Abstract
The hepatitis B virus capsid (core antigen) is able to bind to and activate naïve B cells whereby these become efficient primary antigen-presenting cells for the priming of T cells. We have investigated this interaction by using cryo-electron microscopy, three-dimensional image reconstruction, and molecular modeling to visualize capsids decorated with Fab fragments of a receptor immunoglobulin, and surface plasmon resonance to measure the binding affinity. By both criteria, the mode of binding differs from those of the six monoclonal anti-core antigen antibodies that have been previously characterized. The Fab interacts with two sites, ~ 30 Å apart. One interaction is canonical, whereby the CDR loops engage the tip of one of the 25 Å-long spikes that protrude from the capsid surface. The second interaction is non-canonical; in it, the Fab framework contacts the tip of an adjacent spike. The binding affinity of this Fab for capsids, KD ~ 4 × 10−7 M, is relatively low for an antibody-antigen interaction, but is ~150-fold lower still (~ 2.5 × 10−5 M) for unassembled capsid protein dimers. The latter observation indicates that both of the observed interactions are required to achieve stable binding of capsids by this receptor immunoglobulin. Considerations of conserved sequence motifs in other such molecules suggest that other naïve B cells may interact with HBV capsids in much the same way.
Keywords: virus-host interaction, antigen-antibody interaction, cryo-electron microscopy, B cell receptor, HBV
INTRODUCTION
Despite the availability of an effective prophylactic vaccine and several antiviral therapies, hepatitis B virus (HBV) remains a serious public health concern, particularly in Asia. Of the world’s 400 million chronic carriers one million die annually 1; 2. The nucleocapsid of HBV is exceedingly immunogenic 3. Its building-blocks are dimers of a 183-residue alpha-helical protein, with the two subunits paired through the formation of a four-helix bundle. Capsids are dimorphic and are comprised of either 90 or 120 dimers arranged such that the four-helix bundles project from the surface as 25 Å-long spikes 4. Together these two capsid forms are known as core antigen (HBcAg). The principal antigenic determinant of HBcAg, the so-called “immunodominant loop” (residues 78–83), is located at the apices of the capsid spikes 5; 6. The capsid has been found to bind to the receptor membrane immunoglobulins (mIg) of a high proportion of naïve murine B cells allowing them to present HBcAg to T cells approximately 105-fold more effectively than either macrophages or dendritic cells 7. It has been shown that HBcAg can also bind and activate human B cells in vivo 8. This binding has been proposed to be due to epitopes arrayed on the capsid surface, to be responsible for the capsid’s exceedingly high immunogenicity in mice, and to have a role in human infections 7. Binding involves a short sequence (EDPA) located at the tips of the capsid spikes 9 and a conserved linear motif, either I/LSCKASGYI/SFTS/G or ISCRASQVSTSS, present in the framework region 1 (FR1) complementarity-determining region 1 (CDR-1) junction of the membrane immunoglobin VH and VL domains, respectively10. However, the molecular basis for the interaction of HBcAg with receptor immunoglobin is unknown.
The binding of high-affinity antibodies to viral shells has been visualized by cryo-electron microscopy coupled with image reconstruction and molecular modeling 4; 11. By docking generic monoclonal antibody fragments (Fab), taken from the Protein Data Bank, into electron density maps of Fab-decorated HBcAg particles 12 it has been possible to simulate antibody binding to within a precision of < 2 Å in each dimension 13. The resulting quasi-atomic models have permitted the identification of the residues in six epitopes on HBcAg 4; 14.
To address the question of how HBcAg binds to B cells, capsids decorated with Fabs derived from a monoclonal antibody corresponding to the receptor immunoglobin of a naïve B cell responsive to HBcAg 10 were analyzed by cryo-electron microscopy and image reconstruction. The binding affinity of the antibody was relatively low, as determined by surface plasmon resonance. Nevertheless, we were able to observe two binding interactions per Fab, one mediated by CDRs engaging residues at the top of a spike, and a second, novel one involving the Fab framework making contact with residues at the top of an adjacent spike. This binding mode differs significantly from the interactions previously observed between HBcAg and conventional high-affinity anti-HBcAg antibodies 4; 14.
RESULTS
Localization of Fab 9c8 binding sites
Of the two naïve monoclonal antibodies (5H7 and 9c8) that react only with intact HBV capsid protein, but not closely related proteins 10, only 9c8 decorated capsids, as judged by negative stain electron microscopy (Fig. 1 (a) and (b)). Consequently, 9c8 was selected for further study. HBV capsids were decorated with Fab 9c8 (Fig. 1 (c) and two reconstructions were calculated, one for T=3 and one for T=4 particles. The reconstructions were calculated to a spatial frequency limit of 8 Å, however calculation of the resolution as a function of radius showed that the resolution was slightly higher in the region of the capsid (8 Å) than in that occupied by the Fab (11 Å), where the lower protein density results in a lower signal-to-noise ratio (Fig. 1(d)).
Fig. 1.
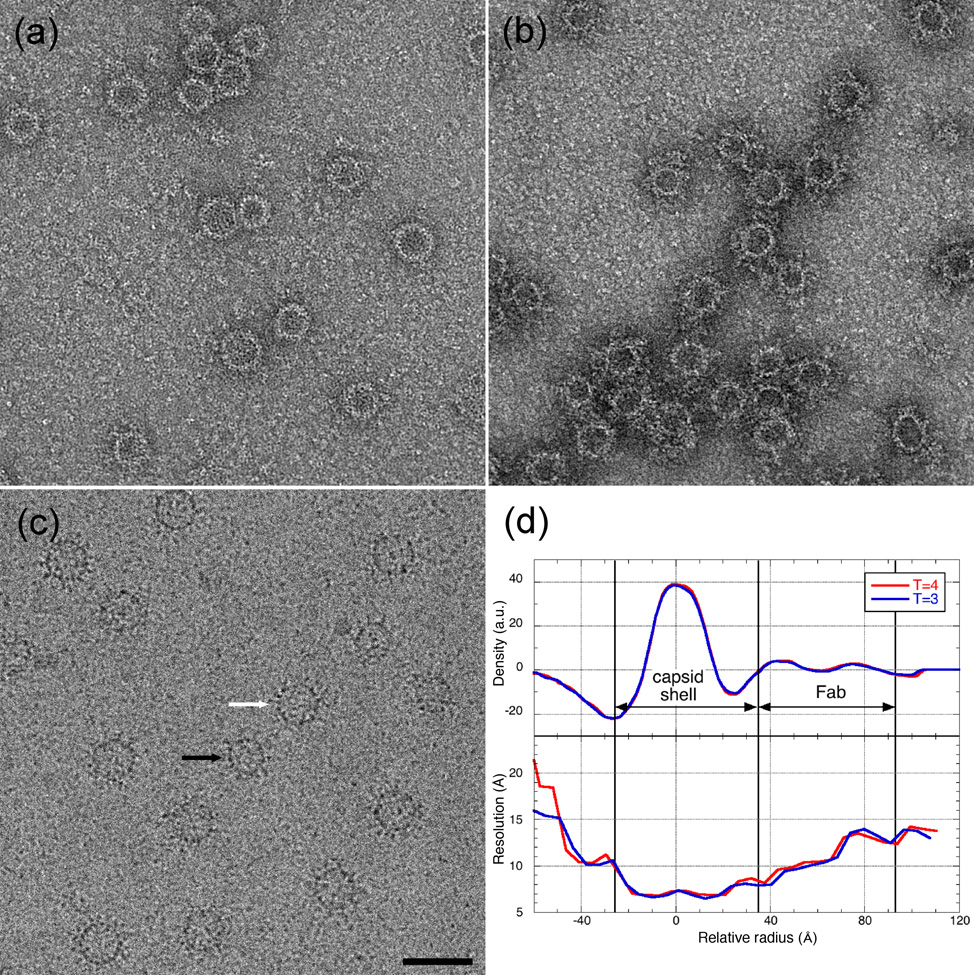
HBV capsids incubated with either Mab 5H7 (a) or Mab 9C8 (b) and visualized in negative stain. Only 9c8 decorated capsids sufficiently for a cryo-EM reconstruction. (c) HBV capsids decorated with Fab 9c8 and visualized by cryo-EM. Black and white arrows point to T=3 and T=4 particles, respectively. Bar = 500 Å. (d) Radial variation of density and resolution of reconstructions of Fab-decorated capsids. The resolution of both the T=3 and T=4 reconstructions is approximately 8 Å in the region of the capsid.
Initial visual inspection of the surface-rendered reconstructions revealed a highly unequal distribution of Fab-related density among the quasi-equivalent sites on a given type of capsid (T=3 or T=4), and suggested that the highest occupancy of Fab occurred in a counter-clockwise orientation about the five-fold axes of symmetry (Fig. 2 (a)–(d)). Central sections taken along the two-fold axes of the maps provide orthogonal views through the capsid subunits and include all three axes of symmetry (Fig. 2 (e), (f)). The sections show that Fab occupancy on the spikes is less than 100%, and particularly so on some of them, confirming the earlier visual impression given by the surface-rendered map. The sections also show that Fab binding occurs at or near the tops of the spikes. To estimate the relative occupancy of the Fabs at the quasi-equivalent sites on the capsids the atomic structure of a surrogate Fab was modeled into the density maps (see below) together with the atomic structures of the capsids. By adjusting the occupancy of Fabs in the models, and comparing central sections with the corresponding ones in the experimental maps, it was possible to evaluate the occupancies at the different sites (Fig. 2 (g), (h)). On T=4 capsids the highest occupancy (~60% as compared to the spike) occurs at the AB dimer. This density originates from the position corresponding to the B monomer and extends in the direction of the three-fold axis (Fig. 2 (b), (d); Fig. 3). In comparison, the occupancy at the quasi-equivalent site on the opposite side of the spike, and corresponding to the A monomer, was only ~5%. Similarly, the occupancy at the two sites on the CD dimer was estimated to be ~25% in both cases. The interpretation of the densities, and the estimation of the occupancies, is complicated by the fact that the observed densities are in fact due to an overlap, on average, of Fabs bound at adjacent, mutually occluding, locations. Nevertheless, by focusing on regions not involved in the overlap, it was possible to assign occupancy levels. The inferred differences in occupancy at quasi-equivalent sites may be due in part to steric factors. For instance, Fabs bound around the T=4 five-fold axis might be expected to experience less mutual crowding when arranged in a counter-clockwise arrangement than in the opposite orientation (Fig. 3). Greater binding around the five-fold axis in this fashion would presumably compete with binding at adjacent locations. For T=3 particles a very similar impression is obtained, with the highest occupancy also occurring on the AB dimer, again with the higher occupancy on the B chain (~60%) and the lowest on the A chain (~5%) (data not shown).
Fig. 2.
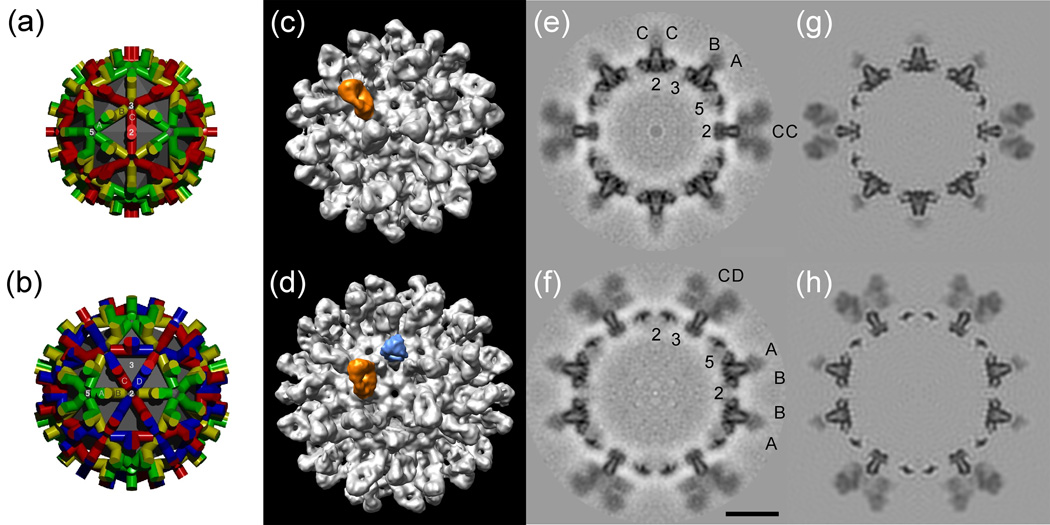
Schematic surface lattices of HBV nucleocapsids, and surface renderings and central sections of the reconstructions of Fab-decorated capsids. (Panels (a) and (b)) Diagrams showing the arrangement of subunits in T=3 and T=4 icosahedral capsids, respectively. The monomeric units are positioned in seven quasi-equivalent environments, of which there are three (A, B, and C) in T=3 capsids, and four (A, B, C, and D) in T=4 capsids. (Panels (c) and (d)) Surface-rendered representations of Fab-decorated T=3 and T=4 capsids viewed along axes of two-fold symmetry. Highlighted are the densities corresponding to Fab (orange) bound at the AB sites on T=3 and T=4 capsids, and the three-fold symmetric average density of Fab bound at the CD sites (light blue). (Panels (e) and (f)) Central sections taken through the reconstructions are shown with the monomeric subunits labeled as in panels (a) and (b), respectively. The 2-, 3-, and 5-fold axes are labeled. (Panels (g) and (h)) Simulations generated by docking atomic structures of the capsid and Fabs into the electron density maps, adjusting the Fab occupancy to match that of the experimental data, and limiting the resulting models to the same resolution as the reconstruction. Bar = 100 Å.
Fig. 3.
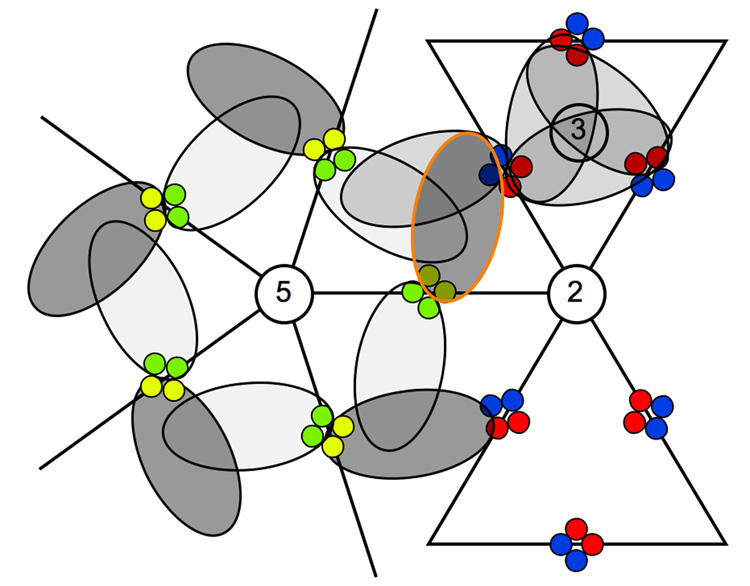
Cartoon showing the approximate arrangement and occupancy of Fab 9c8 bound to the T=4 capsid. The 2-, 3-, and 5-fold axes are labeled. The colored circles represent the four-helix bundles of the spikes, with the A, B, C, and D monomers color-coded as shown in Figure 2. The ellipses represent bound Fab, with darker shading indicating greater occupancy. The Fab highlighted in orange in Fig 2(d) is outlined here in the same color.
Characterization of the Fab binding sites
As no crystal structure is available for antibody 9c8 we selected several IgG2b Fab structures from the PDB to serve as surrogates and fitted them into the well-defined Fab-related electron density associated with the AB dimer. When the Fabs were fitted into the density in either of the two alternative orientations around their pseudo-2-fold axes of symmetry we observed that the better fit was obtained when the heavy chain was positioned close to the A monomer, and the light chain close to the B monomer. The same result was obtained with an automated docking procedure 15. We concluded that this is probably the preferred orientation of Fab 9c8 with respect to the spike.
To clarify how 9c8 interacts with the spike we attempted to model the Fab with a surrogate as similar to it as possible. To this end, the light chain and heavy chain variable domain sequences of 9c8 were used to search the PDB for similar sequences. For the light chain the first 104 residues, and for the heavy chain the first 116 residues, were employed. The PDB entries with the greatest similarity were selected in each case: for the light chain this was Fab 4G2 (PDB: 1UYW) (Martinez-Fleites et al., to be published) with 96.0% identity, and for the heavy chain this was Fab F124 (PDB: 1F11), an antibody coincidentally against hepatitis B surface antigen 16, with 87.5% identity. The sequences of the 4G2 heavy and F124 light chains are not as similar to those of 9c8. The sequences corresponding to the CDRs were identified as described (http://www.bioinf.org.uk/abs/). For the light chain all three CDRs are both in the same position in the sequence and have the identical sequence as in 9c8, except that CDR1 and CDR3 each have single residue differences (Fig. 4). For the heavy chain the CDRs are also in the same positions as their counterparts in 9c8, but the sequences differ slightly, varying from 61.5% to 83.3% identity. Given the high sequence similarities between the 9c8 light and heavy chain variable domains and their counterparts in 4G2 and F124, we assumed that the latter Fabs could be employed as reasonable surrogates for Fab 9c8. Both 4G2 and F124 were docked independently into the electron density at the AB dimer and were observed to be in very close mutual alignment over both their constant and variable domains (C-α, RMSD = 1.1 Å) despite the presence of the chains not similar to those of 9c8. We then modeled the structure of Fab 9c8 by combining the atomic coordinates of the light chain of 4G2 and the heavy chain of F124.
Fig. 4.
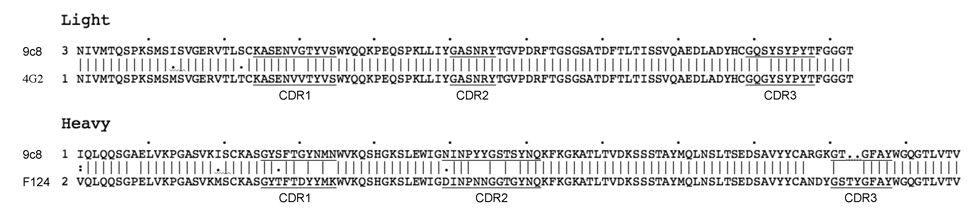
Alignment of surrogate Fab variable domain chains with those of Fab 9c8. The light- and heavy-chain variable domain sequences of 9c8 were employed to search the PDB for similar sequences. The light chain of Fab 4G2 (PDB: 1UYW) had the highest sequence similarity to the light-chain of 9c8, and the heavy chain of Fab F124 (PDB: 1F11) had the highest similarity to that of 9c8. The CDR regions were identified as described in the text and are underlined.
Fab 9c8 binds with highest occupancy to the AB dimer, on both T=3 and T=4 capsids, and consequently it is at this site that it is easiest to interpret the density (Fig. 5 (a), (b)). On both T=3 and T=4 capsids the Fab appears to make three points of contact with the AB dimer; at two points between the light chain and the B monomer, and at one point between the heavy chain and the A monomer. The two light chain contacts with the AB dimer are relatively well resolved in the case of the T=3 capsid, but less so in the T=4 one (Fig. 5 (c), (d)). On both T=3 and T=4 AB dimers the three density continuities between Fab and dimer accommodate the three CDRs that make the closest approach to the spike (Table 1). Heavy chain CDR2 lies largely within the density continuity between the Fab and the A chain, whereas light chain CDRs 1 and 3 lie either within or very close to the density continuity associated with the B chain. Light chain CDR1 appears to make a lateral contact with the B chain whereas light chain CDR3 makes an approach from the top. We conclude that 9c8 probably binds to the capsid spike via light chain CDRs 1 and 3, and heavy chain CDR2.
Fig. 5.
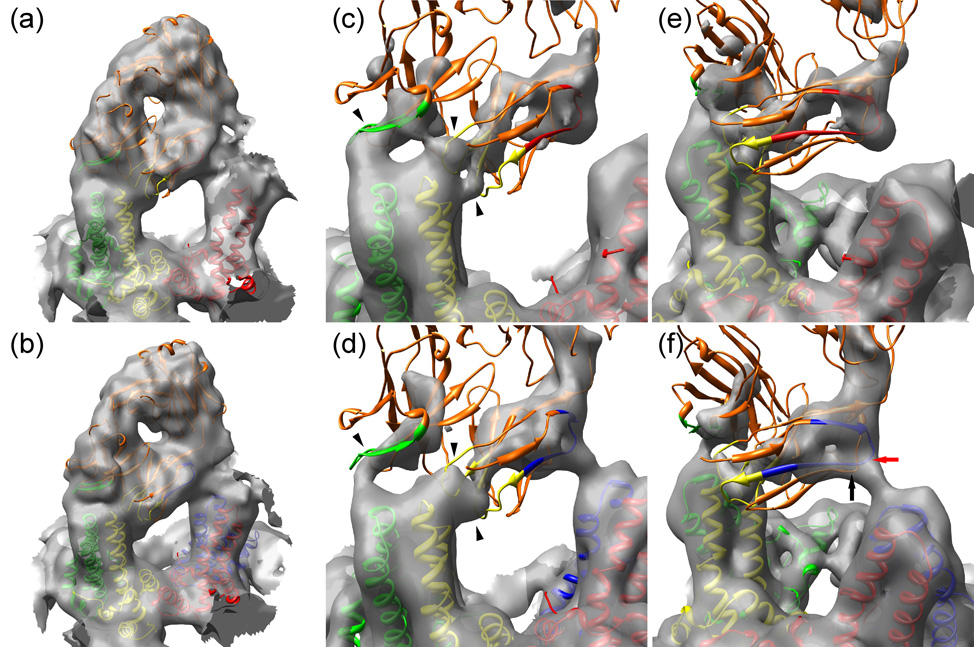
Models of the interactions of Fab 9c8 with HBV capsids. (Panels (a) and (b)) The composite atomic model of Fab 9c8 is shown docked into the electron density at the AB dimer of T=3 (a) and T=4 (b) capsids. The capsid subunits are color-coded as in Fig. 2. The Fab is colored orange with the interacting CDRs highlighted in the same color as the capsid monomer with which they make contact: (heavy chain CDR2, green), and light chain CDRs 1 and 3 (yellow). (Panels (c) and (d)) Closer views of the Fab CDRs engaging the spike tip, and contoured at a high level. The Fab contacts the T=3 AB dimer at three sites (arrowheads) whereas these three points are not as well resolved on T=4 capsids. (Panels (e) and (f)) As in panels (c) and (d) but viewed with the interaction between the Fab framework and the adjacent spike in the foreground on the right. A portion of the FR1 sequences is highlighted, colored the same as the capsid monomer with which they make contact. T=3 capsids do not have a D quasi-equivalent site, and consequently the Fab contacts a C monomer (red) (located in the background but not shown here for clarity), whereas a Fab on a T=4 capsid contacts a D monomer (blue). The points on the FR1 and FR3 sequences making the closest approach to the D monomer are indicated by red and black arrows, respectively.
TABLE 1.
Distances Between Fab Chains and Spike Loopsa
| Fab | Dimer | ||||
|---|---|---|---|---|---|
| Locationb | FR / CDRc | Residued | Monomere | Residuef | Distance (Å) |
| 1 | L CDR 1 | Tyr 34 | B | Asn 74 | 9 |
| 2 | L CDR 1 | Tyr 34 | B | Pro 79 | 10 |
| 3 | L CDR 3 | Tyr 94 | B | Pro 79 | 5 |
| 4 | L CDR 3 | Tyr 96 | B | Pro 79 | 7 |
| 5 | H CDR 2 | Tyr 53 | A | Pro 79 | 10 |
| 6 | L FR 1 | Glu 19 | D | Asp 78 | 11g |
| 7 | L FR 3 | Ser 79 | D | Asp 78 | 14 |
| 8 | L CDR 2 | Gly 52 | B | Pro 79 | 15 |
| 9 | H CDR 1 | Tyr 31 | A | Pro 79 | 14 |
| 10 | H CDR 3 | Tyr 103 | B | Pro 79 | 12 |
Distances at points of closest approach between C-α traces of the light and heavy chains of the pseudo-atomic model of Fab 9c8 and the dimer chains.
Rows 1–5, primary interactions. Rows 6–7, secondary interactions. Rows 8–10, no interactions.
L, light chain. H, heavy chain. CDR, complementarity determining region. FR, framework.
Fab residues at point of closest approach to capsid dimer.
A, B, and D correspond to the monomers at the four quasi-equivalent sites on the T=4 surface lattice, as diagramed in Fig. 2.
Monomer residues at point of closest approach to Fab.
The corresponding distances at the three remaining quasi-equivalent sites are 16, 26, and 33 Å.
Inspection of the three implicated 9c8 CDR sequences, together with the molecular modeling, suggests that hydrogen bonds may be involved in binding the antibody to the antigen. All three CDRs contain tyrosine at or near their point of closest approach to the dimer, and both light chain CDR3 and heavy chain CDR2 contain three tyrosine residues each, with heavy chain CDR2 having two at the presumed point of contact (Fig. 4). Hydrogen bonds may be formed with the glutamic and aspartic acid residues of the EDPA sequences in the loops on the dimer, residues that have previously been implicated in binding 9. Tyrosine residues are common in antibody binding loops, prior to affinity maturation, and give rise to weak interactions 17; 18. This is important because to initiate an immune response B cells should be able to respond to antigens through low-affinity interactions as high-affinity ones may not be present in the initial receptor repertoire. It has also been shown that promiscuous antibody binding involves specific hydrogen bonds rather than hydrophobic interactions 18; 19. Taken together this may mean that naive receptor mIgs (like 9c8) can bind relatively weakly but selectively to antigens (like core) via a network of hydrogen bonds.
9c8 interacts with capsids at a second, unconventional site
In addition to the interactions between the CDR loops and the spike as described above, we also observe what appears to be a second interaction involving an adjacent spike. At a contour level where density connectivity can be observed between the three CDR loops and the AB dimer, there is also a region of comparable density between the side of the Fab and the adjacent CD dimer (Fig. 5 (e), (f)). On T=3 particles the appearance of the secondary interaction is similar to that on T=4 particles, but the tube of density is not continuous, when contoured at the same level, probably on account of slightly lower Fab occupancy (Fig. 5 (e)). With the Fab structure docked in place, the light chain residues 14 – 19 (SISVGE) are juxtaposed with the D-monomer residues 77 – 81 (EDPAS). At the point of closest approach the distance between the C-α carbons of the light chain and the D-monomer is ~11 Å (Table 1) (Note: at the three remaining quasi-equivalent sites the corresponding distances are ~16, 26, and 33 Å; too far to afford stabilization and further explaining the observed preferential binding around the five-fold axes of symmetry). The light chain residues SISVGE are located only four residues amino-terminal to the start of the light chain FR1-CDR1 junction sequence (beginning with residues LSCKAS, Fig. 4) previously identified as being involved in the binding between receptor immunoglobin and core particles 10. Thus the large size of the Fab as compared to the spike spacing, and the low angle at which it interacts with the capsid, brings the entire light chain FR1-CDR1 sequence into close proximity with two adjacent spikes, thereby allowing the CDR1 sequence to engage the residues EDPA in the immunodominant loop of one spike (in this instance the B chain of the AB dimer) and the FR1 sequence to interact with the same loop residues on an adjacent spike (in this case the D chain of the CD dimer). Although the residues on the two spikes are the same, their mode of interaction with the Fab differs. In addition to the FR1 residues, there are also other framework residues that may contribute to the secondary interaction. Residues Ser 78 and Ser 79 in FR3 are located almost immediately adjacent to the SISVGE sequence in FR1, and at a similar distance from the EDPAS sequence on the spike. In summary, the set of potentially hydrogen bonding residues at this interface, as delineated by the electron density, included the following: Ser 14, Ser 16, and Glu 19 (on FR1), Ser 78 and Ser 79 (on FR3), and Glu 77, Asp 78, and Ser 81 (on the D monomer). Serine residues can form hydrogen bonds with minimal steric hindrance, and like tyrosine they are frequently involved in low-affinity binding of the naïve germline antibodies to new antigens 18. Mutation of the spike residue Ala 80 to a large hydrophobic one (Ile 80) has been shown to abrogate binding 9 suggesting that the arrangement of this set of residues is sensitive to disruption. We conclude that this set of closely positioned residues may form multiple hydrogen bonds that contribute to the stability of the binding.
The interpretation of a potential secondary interaction with the CD dimer is not entirely straightforward because the observed electron density distribution is actually an average of the high Fab occupancy on the AB dimer and the lower occupancy on the CD dimer. However, the secondary interaction is indicated for two reasons. First, the density on the T=4 capsid has the form of a single continuous tube clearly not attributable to three CDRs (Fig. 5 (d), (f)); only when contouring at a much higher threshold does evidence of the two lateral densities characteristic of the CDR contacts appear. Second, the angle at which the tube of density emanates from the CD dimer is different from the angle at which a 9c8 Fab binds to a dimer via its CDRs. Additionally, the results from surface plasmon resonance (below) show that the affinity of Fab 9c8 for dimers is significantly lower than for capsids, providing evidence in support of a second site of interaction between Fab and capsid.
Measuring the binding affinity of 9c8 by surface plasmon resonance
From previous competitive inhibition experiments it has been estimated that 9c8 has an affinity several hundred-fold lower than a conventional anti-HBcAg Mab 10. In addition, evidence for a 1:1 Fab:dimer complex was observed by both gel filtration and analytical ultracentrifugation (data not shown), suggesting an affinity in at least the micromolar range. To measure the binding affinity we used surface plasmon resonance, with capsids displayed on immobilized antibody serving as ligand and Fab as the analyte. Binding could be observed but was characterized by a high off-rate. In fact, the off-rate was too high for the equilibrium constant to be determined by conventional sensogram analysis (Fig. 6 (a)). Instead, the affinity (KD) was estimated from a Scatchard analysis of the binding 20 and found to be 3.7 (± 0.8) × 10−7 M (Fig. 6 (b)). The binding was considered to be selective because both non-immune murine monoclonal (Fab 4B5) and human polyclonal Fab controls failed to bind under the same conditions (Table 2).
Fig. 6.
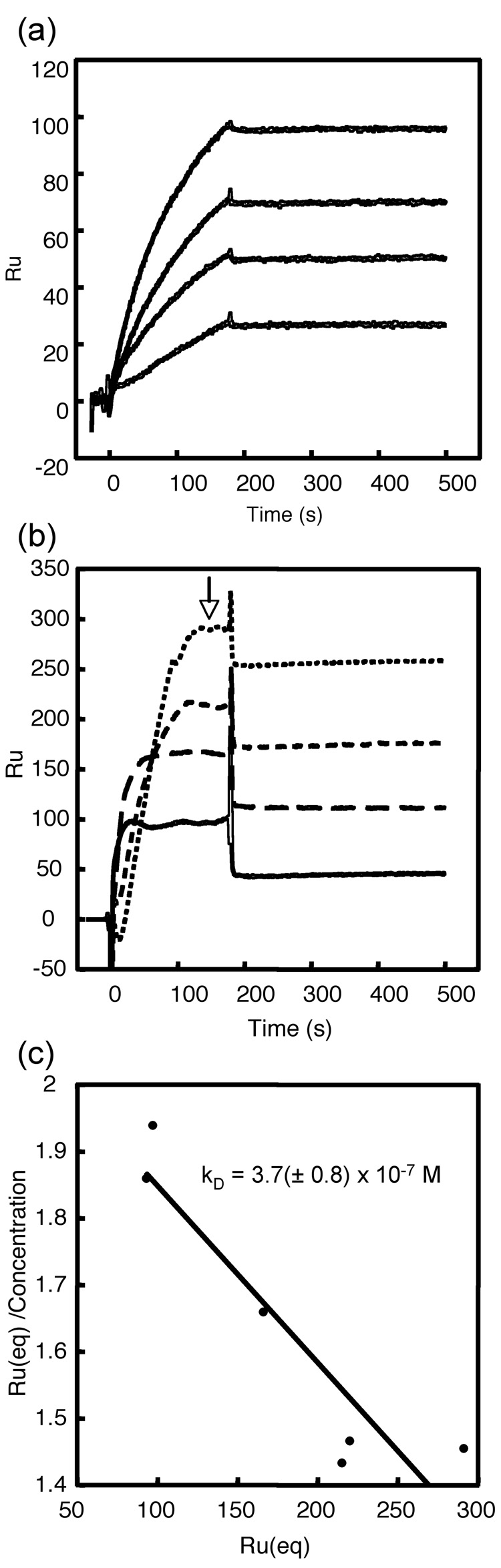
Measurement of the kinetics and affinity of Fabs binding to HBcAg by surface plasmon resonance. (a) Sensograms of F11A4 binding to capsids (bottom to top: 25, 50, 75, and 100 nM analyte). Binding is characterized by an intermediate on-rate but a very low off-rate, accounting for the high affinity of this antibody (KD ~ 1 × 10−10 (M)). (b) Sensograms of Fab 9c8 binding to capsids (bottom to top: 50, 100, 150, and 200 nM analyte). Binding is characterized by an intermediate on-rate but a high off-rate, resulting in a relatively low affinity. Replicates not shown for clarity. Because the high off-rates of 9c8 prevented a conventional simultaneous analysis of the sensograms, the degree of binding at equilibrium (open arrow) as a function of concentration was monitored instead. (c) A Scatchard plot of the equilibrium binding gives a KD of ~ 4 × 10−7 (M).
TABLE 2.
Affinity of Fab 9c8 for cAga
| Capsid |
Dimer | |||
|---|---|---|---|---|
| NaCl (mM)b | Fab 9C8 | Fab 4B5c | Fab (poly) | Fab 9C8 |
| 15 | 147.0d | NB | NB | 1.0 |
| 20 | 49.7 | 5.4 | NB | 1.2 |
| 30 | 17.6 | 5.5 | NB | 1.5 |
| 50 | NB | 5.0 | NB | 3.2 |
| 75 | NB | 5.0 | NB | 4.6 |
| 150 | NB | 7.2 | NB | 11.0 |
Measured by surface plasmon resonance. Antigens captured with immobilized antibody. Fabs are ligands.
Buffer was 10 mM HEPES, pH 7.4 with sodium chloride as indicated.
Control Fabs: Fab 4B5, murine monoclonal anti-sAg; Fab (poly), non-immune human polyclonal.
Surface plasmon resonance response units (arbitrary values). All values are the average of three determinations. NB indicates no binding. Determinations at sodium chloride concentrations <15 mM were complicated by refractive index artifacts (e.g. negative slope) and are not included.
Next we sought to characterize more closely the site of binding on the capsid. To this end we performed competition experiments in which capsid protein dimers (0 or 100 nM) were pre-incubated with 100 nM Fab 9c8 before analyzing the binding to immobilized capsids. We found that the presence of dimers reproducibly caused a ~40% inhibition of binding. This observation suggested that 9c8 interacts with dimer as well as capsid, and that binding involves a region on the dimer that would be presented on the outside of capsids. Both previous biochemical observations 9 and the current reconstruction indicated that binding involves the glutamic and aspartic acid residues at the top of the spike. To test this proposition, capsids in which either of these residues had been mutated were captured with immobilized monoclonal antibody 3120 and probed with Fab 9c8. Compared to the wild-type controls, the mutations E77C and D78C inhibited binding by ~60% and ~40%, respectively. To assess the possibility of a secondary interaction, we compared the binding of Fab 9c8 to both capsids and dimers displayed on immobilized antibody. The affinity of Fab for dimers was ~150-fold lower than for capsids. Taken together, the results indicate that: 9c8 binds to HBcAg in a selective fashion with intermediate affinity (KD ~ 10−7 M); this binding involves areas on dimers present on the outside of capsids including the aspartic and glutamic acid residues in the spike loop(s); and finally, that two adjacent spikes are required for the Fab to achieve full binding.
The reconstruction and molecular modeling show that Fab 9c8 interacts with two adjacent spikes, potentially through the formation of up to five hydrogen bonds. Surface plasmon resonance indicates an affinity on the order of ~10−7 (M), corresponding to a free energy of ~9.5 kcal mol−1. This is in reasonable agreement with the presumed formation of 4 – 5 hydrogen bonds, assuming a liberation of ~2 kcal mol−1 per hydrogen bond.
DISCUSSION
We have presented a model for the low-affinity interaction of hepatitis B virus capsids with the surface receptor immunoglobins of naïve B cells. The model was generated by cryo-EM reconstruction of a complex formed between capsids produced in E. coli and Fab derived from a close-to-naïve B cell. Capsids produced in E. coli have long been used successfully as immunodiagnostic reagents 21; 22, suggesting that all the epitopes are present and properly displayed on their surface, and recent structural studies have in fact confirmed that bacterially produced capsids 12; 23; 24 have essentially the same structure as authentic viral ones 25; 26; 27. The 9c8 Mab also was specifically prepared to resemble the receptor immunoglobin of a single naïve B cell 10. The 9c8 hybridoma has undergone isotype switching since its original isolation, producing IgG2 rather than the original IgM, but this change is known to affect only the Fc portion of the antibody 28. Hence, we consider the observed interaction between HBV capsids and Fab 9c8 to be representative of those naïve B cells that present the VH1 consensus sequence.
It should be noted that two consensus sequences, I/LSCKASGYI/SFTS/G and ISCRASQVSTSS, were originally identified in the VH and VL domains, respectively, of naïve receptors, with the former appearing to mediate the binding of Mab 9c8 10. However, the current results suggest that Fab 9c8 binds primarily via the VL chain rather than the VH chain. The two motifs above have high sequence similarity (58% identity, 75% similarity) 10. Comparison of the 9c8 sequences (Fig 4) reveals the presence of a sequence (LSCKASENVGT) in the light chain FR1-CDR1 region with a high similarity (73%) to the VH1 consensus motif. This may explain the observed orientation of 9c8 binding to the capsid.
It has been shown by a number of different means that cAg binds to and activates approximately 5% of naïve murine B cells, and up to 15% of naïve human B cells, and that this interaction is mediated by conserved linear motifs present in the FR1-CDR1 junctions of the light and heavy chains of the surface receptor immunoglobins 8; 10. As Fab 9c8 binds to HBV capsids via both the conserved sequence at the light chain FR1-CDR1 junction and by a secondary interaction involving residues in the light chain FR1 and FR3 regions (as shown here), and because both of these sequences are presented in the overall context of the conserved structure of the Fab, we consider it likely that other mouse naïve B cells bearing the conserved motif interact with core antigen in much the same way. The similar structure of mouse and human Igs 29 furthermore suggests that a comparable interaction may also occur in humans.
HBV capsids bind 9c8 with a low affinity
The affinity of Fab 9c8 is rather low (KD ~ 10−7 M) though still within the range (10−6 – 10−10 M) where affinity discrimination by B cells occurs 30. The strength of the overall interaction between a capsid and a B cell will be higher, and should vary depending on the number of receptors eventually engaged. Most virus receptors have a low affinity, yet viruses bind effectively even at low concentrations. Given the severe constraints of pleiotropic effects imposed by its tight genomic organization and replication strategy 31 there may be no net benefit to the virus to evolve a higher affinity.
What role might B cell binding play in an actual infection?
There are clear differences between the early adaptive immune responses of individuals with resolved and chronic Hepatitis B infections, with the main one being a robust polyclonal CTL response in the former, and a weak, depleted one in the latter. There are several factors thought to contribute to this condition, including the induction of tolerance by the three viral antigens: HBsAg and HBeAg 32, and HBcAg 10. The involvement of HBcAg-binding B cells in T cell priming is clear from in vivo model systems 9, raising the question of what their role might be in an actual infection. The natural course of HBV infection in immunocompetent adults is viral clearance. This can be explained by an efficient uptake of HBcAg released from intact or lysed hepatocytes, which would assist in the activation of T cells in the subclinical or acute adult infection. On the other hand, in vivo studies 33 have suggested that B cells that have taken up exogenous antigens have the ability to inactivate antigen-specific CTLs, which should promote viral persistence. Alternatively, HBcAg may function as a B cell superantigen. Comparison with the six known B cell superantigens (reviewed in 34) shows that HBcAg engages the Ig variable domain at essentially the same location as Peptostreptococcus magnus protein L (Ppl), the only other VL-specific superantigen. The affinity between superantigens and naïve B cells is in the micromolar range 34. The affinity between HBcAg and Fab 9c8 (4 × 10−7 M) is similar to that between Ppl and Fab 2A2 (0.13 × 10−6 M) 35, but its potential valency is considerably higher. The level of free HBcAg in circulation is most likely low during a chronic infection when the levels of anti-HBcAg IgG are high, therefore HBcAg probably exerts its effect in the early acute infection.
Alternative receptors for HBV capsids
HBcAg can also attach to cells in other ways. For example, proteoglycan-mediated binding has been observed in a variety of cell types 36; 37, however, the significance of this type of binding for HBV infections in humans is unclear. In contrast, numerous in vivo experiments have clearly demonstrated the requirement for B cells in the efficient priming of both CD4+ T-cells 7 and cytotoxic T lymphocytes 9. If proteoglycans, or other ubiquitously distributed cellular receptors, were sufficient alone for binding, uptake, and presentation, then this requirement for B cell involvement would not have been observed.
MATERIALS & METHODS
Preparation of core protein dimers and capsids
The HBcAg construct Cp149.3CA was purified and assembled as described previously 38. The mutations E77C and D78C were introduced to test antibody binding at the top of the spike. The mutation G123A was employed to prevent assembly of dimers into capsids (Stahl et al., unpublished observations).
Preparation of Antibodies and Fabs
Mabs 5H7 and 9C8 were specifically generated to be representative of naïve B cell receptors, as described previously 10. Mab 9c8 was observed to have a higher affinity for HBV capsids and was therefore selected for closer study. Fab 9c8 was prepared by standard procedures using immobilized papain and Protein A (Pierce). Four other murine monoclonal antibodies were also employed: Mab 4B5 binds hepatitis B surface antigen (HBsAg) (P. T. W., unpublished), Mab F11A4 39 and Mab 3120 13 bind HBcAg, and Mab e6 binds hepatitis B e-antigen (HBeAg) 40. Polyclonal human antibody with no specificity for HBV proteins was also used. Fabs were generated from 4B5 and the human antibody as described for 9c8.
Cryo-electron microscopy
Fab 9c8 was added to capsids at a ratio of four Fabs per dimer, and the preparation was concentrated by ultrafiltration to 3.5 mg/ml with respect to capsid protein. Focal pairs of micrographs were recorded at 50,000x magnification under low dose conditions (~10 e/Å2 per exposure), as described previously 14. The first exposure was recorded such that the first zero of the contrast transfer function was at ~ (18.5 Å)−1, while for the second exposure the first zero was at ~ (23.5 Å)−1.
Image Reconstruction and Molecular Modeling
Reconstructions were calculated essentially as described previously 14. Briefly, 12 focal pairs were scanned at a rate corresponding to 1.40 Å/pixel at the specimen. Image processing was performed with Bsoft 41. A total of 2942 particles were picked manually and sorted automatically according to diameter to yield 1355 (T=3) and 1587 (T=4) particles. Origins and orientations were determined by comparison with pre-existing maps of unlabeled T=3 and T=4 capsids 24. PFT2 and EM3DR2 42; 43; 44 were then used iteratively until no improvements were observed in the resolution. Reconstructions included all the particles with real-space correlation coefficients higher than a threshold calculated by decreasing the average value by the standard deviation, which yielded 1246 particles for T=3 and 1363 particles for T=4. Density maps were calculated to a spatial frequency limit of 8 Å with full CTF correction applied. A resolution of 10.0 Å was estimated for both reconstructions by Fourier shell correlation 45 and a threshold of 0.5. Resolution as a function of radius was estimated by calculating the Fourier shell correlation of consecutive shells, 28 Å thick and 5.6 Å apart, as described 14.
Molecular Modeling
Dimers of capsid protein (PDB: 1QGT) were first docked into the density maps of the T=3 and T=4 reconstructions 46. Fab 9c8 was modeled by combining the light and heavy chains from two different Fabs with high sequence similarity to those of 9c8. Specifically, Fab 4G2 (PDB: 1UYW) and Fab F124 (PDB: 1F11) were fitted into the T=3 and T=4 reconstructions, using the binding site with the highest occupancy for the docking analysis. All the docking procedures were performed manually using Chimera 47, and the results checked by automated correlation-based fitting with colores in the SITUS program 15. Because of the presence of a pseudo-2-fold symmetry axis in the Fabs, for each fitting determined, the alternative solution corresponding to a 180° rotation was tested, and the solution with the higher correlation coefficient was chosen. Finally, the atomic coordinates of the light chain from Fab 4G2 and those of the heavy chain from Fab F124 were combined to generate a model for 9c8.
An initial estimate of Fab fractional occupancies was obtained by identifying the highest densities in the variable domains of bound Fabs, and relating these values to the peak capsid density, after background subtraction. Density maps of Fab-labeled capsids were computed using resolutions and Fab occupancies as previously estimated. The coordinates of the Fabs and the dimer of the capsid protein as modeled into an asymmetric unit were converted to density and extended to all the other units with the programs bsf and bsym in Bsoft. The initial estimates of Fab occupancies were iteratively refined by comparing sections of simulated maps with those of the corresponding reconstructions, and adjusting them accordingly. All surface representations were produced using Chimera.
Determination of binding affinities
Binding affinities were determined by surface plasmon resonance (BIAcore) by immobilizing antibody on the sensor surface, capturing antigen, and observing the binding of Fab to the displayed antigen. The capture antibodies included Mab F11A4 39, Mab 3120 48, and Mab e6 40. Cp149.3CA capsids were captured with Mab F11A4, whereas capsids bearing the E77C or D78C mutations were captured with Mab 3120, which binds between spikes 13 and is therefore unaffected by these mutations. Unassembled dimers were captured with Mab e6, which binds to a conformational epitope in the vicinity of the C-terminus and therefore does not obstruct regions on the spike. The binding of Fab 9c8 was compared to that of two controls, Fab 4B5 (obtained from murine monoclonal anti-sAg) and an unfractionated mixture of Fab generated from human non-immune IgG. Binding of Fabs to the displayed capsids and dimers was determined in 10 mM HEPES, pH 7.4 buffer containing concentrations of sodium chloride ranging from 15 to 150 mM. The binding affinity of 9c8 was obtained by Scatchard analysis 20. Competition experiments were performed by mixing either 0 or 100 nM dimer with 100 nM Fab 9c8, incubating for 30 min at room temperature, and then measuring binding of Fab to capsids as described above.
ACKNOWLEGMENTS
We wish to thank Dr. Bernard Heymann for guidance with Bsoft and computational support, Drs. Bridget Ferns and Richard Tedder for providing Mabs F11A4 and e6, Dr. Audray Harris for helpful discussions, and Joshua Kaufman and Ira Palmer for technical assistance. This research was supported by the NIAMS Intramural Research Program of the NIH, and by the Swedish Cancer Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Liaw YF. Management of chronic hepatitis B: an evolving issue. Liver Int. 2006;26 Suppl 2:1–2. [Google Scholar]
- 2.Lesmana LA, Leung NW, Mahachai V, Phiet PH, Suh DJ, Yao G, Zhuang H. Hepatitis B: overview of the burden of disease in the Asia-Pacific region. Liver Int. 2006;26 Suppl 2:3–10. [Google Scholar]
- 3.Vanlandschoot P, Cao T, Leroux-Roels G. The nucleocapsid of the hepatitis B virus: a remarkable immunogenic structure. Antiviral Res. 2003;60:67–74. doi: 10.1016/j.antiviral.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Steven AC, Conway JF, Cheng N, Watts NR, Belnap DM, Harris A, Stahl SJ, Wingfield PT. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv Virus Res. 2005;64:127–165. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- 5.Salfeld J, Pfaff E, Noah M, Schaller H. Antigenic determinants and functional domains in core antigen and e antigen from hepatitis B virus. J. Virol. 1989;63:798–808. doi: 10.1128/jvi.63.2.798-808.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sällberg M, Ruden U, Magnius LO, Harthus HP, Noah M, Wahren B. Characterisation of a linear binding site for a monoclonal antibody to hepatitis B core antigen. J. Med. Virol. 1991;33:248–252. doi: 10.1002/jmv.1890330407. [DOI] [PubMed] [Google Scholar]
- 7.Milich DR, Chen M, Schodel F, Peterson DL, Jones JE, Hughes JL. Role of B cells in antigen presentation of the hepatitis B core. Proc. Natl. Acad. Sci. USA. 1997;94:14648–14653. doi: 10.1073/pnas.94.26.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao T, Lazdina U, Desombere I, Vanlandschoot P, Milich DR, Sallberg M, Leroux-Roels G. Hepatitis B virus core antigen binds and activates naive human B cells in vivo: studies with a human PBL-NOD/SCID mouse model. J Virol. 2001;75:6359–6366. doi: 10.1128/JVI.75.14.6359-6366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazdina U, Alheim M, Nystrom J, Hultgren C, Borisova G, Sominskaya I, Pumpens P, Peterson DL, Milich DR, Sallberg M. Priming of cytotoxic T cell responses to exogenous hepatitis B virus core antigen is B cell dependent. J Gen Virol. 2003;84:139–146. doi: 10.1099/vir.0.18678-0. [DOI] [PubMed] [Google Scholar]
- 10.Lazdina U, Cao T, Steinbergs J, Alheim M, Pumpens P, Peterson DL, Milich DR, Leroux-Roels G, Sallberg M. Molecular basis for the interaction of the hepatitis B virus core antigen with the surface immunoglobulin receptor on naive B cells. J. Virol. 2001;75:6367–6374. doi: 10.1128/JVI.75.14.6367-6374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith TJ. Structural studies on antibody-virus complexes. In: Chiu W, Johnson JE, editors. Adv. Prot. Chem.: Virus Structure. Vol. 64. San Diego: Academic Press; 2003. pp. 409–454. [DOI] [PubMed] [Google Scholar]
- 12.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol. Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 13.Conway JF, Watts NR, Belnap DM, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Characterization of a conformational epitope on hepatitis B virus core antigen and quasi-equivalent variations in antibody binding. J. Virol. 2003;77:6466–6473. doi: 10.1128/JVI.77.11.6466-6473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris A, Belnap DM, Watts NR, Conway JF, Cheng N, Stahl SJ, Vethanayagam JG, Wingfield PT, Steven AC. Epitope diversity of hepatitis B virus capsids: quasi-equivalent variations in spike epitopes and binding of different antibodies to the same epitope. J Mol Biol. 2006;355:562–576. doi: 10.1016/j.jmb.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 15.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 16.Saul FA, Vulliez-Le Normand B, Passafiume M, Riottot MM, Bentley GA. Structure of the Fab fragment from F124, a monoclonal antibody specific for hepatitis B surface antigen. Acta Crystallogr D Biol Crystallogr. 2000;56:945–951. doi: 10.1107/s0907444900008088. [DOI] [PubMed] [Google Scholar]
- 17.Padlan EA. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for ligand binding. Proteins: Struct., Funct., Genet. 1990;7:112–124. doi: 10.1002/prot.340070203. [DOI] [PubMed] [Google Scholar]
- 18.Clark LA, Ganesan S, Papp S, van Vlijmen HW. Trends in antibody sequence changes during the somatic hypermutation process. J Immunol. 2006;177:333–340. doi: 10.4049/jimmunol.177.1.333. [DOI] [PubMed] [Google Scholar]
- 19.James LC, Tawfik DS. The specificity of cross reactivity: promiscuous antibody binding involves specific hydrogen bonds rather than nonspecific hydrophobic stickiness. Protein Sci. 2003;12:2183–2193. doi: 10.1110/ps.03172703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lea SM, Powell RM, McKee T, Evans DJ, Brown D, Stuart DI, van der Merwe PA. Determination of the affinity and kinetic constants for the interaction between the human virus echovirus 11 and its cellular receptor, CD55. J Biol Chem. 1998;273:30443–30447. doi: 10.1074/jbc.273.46.30443. [DOI] [PubMed] [Google Scholar]
- 21.Stahl S, MacKay P, Magazin M, Bruce SA, Murray K. Hepatitis B virus core antigen: synthesis in Escherichia coli and application in diagnosis. Proc. Nat'l. Acad. Sci. USA. 1982;79:1606–1610. doi: 10.1073/pnas.79.5.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen BJ, Litton PA, Mortimer PP, Simmons P. Hepatitis B core antigen synthesised in Escherichia coli: its use for antibody screening in patients attending a clinic for sexually transmitted diseases. J Hyg (Lond) 1984;93:225–232. doi: 10.1017/s0022172400064743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 24.Conway JF, Cheng N, Zlotnick A, Wingfield PT, Stahl SJ, Steven AC. Visualization of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;386:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- 25.Dryden KA, Wieland SF, Whitten-Bauer C, Gerin JL, Chisari FV, Yeager M. Native hepatitis B virions and capsids visualized by electron cryomicroscopy. Mol Cell. 2006;22:843–850. doi: 10.1016/j.molcel.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA. A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci U S A. 2005;102:15821–15826. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seitz S, Urban S, Antoni C, Bottcher B. Cryo-electron microscopy of hepatitis B virions reveals variability in envelope capsid interactions. Embo J. 2007;26:4160–4167. doi: 10.1038/sj.emboj.7601841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder HW., Jr Similarity and divergence in the development and expression of the mouse and human antibody repertoires. Dev Comp Immunol. 2006;30:119–135. doi: 10.1016/j.dci.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Batista FD, Neuberger MS. B cells extract and present immobilized antigen: implications for affinity discrimination. Embo J. 2000;19:513–520. doi: 10.1093/emboj/19.4.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 32.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett SR, Carbone FR, Toy T, Miller JF, Heath WR. B cells directly tolerize CD8(+) T cells. J Exp Med. 1998;188:1977–1983. doi: 10.1084/jem.188.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silverman GJ, Goodyear CS. Confounding B-cell defences: lessons from a staphylococcal superantigen. Nat Rev Immunol. 2006;6:465–475. doi: 10.1038/nri1853. [DOI] [PubMed] [Google Scholar]
- 35.Graille M, Stura EA, Housden NG, Beckingham JA, Bottomley SP, Beale D, Taussig MJ, Sutton BJ, Gore MG, Charbonnier JB. Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure. 2001;9:679–687. doi: 10.1016/s0969-2126(01)00630-x. [DOI] [PubMed] [Google Scholar]
- 36.Cooper A, Shaul Y. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsi-like core particles. J Biol Chem. 2006;281:16563–16569. doi: 10.1074/jbc.M601418200. [DOI] [PubMed] [Google Scholar]
- 37.Vanlandschoot P, Van Houtte F, Serruys B, Leroux-Roels G. The arginine-rich carboxy-terminal domain of the hepatitis B virus core protein mediates attachment of nucleocapsids to cell-surface-expressed heparan sulfate. J Gen Virol. 2005;86:75–84. doi: 10.1099/vir.0.80580-0. [DOI] [PubMed] [Google Scholar]
- 38.Wingfield PT, Stahl SJ, Williams RW, Steven AC. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry. 1995;34:4919–4932. doi: 10.1021/bi00015a003. [DOI] [PubMed] [Google Scholar]
- 39.Ferns RB, Tedder RS. Human and monoclonal antibodies to hepatitis B core antigen recognise a single immunodominant epitope. J. Med. Virol. 1986;19:193–203. doi: 10.1002/jmv.1890190213. [DOI] [PubMed] [Google Scholar]
- 40.Ferns RB, Tedder RS. Monoclonal antibodies to hepatitis Be antigen (HBeAg) derived from hepatitis B core antigen (HBcAg): their use in characterization and detection of HBeAg. J. Gen. Virol. 1984;65:899–908. doi: 10.1099/0022-1317-65-5-899. [DOI] [PubMed] [Google Scholar]
- 41.Heymann JB. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 42.Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- 43.Crowther RA, DeRosier DJ, Klug A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proceedings of the Royal Society of London. Series A, Mathematical and Physical Sciences. 1970;317:319–340. [Google Scholar]
- 44.Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles -the uncommon line. J. Struct. Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- 45.Harauz G, van Heel M. Exact filters for general geometry three-dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- 46.Belnap DM, Watts NR, Conway JF, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Diversity of core antigen epitopes of hepatitis B virus. Proc. Nat'l. Acad. Sci. USA. 2003;100:10884–10889. doi: 10.1073/pnas.1834404100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K, Machida A, Funatsu G, Nomura M, Usuda S, Aoyagi S, Tachibana K, Miyamoto H, Imai M, Nakamura T, Miyakawa Y, Mayumi M. Immunochemical structure of hepatitis B e antigen in the serum. J. Immunol. 1983;130:2903–2907. [PubMed] [Google Scholar]


