements the recruitment pathways reliant on acetylation and SWR1 (C.1, C.2).
The relevance of the acetylatable lysines at the N-terminal tail of H2A.Z to its chromatin deposition was demonstrated by analyzing the chromatin distribution in mutants for the four H2A.Z N-terminal lysines (K3R, K8R, K10R, K14R) and a mutant lacking the whole N-terminal. It was shown that the requirement of H2A.Z for growth in different media (i.e.; different carbon sources or in the presence of different genotoxic components) as well as for telomeric silencing is not dependent on the acetylatable lysines [22∗∗]. Comparisons between bulk H2A.Z distribution and the occurrence of these mutants in chromatin revealed that the latter were preferentially absent from repressed genes and from genes undergoing the transition from activation to repression, suggesting that acetylation of H2A.Z regulates its deposition into yeast chromatin [22∗∗]. These finding would support a physical and a functional connection between NuA4 and SWR1, indicating that part of the assembly process of H2A.Z into chromatin would require its acetylation, Fig. (1c). Although these results are in disagreement with those described above [27∗∗], it has been recently shown that H2A.Z can be also assembled into chromatin in a SWR1-independent manner [31] Fig. (1c). This additional pathway for H2A.Z deposition could reconcile these discrepancies.
Acetylation of lysine residues at N-terminal tails of other core histones have also been shown to be relevant for efficient recruitment of H2A.Z into chromatin. For instance, strains bearing deletions in the Eaf1 and Elp3 HATs, which are specific for H3 and H4 acetylation, resulted in reproducible defects in the H2A.Z levels present in chromatin. Conversely, yeast strains carrying lysine to arginine mutations at H3 and H4 N-terminal regions have a similar effect [16∗∗]. Furthermore, a positive correlation between H2A.Z-K14Ac and other acetylation hallmarks of gene activity (i.e.; H3-K14Ac, H3-K18Ac), as well as a negative correlation with modifications anticorrelated with transcription (i.e.; H4-K16Ac) were established [17∗∗, 22∗∗]. Notwithstanding, it is unlikely that a single modification ‘code’ is responsible for directing global H2A.Z, as not all the yeast IGRs occupied by H2A.Z contain all correlated marks. Thus, there is the implicit possibility that different promoter contexts might impose a different reliance on specific modifications for deposition [17∗∗].
The catalytic subunit of SAGA, Gcn5, that (in conjunction with other HATs) acetylates H3K14 and other lysines, and Bdf1 (the SWR1 component responsible for recognizing the acetylation sites) have been shown to be linked to the deposition of yeast H2A.Z at many loci. Hence, transcription factors may operate in conjunction with HATs and Bdf1 to recruit SWR1 to certain active promoters. However, as stated above, the situation may be more complex and possibly other HATs (such as NuA4) may be involved, as H2A.Z deposition also takes place in cells lacking Bdf1 and Gcn5 [17∗∗]. Also, because acetylation of nucleosomes at promoter regions is generally correlated with gene activity, the requirement of histone tail acetylation and the targeting of SWR1 by subunits recognizing such modifications does not account for the H2A.Z deposition that has been observed at inactive genes in yeast euchromatin [16∗∗].
A recent work has shown that deacetylation of H4-K16 is necessary for the association of Bdf1 with chromatin in vivo [32]. Interestingly, hypoacetylation has been observed in the two H2A.Z nucleosomes flanking the NFR [33], which is in perfect agreement with the H2A.Z localization in these nu-cleosomes and supports the role of Bdf1 in promoting H2A.Z deposition.
Alternative Pathways for H2A.Z Recruitment
A hypothesis has been put forward that H2A.Z is deposited in inactive as well as active genes through recognition of specific DNA elements at promoters that determine its deposition. This issue was addressed by systematically mutating a typical promoter containing two positioned H2A.Z nu-cleosomes, corresponding to the yeast SNT1 gene. A drastic reduction in H2A.Z enrichment was observed in mutants of the NFR upstream of the ORF region, and refinement of these mutagenic analyses revealed the presence of two redundant signals required for H2A.Z deposition [16∗∗]. They consist of a 10 bp-long segment that contains a binding site for the regulatory factor Reb1 (RNA polymerase I enhancer binding protein) and an adjacent tract of seven dT:dA base pairs. This binding motif is the single most conserved motif found in yeast promoters, and is even more conserved across species than the TATA-box [34]. The tract of dT:dA base pairs is common in yeast promoters, particularly in NFRs [21∗] and it has been related to nucleosome depletion from promoters [25, 26]. The insertion of a 22 bp segment containing both elements in the middle of the inactive PRM1 gene resulted in a robust H2A.Z enrichment. It additionally caused a de-localization of the nucleosome pattern in the PRM1 gene, and resulted in the formation of an NFR, Fig. (1c). In addition, the insertion of this 22 bp segment induced the appearance of two positioned H2A.Z nucleosomes. Furthermore, acetylation levels at H4 in these nucleosomes did not show differences from those observed in the wild type PRM1 genes, suggesting that this deposition mechanism is independent from (but complementary to) that involving acetylation and Bdf1 [16∗∗].
In humans, an equivalent of the yeast NuA4 complex has been described [35, 36], which contains the expected yeast homologs and also subunits homologous to those in the SWR1 complex from yeast, known to incorporate H2A.Z into nucleosomes [19, 20, 37]. Thus, as suggested by Bruce and colleagues, the larger human complex appears to combine the H4/H2A HAT activities of NuA4 with the remodeling and H2A.Z exchange capabilities of SWR1, allowing H2A.Z to be deposited into nucleosomes in an acetylated form [11∗∗].
Although the H2A.Z deposition problem is still obscure, much less is known about the fate of the H2A.Z nu-cleosomes upon gene activation. A possible scenario is that in which a chromatin remodeling complex upon recognition of a defined histone PTM pattern could actively eject the H2A.Z-containing nucleosomes from the NFR. Indeed, acetylation of histones has been known to weaken the affinity with which H2A.Z binds to chromatin [23∗].
EFFECT OF H2A.Z OCCUPANCY AND ACETYLATION ON HETEROCHROMATIN SPREADING
Yeast strains lacking H2A.Z display the physical spreading of silencing factors (such as the silent information regulator -Sir- proteins) from certain telomeres into telomere-proximal genes [37]. It is interesting to note that cells bearing H4K16 substitutions also display Sir-dependent silencing of telomere-proximal genes [7, 8] even in the presence of H2A.Z at these loci. This would indicate the occurrence of a spread of Sir proteins through H2A.Z-occupied genes in the adjacent telomeric region in absence of H4 acetylation, Fig. (2). All this depicts H2A.Z as one of the many factors contributing to heterochromatin boundary maintenance, a process in which histone H4 acetylation may also play a critical role [17∗∗].
The effect of H2A.Z acetylation itself in heterochromatin spreading has been recently studied in different yeast genes located close to the telomeres and to the silent mating-type (HMR) locus. Unacetylatable H2A.Z mutants (where the four N-terminal lysines are replaced by arginines) were shown to be defective in boundary formation, as reflected by the reduction in the expression levels of genes close to te-lomeric heterochromatin. This repression was entirely dependent on Sir proteins, as a deletion of Sir2 in the H2A.Z-R mutants rescued the wild-type expression levels [27∗∗]. These authors conclusively demonstrated that such a loss in boundary function was due to the loss of H2A.Z acetylation, and not due to mislocalization of H2A.Z, thus identifying the first specific function for H2A.Z acetylation in vivo. Furthermore, cells deficient in NuA4 were more severely defective in boundary function than either H2A.Z-deficient mutants or unacetylatable H2A.Z mutants, underscoring the important contribution of acetylation in this process,(Fig. 2) Fig. (2). In contrast to telomeres, the boundaries blocking the spread of silencing at the HMR locus were unaffected in the unace-tylatable mutants, probably due to the dual or different boundary mechanisms operating at this locus. The chief contribution of H2A.Z acetylation in blocking heterochromatin spreading has also been confirmed in other organisms such as in chicken, where the presence of acetylated H2A.Z at sites flanking the 16 kb heterochromatin domain at the β-globin locus suggests a link to barrier activity 11∗∗].
Fig. (2).
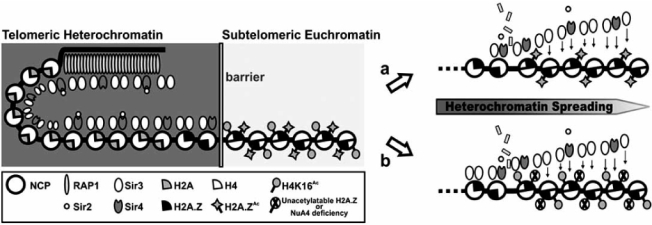
A model for yeast telomeric heterochromatin adapted from [39]. Heterochromatin formation and maintenance involve the RAP-1 containing telosome, different Sir proteins and the interaction of these elements with histone H4. H2A.Z participates in preventing the spread of telomeric heterochromatin to subtelomeric euchromatin regions by creating a barrier effect, and this boundary function is highly dependent on acetylation. For instance, the absence of H4-K16Ac results in the disruption of this barrier even in the presence of acetylated H2A.Z (pathway a [7, 8, 17∗∗]). Furthermore, absence of H2A.Z acetylation or NuA4 deficiency (pathway b) also break this barrier even in the presence of H4-K16Ac [11∗∗, 27∗∗]. Thus, the boundary function played by H2A.Z and H4 histones is mainly regulated by their acetylation rather than by their mislocalization at subtelomeric regions.
A work has recently addressed the issue of the H2A.Z and H2A.Z-K14Ac distribution in relation to the distance from telomeres. Surprisingly, in this case the authors found that while H2A.Z is present at average levels in regions adjacent to subtelomeric heterochromatin, it remains hypoace-tylated for upto 50 kb from the end of chromosomes [22∗∗]. These results are in striking disagreement with that described above, as the absence of H2A.Z acetylation close to telo- meres results in a defective chromatin boundary function [27∗∗].
Therefore, as with other functional and structural data dealing with histone H2A.Z in the past [38], the role of H2A.Z acetylation still remains controversial. Several critical questions remain unsolved, including what is the H2A.Z acetylation partner and what are the HDACs involved in the dynamic deacetylation of H2A.Z? Nevertheless, the very recent papers we have reviewed here, made an important contribution to unraveling the biological significance and the genome-wide distribution of this important histone H2A variant.
ACKNOWLEDGEMENTS
The authors thank Lindsay J. Frehlick for critical reading and discussion of this manuscript. This work was supported by grants from the Canadian Institutes of Health Research (CIHR), grant number MOP-57718 (to J.A.) and by a Postdoctoral Marie Curie International Fellowship within the 6th European Community Framework Programme (to J.M.E.-L).
ABBREVIATIONS
- ChIP
Chromatin Immunoprecipitation
- ESI-MS
Electrospray Ionization Mass Spectrometry
- HATs
Histone Acetyl-Transferases
- HDACs
Histone Deacetylases
- IGRs
Inter-Genic Regions
- MMS
Methyl methanosulfonate
- NCP
Nucleosome core particle
- NFRs
Nucleosome Free Regions
- ORFs
Open Reading Frames
- PTMs
Post-Translational Modifications
- SIR
Silent Information Regulator
REFERENCES
- 1.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Owen-Hughes T. Colworth memorial lecture. Pathways for re-modelling chromatin. Biochem Soc Trans. 2003;31:893–905. doi: 10.1042/bst0310893. [DOI] [PubMed] [Google Scholar]
- 4.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 5.Dryhurst DD, Thambirajah AA, Ausió J. New twists on H2A.Z: a histone variant with a controversial structural and functional past. Biochem Cell Biol. 2004;82:490–497. doi: 10.1139/o04-043. [DOI] [PubMed] [Google Scholar]
- 6.Calestagne-Morelli A, Ausio J. Long-range histone acetylation: biological significance, structural implications, and mechanisms. Biochem Cell Biol. 2006;84:518–527. doi: 10.1139/o06-067. [DOI] [PubMed] [Google Scholar]
- 7.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 8.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine 16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 9.Ng HH, Ciccione DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci USA. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos-Rosa H, Bannister AJ, Dehe PM, Geli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem. 2004;279:47506–47512. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- 11.Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in hyperacetylated form is a feature of active genes in chicken. Nucl Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larochelle M, Gaudreau L. H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 2003;22:4512–4522. doi: 10.1093/emboj/cdg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 14.Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, Emili A, Buratowski S, Hieter P, Greenblatt JF. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc Natl Acad Sci USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhillon N, Oki M, Szyjka SJ, Aparicio OM, Kamakaka RT. H2A.Z functions to regulate progression through the cel cycle. Mol Cell Biol. 2006;26:489–501. doi: 10.1128/MCB.26.2.489-501.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z.marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi1/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, Richards DP, Wu X, Emili A, Hughes TR, Buratowski S, Greenblatt JF. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 20.Mizuguchi G, Shen X, Landry J, Hu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 21.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 22.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thambirajah AA, Dryhurst DD, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z sabilizes chromatin in a way that is dependent on core histone acetylation. J Biol Chem. 2005;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 24.Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J Biol Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 27.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2A.X and H2A.Z. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 30.Jackson JD, Gorovsky MA. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucl Acids Res. 2000;28:3811–3816. doi: 10.1093/nar/28.19.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu WH, Alami S, Luk E, Wu CH, Sen S, Mizuguchi G, Wei D, Wu C. Swc2 is a widely conserved H2AZ-binding module essential for ATP-dependent histone exchange. Nat Struct Mol Biol. 2005;12:1064–1071. doi: 10.1038/nsmb1023. [DOI] [PubMed] [Google Scholar]
- 32.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global hisone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:E328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elemento O, Tavazoie S. Fast and sysematic genome-wide discovery of conserved regulatory elements using a non-aligment based approach. Genome Biol. 2005;6:R18. doi: 10.1186/gb-2005-6-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 36.Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ausió J. Histone variants: the structure behind the function. Brief Funct Genomic Proteomic. 2006;5:228–243. doi: 10.1093/bfgp/ell020. [DOI] [PubMed] [Google Scholar]
- 39.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]


