Abstract
DNA damage is a form of cell stress and injury that has been implicated in the pathogenesis of many neurologic disorders, including amyotrophic lateral sclerosis, Alzheimer disease, Down syndrome, Parkinson disease, cerebral ischemia, and head trauma. However, most data reveal only associations, and the role for DNA damage in direct mechanisms of neurodegeneration is vague with respect to being a definitive upstream cause of neuron cell death, rather than a consequence of the degeneration. Although neurons seem inclined to develop DNA damage during oxidative stress, most of the existing work on DNA damage and repair mechanisms has been done in the context of cancer biology using cycling non-neuronal cells but not nondividing (i.e. postmitotic) neurons. Nevertheless, the identification of mutations in genes that encode proteins that function in DNA repair and DNA damage response in human hereditary DNA repair deficiency syndromes and ataxic disorders is establishing a mechanistic precedent that clearly links DNA damage and DNA repair abnormalities with progressive neurodegeneration. This review summarizes DNA damage and repair mechanisms and their potential relevance to the evolution of degeneration in postmitotic neurons.
Keywords: Alzheimer disease, Amyotrophic lateral sclerosis, Apoptosis, Aprataxin, Cortical neuron, Motor neuron, p53
INTRODUCTION
DNA damage contributes to the mechanisms of aging and disease (1). It has broad relevance to human pathobiology with its involvement in birth defects, cancer, premature aging syndromes, and certain neurologic disorders. Four decades have passed since Cleaver, in his seminal work, recognized the connection between defective repair of DNA damage, cancer, and neurologic disease in children with xeroderma pigmentosum (XP) (2). The number of genes in the human genome whose products are involved directly in DNA repair is now considered more than 125 (3). The neurologic phenotype of XP patients includes ataxia, micro-encephaly, deafness, learning disability, and peripheral neuropathy; the neuropathologic findings include loss of large sensory fibers and dorsal root ganglion cells, cerebellar and cerebral atrophy, and neuronal degeneration (4). Mutations in genes involved in the nucleotide excision repair (NER) pathway cause most forms of XP (3, 4).
Other human diseases that are associated with abnormalities in NER genes are Cockayne syndrome (CS) and trichothiodystrophy (TTD). These disorders have clinical manifestations that overlap with XP but are not associated with an increased susceptibility to cancer (5). Ataxia telangiectasia (AT) is another tragic childhood disease with pleiotropic clinical manifestations characterized by increased risk for cancer (e.g. acute lymphocytic leukemia and lymphoma) and neurologic abnormalities, including progressive impairment of gait and speech, oculomotor apraxia, and cerebellar atrophy with Purkinje cell degeneration (5). Ataxia telangiectasia is caused by mutations in the ataxia telangiectasia mutated (ATM) gene, which encodes a serine-threonine protein kinase required for cellular responses to DNA double-strand breaks (DSBs) (5).
In contrast to XP, CS, TTD, and AT, which are multisystemic disorders (5), some defects in human DNA repair processes are manifested primarily in neural tissues. These include the neurologic disorders ataxia with oculomotor apraxia Types 1 and 2 (AOA-1 and AOA-2) (6, 7) and spinocerebellar ataxia with axonal neuropathy 1 (SCAN-1) (8). The newly recognized connection between defined molecular defects and apparently selective neural diseases offers an unprecedented rationale to examine the relationships between DNA damage, DNA repair, and neurodegeneration. Historically, it has been difficult to pinpoint DNA damage in the specific pathogenic mechanisms of human neurodegenerative disease because most data indicate associations, rather than causal mechanisms.
DNA DAMAGE OCCURS IN MANY FORMS
DNA damage is defined as any modification of DNA that changes its coding properties or normal function in transcription or replication (1, 9). DNA lesions can occur in many different forms, including apurinic/apyrimidinic (AP) sites (abasic sites), adducts, single-strand breaks (SSBs), DSBs, DNA-protein cross-links, and insertion/deletion mismatches (9).
Apurinic/apyrimidinic sites are common lesions in DNA and are formed either spontaneously or as intermediates during the course of the normal repair of oxidized, deaminated, or alkylated bases (1). Apurinic/apyrimidinic sites are 1 of the major types of damage generated by reactive oxygen species (ROS). Hydroxyl radical attack on the deoxyribose moiety can cause the release of free bases from DNA (10). Apurinic/apyrimidinic sites can be mutagenic or they can cause cell death (10, 11). It has been estimated that endogenous ROS can cause approximately 50,000 to 200,000 AP sites per mammalian cell per day, and that brain cells contain the most AP sites (10). In some organs, the level of AP sites seems to be age dependent, with higher levels found in older animals (10).
Frequently studied oxidized base lesions are 7,8-dihydro-8-oxoguanine (8-oxoG), 8-hydroxy-2-deoxyguanosine (OHdG), and 5-hydroxyuracil. 8-hydroxy-2-deoxyguanosine can be generated from hydroxyl radicals formed by either the Fenton reaction involving homolytic cleavage of hydrogen peroxide (H2O2) catalyzed by Fe2+ or by the decomposition of peroxynitrite (ONOO−), which is formed by the combination of superoxide and nitric oxide (10, 12). Specific sites in a DNA sequence can preferentially accumulate OHdG (12), and it is possible that DNA damage occurs preferentially in some promoter regions in the aging human brain genome (13). Because they mispair with adenine during DNA replication and transcription, 8-oxoG and OHdG are mutagenic DNA lesions. In well-characterized animal models, we found the accumulation of OHdG lesions in preapoptotic neurons during retrograde degeneration (14, 15) and in prenecrotic neurons during ischemic neurodegeneration (16). 5-hydroxyuracil, which arises from the oxidation of cytosine to unstable cytosine glycol that undergoes deamination, is also premutagenic because it gives rise to C-to-T transitions. Hydrolytic deamination of cytosine occurs at a rate of approximately 100 to 500 times per cell per day (17), and most DNA polymerases fail to recognize these mismatches, resulting in base transversions. Thus, OHdG can interfere with the fidelity of the transcription process and may therefore mediate a slow, insidious DNA injury process in long-lived noncycling neurons (9).
Another major type of DNA damage is the SSB in which there is a loss of a purine or pyrimidine base and the deoxyribose with a cut in the phosphodiester backbone of 1 strand of the double helix. Measurements of DNA-SSBs are a very sensitive assessment of genome integrity (18). DNA-SSBs can be identified directly in neurons using antibody-based, end labeling, and DNA elution approaches such as the comet assay (see succeeding discussion). In animal and cell models, we have consistently found that DNA SSBs are formed early in the progression of both apoptotic and nonapoptotic forms of neurodegeneration in vivo and in vitro (19-24). The time at which DNA-SSBs are formed in our neurodegenerative disease models suggests that they are upstream activators of a p53-dependent apoptosis cascade (20, 24).
The mirror counterpart lesion of the SSB is the DSB. The number of spontaneously occurring DSBs in mammalian cells is approximately 9 per day (9). ONOO− is a potent inducer of DNA-DSBs in neurons (21). DNA-DSBs in neurons might be particularly toxic because they can be repaired only by physical recombination with another homologous DNA molecule (9).
DNA-protein cross-links are induced by various chemical agents (9), many of which are suspected or known carcinogens (e.g. arsenic and chromate) or are used as cytostatic drugs in chemotherapy (e.g. cisplatin). DNA-protein cross-links interfere with DNA replication and transcription.
MECHANISMS OF DNA DAMAGE: ROS-BASED MECHANISMS ARE CRITICAL IN NEURONS
Endogenous and environmental agents can cause DNA damage in cells (1, 10, 18). The endogenous agents, ROS and reactive nitrogen species (RNS), are generated by cellular metabolism and other factors are temperature, errors in DNA replication and repair, and methylation. Intrinsically generated DNA lesions occur as mismatched base pairs, base structure alterations such as tautomeric shifts and deamination, base adducts (e.g. hydroxylation), and base deletions causing AP sites, SSBs, and DSBs. The genotoxic actions of ROS/RNS (e.g. hydroxyl radicals, H2O2, and ONOO−) are dramatic (10, 21). Metabolism-generated ROS can cause approximately 10,000 lesions per day in the genome of a human nonneuronal cell, and purine base turnover in DNA, resulting from hydrolytic depurination and subsequent repair, is approximately 2,000 to 10,000 bases per day (9). In nonproliferating, long-living cells such as neurons, approximately 108 purines are lost because of spontaneous depurination during a life span (9). The estimated frequency occurrence of spontaneous (endogenous) DNA-SSBs in a mammalian cell is approximately 20,000 to 40,000 and can be much higher depending on diet, lifestyle, and tissue type. We have found that the neuronal genome, specifically the motor neuron genome, is very sensitive to ROS and RNS because they induce the formation of AP sites, SSBs, and DSBs (21). The different forms of DNA damage can be converted from 1 form to another type. For example, hydroxyl radical and ONOO− attack on DNA induces base adducts and then strand breaks because AP sites are converted into SSB if they are not repaired (9, 21). Hydroxyl radical and ONOO− are also potent mutagens in human cells (18).
In addition to the intrinsically generated lesions to DNA, dietary mutagenic chemicals, ultraviolet and ionizing radiation, and heavy metals are environmental agents that damage the genome, causing DNA cross-links, adducts, and oxidative cleavage (18). Some of these exogenous insults may have lower relevance than others as DNA-damaging stressors for the CNS, but microenvironmental insults in the brain such as the presence of β-amyloid might instigate DNA damage through the formation of purine dimers (25).
SINGLE-CELL ANALYSIS OF EARLY LOW-LEVEL DNA DAMAGE IN NEURONS
Single-cell gel electrophoresis, also called “comet” assay, is considered the most sensitive quantitative method for measuring early damage to genomic DNA of eukaryotic cells on a single-cell basis (18). By adjusting pH conditions, the types of DNA lesions that are detectable by comet assay include AP sites (alkali-labile sites), SSBs, DSBs, and DNA-protein cross-links (18). Three different pH conditions (i.e. pH ≥ 13, pH 12, and pH 7.4) can be used differentially to detect distinct DNA lesions. Loss of a purine or pyrimidine base from the DNA sugar-phosphate backbone facilitates an alkali-catalyzed β-elimination of the 3′-phosphate (18), and at a pH level of 13 or greater, alkali-labile sites are converted to SSBs for AP site detection (18). A pH level of 12 is appropriate for SSB detection (Fig. 1F, G) because hydrogen bonds destabilize at this pH, and double-stranded DNA separates into individual single strands, rendering shorter single-stranded DNA (resulting from SSBs) more easily eluted from the nucleus (18). Neutralized conditions (pH 7.4) are used to detect DSBs because DNA remains double stranded under this condition, and regions containing DSBs migrate more readily in an electrophoretic field.
FIGURE 1.
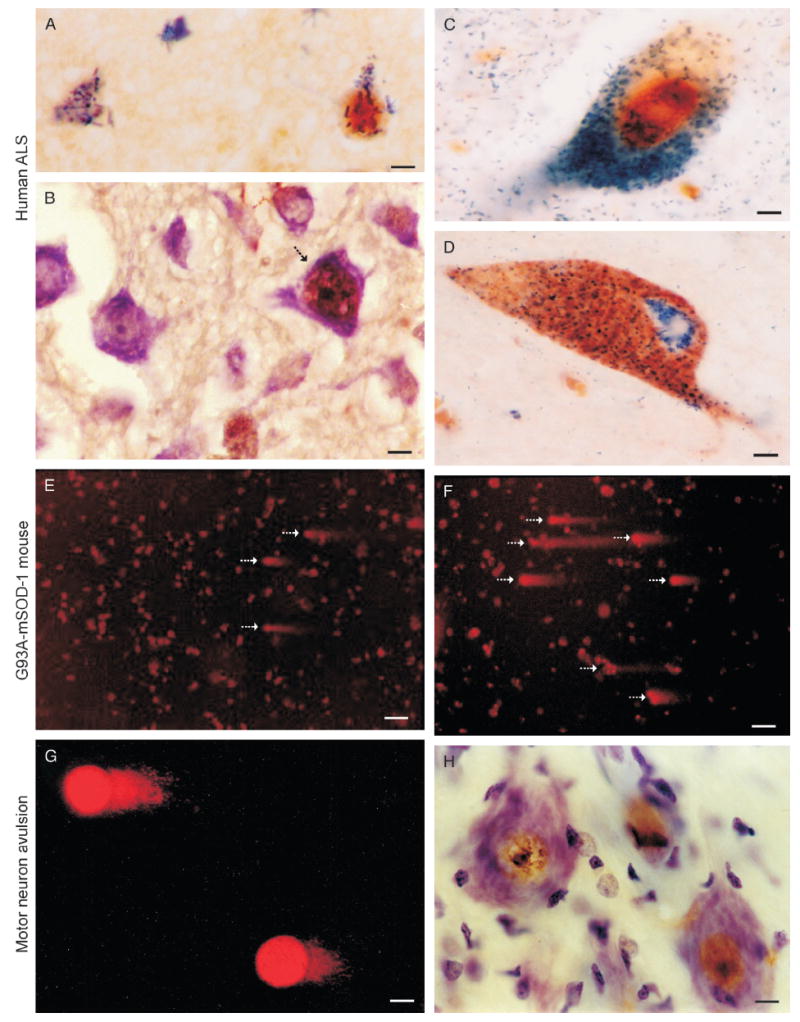
DNA damage and neuronal degeneration in human amyotrophic lateral sclerosis (ALS) and mouse models of motor neuron degeneration. (A) DNA damage in the form of 8-hydroxy-2-deoxyguanosine (OHdG) immunoreactivity (blue-green crystals) and p53 immunoreactivity (brown) colocalize in the nucleus of subsets of upper motor neurons (cell at right) in human ALS cerebral cortex. Other neurons (cell at left) have accumulated OHdG immunoreactivity within the cytoplasm, possibly corresponding to DNA damage within mitochondria, but are not positive for p53. Scale bar = 10 μm. (B) Activated p53, seen as phosphoserine392-p53 immunoreactivity (brown labeling), accumulates in subsets of cortical motor neurons (arrow) in human ALS. Other neurons in the field seen by the cresyl violet counterstain show no immunoreactivity. Scale bar = 10 μm. (C) In human ALS spinal motor neurons, nuclear OHdG-DNA damage (brown) and active caspase 3 (blue-green labeling) colocalize. Scale bar = 5 μm. (D) Mitochondrial accumulation identified by cytochrome c oxidase subunit 1 immunoreactivity (brown labeling) occurs in the perikaryon of human ALS spinal motor neurons with active caspase 3 (green in cytoplasm and nucleus). Many mitochondria are positive for active caspase 3. Scale bar = 5 μm. (E, F) Comet assay on isolated lumbar spinal cord motor neurons from G93A/mutant superoxide dismutase 1 mice at 6 (F) and 8 weeks (G) of age reveals the accumulation of DNA single-strand breaks (SSBs) in subsets of cells (arrows; see original publications for assay details; 19–21). Scale bars = (F) 60 and (G) 40 mm. (G, H) Spinal motor neurons with avulsed axons accumulate DNA-SSBs (detected by comet assay, note the short tails in G) very early in response to injury and coinciding with nuclear accumulation of p53 (brown nuclear labeling in H). Note that the p53 accumulation within the nucleus can be seen at chromatin strands. Scale bars = (G, H) 6 μm. mSOD-1, mutant superoxide dismutase 1.
We have profiled DNA strand-break lesions directly in mammalian motor neurons and cortical neurons in paradigms of neurodegeneration (14, 19-24; Fig. 1). We examined the hypothesis that DNA damage occurs in injured neurons at a time that might serve as a trigger for the degeneration in vitro and in vivo. In vitro, primary motor neurons were exposed to ROS and RNS and profiled for DNA-SSBs and DNA-DSBs using the comet assay (19-21). Healthy control neurons have very few comets, indicating that steady-state levels of DNA-SSBs and DNA-DSBs are low. H2O2, nitric oxide donors (NONOate and sodium nitroprusside), and ONOO− all induce prominent DNA damage in motor neurons (19, 21); ONOO− quickly induces DNA-SSBs and DNA-DSBs in motor neurons in vitro.
Several different in vivo models of neurodegeneration also show that DNA damage is a common antecedent of motor neuron death regardless of whether it is apoptotic or necrotic like (Fig. 1). For example, motor neurons destined to undergo apoptosis induced by axon avulsion accumulate DNA-SSBs at a time corresponding to p53 activation and nuclear import (19, 20; Fig. 1H). In transgenic mice expressing human mutant superoxide dismutase 1, the severe degeneration of motor neurons is preceded by accumulation of DNA-SSBs (Fig. 1F, G). Interestingly, however, nuclear import of p53 is blocked (23). In this mouse amyotrophic lateral sclerosis (ALS) model, DNA-DSBs accumulate late in motor neurons when degenerative structural changes are prominent (23). When cultured mouse cortical neurons have acutely inactivated DNA topoisomerase 1 (Topo-1), they rapidly accumulate DNA-SSBs and show simultaneously p53 activation prior to apoptosis (24). These studies demonstrate that DNA damage, particularly SSBs, can rapidly accumulate in terminally differentiated neurons that are undergoing oxidative stress and Topo-1 dysfunction prior to or along with the engagement of the molecular mechanisms of cell death. This work also demonstrates that the comet assay is a feasible method for profiling DNA lesions in single neurons. Therefore, these approaches may allow the study of neurodegeneration to move in new directions, particularly with regard to understanding any upstream causal roles of DNA damage in neuronal dysfunction/death.
A puzzling issue is that when wild-type neurons do accumulate DNA damage, particularly SSBs, the p53 response is robust and rapidly mediates cell death. In chronic progressive neurodegenerative disease, however, if populations of neurons selectively accumulate DNA damage as part of the mechanistically relevant pathobiology, why is the neurodegeneration apparently so slow? Neurons born with a DNA repair defect would be expected to display a developmental phenotype as seen with DNA repair protein-deficient apurinic/apyrimidinic endonuclease (APE) null mice (26), but mice with a deletion of DNA glycosylases are viable (27). The DNA repair defect might be acquired slowly over time (i.e. aging) in neurons, with the amount of DNA damage accumulating initially at subthreshold levels for engaging cell death mechanisms. Alternatively, the DNA damage might accumulate asynchronously in subsets of disease-vulnerable postmitotic neurons during aging, and they might then be eliminated individually slowly over time through p53-mediated mechanisms (28) (Fig. 1A, B).
DNA REPAIR: BASE-EXCISION REPAIR AND NER
Aerobic cells constantly sustain damage to DNA (1). This damage originates from environmental agents and endogenous processes. Mammalian cells have elaborate DNA repair mechanisms (3, 5, 29, 30). Repair of DNA during transcription is termed transcription-coupled repair, and DNA repair occurring independently of transcription is termed global genomic repair. DNA repair in human genes might be domain selective with faster repair rates at sequences near the transcription initiation site (31). DNA repair events can be divided into 3 classes: 1) damage reversal directly without involving hydrolysis of the phosphodiesterase bond (e.g. demethylation); 2) damage repair by excision; and 3) damage repair by recombination. This latter form of repair may not be significant for postmitotic neurons because recombination repair is essentially a postreplication process that corrects errors introduced by active DNA synthesis. However, we cannot yet rule out the possibility of unscheduled DNA synthesis in neurons in neurodegenerative diseases.
Some forms of DNA damage are directly reversed enzymatically. This repair applies to alkylated bases and involves O6-methylguanine-DNA methyltransferase, which removes alkyl groups from guanine and thymine (5, 9). This DNA repair process is a form of enzyme suicide because 1 O6-methylguanine-DNA methyltransferase molecule reverses 1 lesion and then becomes permanently inactivated and degraded.
DNA-base excision repair (BER) is believed to be the major pathway for repairing deaminated bases and bases with oxidative damage generated by ROS and can be used to repair alkylated bases (29, 30). Base excision repair has 4 main steps (i.e. base removal, AP site incision, synthesis, and ligation) and involves 4 major classes of DNA repair enzymes: DNA glycosylases, APE, DNA polymerases, and DNA ligases.
Base excision repair begins with base recognition that is then followed by removal of a damaged purine or pyrimidine by the activity of a DNA N-glycosylase (29, 30). This damaged base excision is achieved by either bifunctional or monofunctional DNA glycosylase/AP lyases that are distinct in structural features and reaction mechanisms. These enzymes possess activities to excise a damaged base by cleavage of the N-glycosidic bond, thus creating an AP site, but monofunctional enzymes do not have the ability to nick the DNA backbone on the 3′ side of the AP site through AP lyase activity (β-elimination or β,δ-elimination) to generate an SSB with 3′-α,β-unsaturated aldehyde and 5′-phosphate termini or 3′- and 5′-phosphate termini. Most DNA glycosylases have broad substrate specificities but can have preferences for either purines or pyrimidines. The major bifunctional glycosylase for purines is 8-oxoguanine DNA glycosylase 1, which removes OHdG and 8-oxoG. Most oxidized pyrimidines are removed by endonuclease III-like protein, uracil DNA glycosylase (UNG), and Nei-like DNA glycosylase (NEIL-1 and NEIL-2). In human cells, there are at least 2 isoforms of OGG1 (α and β) that arise from alternative splicing of products of the OGG1 gene (32). α-OGG1 is a 345-amino acid (~39 kd) protein that localizes to the nucleus and mitochondria, whereas β-OGG1 is a 424-amino acid protein (47 kd) that seems to localize exclusively to mitochondria (32). Because of the absence of an αO helix domain, β-OGG1 seems to lack glycosylase activity (33), although this seems controversial (34). Endonuclease III-like protein 1 is a 312-amino acid (~34 kd) protein that localizes to the nucleus. OGG1 and endonuclease III-like protein require duplex DNA for efficient repair. Targeted deletion of OGG1 or endonuclease III-like protein genes causes no overt phenotype in mice (27, 35), but OGG1 null mice have 20-and 40-fold increases in 8-oxoG levels in liver nuclear and mitochondrial DNA, respectively (36). Endonuclease III-like protein 1 excises 5-hydroxyuracil inefficiently, whereas oxidized uracil and cytosine derivatives are removed from human DNA by UNG (37). Uracil DNA glycosylase 1 (mitochondrial) and UNG2 (nuclear) are present in human cells arising from alternative splicing of products of the UNG gene. Uracil DNA glycosylase proteins are functional as monomers (~35 kd). Mice deficient in UNG display no obvious phenotype under physiologic conditions (38), which may be compensated partly by single-strand selective mono-functional UDG-1, but in ischemic challenge, they have more brain damage than control mice, suggesting possible functions in response to oxidative stress (17). When substrate lesions are in single-stranded DNA or replication bubbles, OGG1 and NTH-1 are ineffective for repair, so NEIL-type enzymes are used for excision, suggesting their preferential involvement in repairing DNA sequences during transcription or replication (39). Nei-like DNA glycosylase 1, NEIL2, and NEIL3 are products of distinct genes; NEIL-1 is 390 amino acids (~44 kd), NEIL-2 is 332 amino acids (~37 kd), and NEIL-3 is 605 amino acids (68 kd) (29). All have 2 to 3 isoforms, with NEIL-1 having apparent nuclear and mitochondrial localizations (40). Nei-like DNA glycosylase 1 and NEIL-2 interact stably with polynucleotide kinase 3′-phosphatase (PNKP), DNA polymerase β (Polβ), DNA ligase III, and x-ray repair cross-complementing protein 1 (XRCC-1) (41). Nei-like DNA glycosylase 1- and NEIL-2-mediated repair of oxidized bases is PNKP dependent but APE independent (41). Nei-like DNA glycosylase 1 null mice display a metabolic syndrome with mitochondrial DNA damage (42).
After the glycosylase reaction, the 3′ fragmented sugar terminus is removed from DNA through incision on the 5′ side of the sugar remnant by APE (also known as HAP1 and redox factor 1), resulting in the loss of a base and an SSB with 5′-phosphate and 3′-OH termini (30). Apurinic/apyrimidinic endonuclease also cleaves intact AP sites by making a single nick 5′ to the AP site to create 3′-OH and 5′-deoxyribose phosphate termini that are removed by the activity of 3′-OH and 5′-deoxyribose phosphatase proteins such as DNA Polβ. The 3-phosphate removal activity of APE is, however, approximately 70-fold lower than the AP endonuclease activity of APE (43), suggesting that other enzymes are important for the processing of damaged 3′ ends (44). Apurinic/apyrimidinic endonuclease is a 317-amino acid (~37 kd) multifunctional protein localized to the nucleus (45). In addition to its role in BER, APE is required for the redox activation of transcription factors (e.g. p53) spontaneously oxidized at cysteine residues in DNA binding domains to establish DNA binding activity (46). Deletion of APE in mice causes embryonic lethality (26). Cells from mice with APE haploinsufficiency respond poorly to oxidative stress (47). The repair of the SSB (originally an AP site) is accomplished by Polβ, removing the phosphate backbone of the AP site and allowing the addition of a normal and correct nucleotide, and then adenosine triphosphate-dependent DNA ligase (Ligase [Lig] I or III) subsequently seals the nick to complete the repair (30).
DNA helix-distorting lesions are repaired by NER, which removes a short single-stranded DNA segment (25–30 nucleotides) that includes the lesion (48). Nucleotide excision repair in mammalian cells involves 9 major proteins that are named from the human diseases associated with deficiencies in the proteins (XP and CS) (4). The process of NER is categorized into global genome-NER and transcription-coupled-NER (49). Global genome-NER acts on DNA damage in inactive and active genes throughout the genome. Transcription-coupled-NER specifically targets DNA lesions in genes that are actively being transcribed and uses stalled RNA polymerase and CS protein A and CS protein B proteins (instead of XP complementation Group A protein (through XPG)-XPC-human homolog of Rad23, protein B complex used in global genome-NER) in DNA damage recognition (50). Thus, the 2 pathways differ in the proteins that recognize the DNA lesions, but the downstream components are similar and can be divided as follows: i) unwinding of the double helix at the lesion site localized XPC-human homolog of Rad23, protein B by basal transcription factor TFIIH and the helicases XPB and XPD, allowing XP complementation Group A protein (through XPG) to bind so that other repair proteins can be loaded onto the damaged site; ii) single-strand incision at both sides of the lesion and excision of the lesion-containing single-stranded DNA fragment by the endonuclease XPF/ERCC1; iii) DNA repair synthesis by DNA polymerase δ and ε to replace the gap; and iv) ligation of the remaining single-stranded nick.
DNA-SSB SENSOR PROTEINS
Poly (adenosine diphosphate-ribose) polymerases (PARP-1 and PARP-2) are proteins that recognize DNA SSB (51). These enzymes transfer adenosine diphosphate-ribose moieties from nicotinamide adenine dinucleotide to histone proteins and several nuclear proteins involved in DNA metabolism. Agents that cause DNA-SSB formation lead to the activation and early recruitment of PARP-1 and PARP-2 to sites of the break. Poly (adenosine diphosphate-ribose) polymerases form complexes with XRCC-1, aprataxin (APTX), Polβ, and Lig III (51).
X-ray repair cross-complementing protein 1 is a 633-amino acid (~70 kd) protein that functions in homodi-meric form as a scaffold for recruiting BER proteins to the SSB (52). X-ray repair cross-complementing protein 1 interacts physically with several BER proteins (OGG1, NEILs, APE, PNKP, DNA Polβ, and Lig III) to form a multimeric complex and can coordinate and activate some BER proteins (i.e. OGG1 and APE) (52, 53).
DNA REPAIR IN MITOCHONDRIA
Mitochondrial DNA sustains higher steady-state damage compared with nuclear DNA (34). This greater lesion accumulation is likely to be caused by greater local levels of ROS and lack of chromatin protection afforded by histones. Active BER proteins, all of which are encoded by nuclear DNA, are present in mitochondria (54), although at lower levels than in nuclei. Nuclei and mitochondria use variant proteins for BER (55). Splice variants or truncation products of nuclear OGG1, NTH1, APE, and Lig IIIβ are present in mitochondria, whereas DNA polymerase γ is thought to be unique for mitochondrial BER. Interestingly, import mechanisms for OGG1 into mitochondria may undergo age-related perturbations (34).
DNA REPAIR GENE ABNORMALITIES IN SOME HUMAN NEURODEGENERATIVE DISEASES
A recent appreciation has appeared that envisions neuron vulnerability to degeneration dictated by a propensity for progressive accumulation of unrepaired DNA lesions (56), although the concept regarding DNA damage, DNA repair, and neurologic disease had been proposed earlier (2, 57). As previously described, abnormalities in DNA repair or DNA damage response networks have been identified in human childhood- and adult-onset neurologic diseases, including XP, CS, TTD, and AT. Xeroderma pigmentosum, CS, and TTD are caused by defective NER, whereas AT is caused by a defective DNA-DSB response. These diseases have been reviewed in detail elsewhere (4, 5, 48, 50). More recently, the disease-causing genes for AOA-1, AOA-2, and SCAN-1 have been identified (6-8). The defective proteins involved in AOA-1 and SCAN-1 are enzymes that function in DNA end-processing in the repair of DNA-SSBs. The defective protein involved in AOA-2 and juvenile ALS is a DNA/RNA helicase that participates in various aspects of DNA repair, DNA transcription, and processing of noncoding RNAs (58).
Ataxia with oculomotor apraxia Type 1 is an autosomal recessive neurodegenerative disorder characterized by early-age-onset and progressive ataxia, ocular motor apraxia, absence of tendon reflexes, loss of distal position and vibration sense, and pyramidal weakness of the legs (6). The neuropathologic findings in AOA-1 include loss of Purkinje cells, degeneration of posterior columns and spinocerebellar tracts in spinal cord, and degeneration of peripheral nerve axons. Ataxia with oculomotor apraxia Type 1 is caused by mutations in the gene encoding APTX, a member of the histidine triad family of nucleotide hydrolases and transferases (6). Short and long forms of the protein are generated by alternative splicing. The major form of APTX is the long form consisting of 342 amino acids (~39 kd). Aprataxin is a nuclear protein containing a histidine triad motif, a DNA-binding C2H2 zinc-finger motif, and a PNKP motif. The AOA-1-linked mutations cause APTX loss of function and are confined largely to the histidine triad domain. Aprataxin interacts with several DNA repair proteins, including XRCC-1, Polβ, DNA Lig III, PARP-1, and p53 (59). It functions in the end-processing of DNA-SSBs (44, 60, 61). Human APTX specifically removes 3′-phosphate, 5′-phosphate, and 3′-phosphoglycolate ends, but not 3′-α,β-unsaturated aldehyde ends, of DNA-SSBs (44, 61). It has been postulated that mutant APTX-associated neurologic disorders are caused by gradual accumulation of nonrepaired DNA-SSBs resulting from defective DNA end-processing and abortive DNA ligation (61).
Ataxia with oculomotor apraxia Type 2 is caused by autosomal recessive mutations in the gene encoding senataxin (SETX) (7). Dominant missense mutations in the Setx gene have been also linked to juvenile ALS (ALS4) (58). Senataxin is a large protein (2,677 amino acids; ~303 kd) containing a superfamily I DNA/RNA helicase domain in the C-terminus (7, 58). The SETX helicase domain shares homology to other human proteins known to have helicase activity, notably immunoglobulin mu-binding protein 2. Interestingly, infantile spinal muscular atrophy with respiratory distress Type 1, manifesting as weakness and difficulty with respiration at age 1 to 6 months, results from recessive mutations in immunoglobulin mu-binding protein 2. Currently, information about the functions of SETX is very limited. The presence of the helicase domain is a strong indicator that 1 function of SETX is nucleic acid processing. Senataxin is expressed in a variety of tissues (7, 58). In mouse brain, SETX is enriched in cerebellum and hippocampus and is present primarily in neurons where it has a cytoplasmic and nuclear localization (62). It is presently unclear how mutations in SETX cause neurodegeneration in AOA-2 and juvenile ALS. In nonneural cells with reduced SETX, repair of H2O2-induced DNA-DSBs is defective, but repair of DNA-SSBs and damage caused by irradiation seems normal (63). These data suggest that SETX functions in DNA-oxidative damage response.
Spinocerebellar ataxia with axonal neuropathy 1 is caused by autosomal recessive mutations in the gene encoding tyrosyl-DNA phosphodiesterase 1 (TDP1) (8). Tyrosyl-DNA phosphodiesterase 1 is a 608–amino acid (~68 kd) nuclear protein that functions in repairing covalent Topo-1-DNA complexes (64). Topological changes in the DNA helix are required in many DNA functions, including replication, transcription, and repair. In this process, Topo-1 transiently breaks a DNA strand and forms a covalent linkage between the active-site tyrosine and the 3′-phosphate of the broken DNA strand, thereby permitting rotation of the DNA double helix on the intact phosphodiester bonds opposite the Topo-1-mediated cleavage (64). Once the DNA has been relaxed, the strand is aligated subsequently by nucleophilic attack of the 5′-hydroxyl on this phosphodiester bond to regenerate intact duplex DNA. Sometimes, the nucleophilic attack of the 5′-hydroxyl does not occur, and the Topo-1 forms a stalled covalent complex with DNA. Efficient repair of these stalled complexes requires TDP1, which catalyzes the hydrolysis of the phosphodiester bond between the tyrosine residue and the DNA 3′-phosphate. Tyrosyl-DNA phosphodiesterase 1 can also remove glycolate from ssDNA containing a 3′-phosphoglycolate, suggesting a role in repair of free radical-mediated DNA damage. Tyrosyl-DNA phosphodiesterase 1 interacts with XRCC-1, PNKP, and DNA Lig III (65, 66). Loss-of-function mutations in TDP1 resulting in defective DNA SSBs repair are believed to cause SCAN-1 (65). Nonneuronal SCAN-1 cells are defective in repair of transcription-dependent Topo-1 cleavage complexes (67). The results of TDP1 gene ablation experiments are discrepant. One study reported an age-dependent and progressive atrophy of cerebellum in TDP1−/− mice (68), whereas another study did not detect an overt phenotype (69). Both studies, however, found a deficient ability of TDP1 null cells to repair DNA-SSBs in response to Topo-1 inhibition or oxidative stress.
Abnormalities in DNA repair have also been implicated in ALS, the third most common adult-onset neurodegenerative disease. This is particularly interesting in light of the identification of the gene causing ALS4. Missense mutations in the APE gene have been identified in sporadic and familial ALS (70), but other studies have not identified prominent contributions of APE mutations to ALS (71). We evaluated the expression and function of the Class II APE in ALS CNS (72). This protein is also of interest in ALS because APE functions as a redox factor (redox factor 1), which facilitates the DNA binding of transcription factors through redox modulation (26). Apurinic/apyrimidinic endonuclease protein levels and repair activity are increased in the motor cortex in ALS patients, supporting the hypothesis that DNA damage is an upstream mechanism for neurodegeneration (72).
A Ser326Cys polymorphism in OGG1 is associated with sporadic ALS but not with AD (73). This polymorphism is significant because this form of hOGG1 has reduced capacity to repair oxidatively damage DNA (74). Polymorphisms in NER genes (ERCC2 and ERCC4) are not associated with sporadic AD (75). In a recently published seminal study, Kovtun et al showed that in the process of removing base lesions, OGG1 drives age-dependent trinucleotide (CAG) expansion associated with Huntington disease in somatic cells (76). Thus, independent of mutations in DNA repair genes and germ cell transmission, error-prone DNA repair processes might result in somatic cell mosaics and cause major human neurodegenerative diseases.
ELEVATED DNA DAMAGE IS FOUND IN SEVERAL NEURODEGENERATIVE DISORDERS
Several examples highlight the possibility of exaggerated DNA damage in human neurologic disorders. By biochemical assay on isolated nuclei, the level of DNA strand breakage is 2-fold higher in the AD cerebral cortex compared with control cortex (77). Increased DNA lesions have also been detected in the brains of PD patients compared with age-matched controls (78). DNA damage can also be involved in the pathogenesis of ALS (57, 72). DNA damage in ALS may be caused by oxidative stress from mitochondrial or superoxide dismutase 1 dysfunction (23), and OHdG adducts are elevated in postmortem CNS tissue extracts from individuals with ALS (79). We have found OHdG-DNA and ssDNA-lesions specifically in vulnerable neurons in individuals with ALS (Fig. 1A) (80). Many of these motor neurons with DNA lesions have activated p53 and activated caspase 3 (Fig. 1A–C). We have additional evidence that, surprisingly, nuclear DNA accumulates more AP sites than mitochondrial DNA in motor neuron regions (K. McClendon and L.J. Martin, unpublished observations), suggesting that nuclear DNA repair is defective, whereas mitochondrial DNA repair sustains efficiency in the disease. This evidence is consistent with chromosomal DNA damage being a strong signal for apoptosis. The finding that p53 is overactive in ALS (28) further implicates DNA damage as an upstream pathogenic event in ALS consistent with APE data in ALS (72).
In PD, several different DNA repair enzymes are upregulated in the substantial nigra (81). In Down syndrome cerebral cortex, several DNA repair proteins are increased at the transcriptional level (82), but the mechanisms of DNA damage accumulation in neurons are not understood. Possible mechanisms for elevated levels of DNA damage include more spontaneously generated DNA damage by ROS and RNS, defective DNA repair, or even increased DNA repair.
DNA REPAIR IN THE NERVOUS SYSTEM
DNA repair enzymes have been studied extensively in the context of cancer pathobiology in nonneural tissues, but less is known regarding these enzymes in the nervous system in general and specifically in neurons. Different tissue types are endowed with different repair capacities for particular DNA lesions (83) and with different detoxification capacities for neutralizing effects of DNA-damaging agents. Different cell types also vary in the propensity for generating DNA strand breaks. Few studies have examined the expression of DNA repair proteins in different brain cell types, and the complete complement of DNA repair enzymes in different types of neurons is unknown. DNA glycosylase subtypes in nuclei and mitochondria have different levels of activity in different mouse brain regions, some of which display reduced levels in the aged brain (84). The activity of 8-oxoguanine DNA glycosylase 1 and the presence of 8-oxoguanine DNA glycosylase 1 mRNA and protein in quail and rat brain neurons and glia seem to be different (85). Apurinic/apyrimidinic endonuclease is expressed differentially throughout the human brain, and high levels are found in hippocampal and motor neurons (86). DNA polymerase A seems to be a main DNA polymerase in the brain, comprising approximately 99% of the DNA polymerase activity (87), and its activity in rat brain decreases with age (88). DNA polymerase β null mice have a neonatal lethal phenotype with prominent neurodegeneration (89). Recently, Polβ has been implicated in AD pathogenesis through a mechanism involving β-amyloid stimulated loading into DNA replication forks (90).
In CNS injury paradigms, DNA repair enzymes seem to be regulated differentially, and their loss of function can enhance neurodegeneration. DNA damage, faulty DNA repair, and mutagenesis can occur in mouse cerebral cortex after transient forebrain ischemia (91). Uracil DNA glycosylase null mice have greater infarct size than wild-type mice after focal cerebral ischemia, and cultured cerebellar neurons from UNG−/− mice have increased sensitivity to oxygen/glucose deprivation (17). After transient focal and global cerebral ischemia, APE protein levels are reduced (92). Cerebellar granule neuron cultures made from APE+/− mice show increased sensitivity to ROS (47), as do wild-type hippocampal and dorsal root ganglion neurons with knocked-down APE (93).
PROTEIN KINASE PATHWAYS TRANSDUCE DNA DAMAGE SIGNALS
The DNA damage response is a hierarchical process. Nonneuronal cells that have sustained DNA damage from ROS/RNS and other genotoxic agents can undergo cell cycle arrest to implement repair mechanisms or undergo apoptosis by engaging molecular cascades involving expression or activation of p53, Bax, and caspases (94). The signal transduction mechanisms in neurons that link DNA damage to apoptosis are not well characterized, however, and the sensors of DNA damage in neurons are largely unknown (24). DNA damage sensor proteins have been identified in nonneuronal cells (94). The signal for DNA damage is transduced by DNA-dependent protein kinase (DNA-PK) and ATM, which are 2 members of the phosphatidylinositol 3-kinase family (95, 96). DNA-DSBs are recognized and bound by the heterodimeric regulatory subunits of DNA-PK (Ku70/80), thereby recruiting the catalytic DNA-PK that phosphorylates p53 (96). In an in vitro system, DNA-PK-mediated phosphorylation of p53 at serine15 alleviates the inhibition by murine double minute 2 (Mdm-2) and correlates with the stabilization of p53 and its activation as a transcription factor (97). Other in vitro studies have shown that DNA-DSBs result in phosphorylation of p53 and stabilization through a process mediated by ATM (95). Ataxia telangiectasia mutated and ATM/Rad3-related kinase (ATR) are other kinases that become activated in response to DNA damage (94). Identified substrates for ATM and ATR are p53, Mdm-2, and Rad17. Ataxia telangiectasia mutated is required for the phosphorylation of p53 at serine15 and serine20. We have identified ATM-dependent phosphorylation of p53 in cortical neurons very early, that is, within 1 hour, in the process of DNA damage-induced apoptosis in vitro (24). This interaction is consistent with data in the developing nervous system showing ATM-dependent apoptosis requires p53 phosphorylation and caspase 3 activation (98).
p53 ACTIVATION IS DOWNSTREAM OF THE DNA DAMAGE RESPONSE IN NEURONS
p53 is a key molecule that functions in DNA damage and growth control in cycling cells. p53 is activated by genotoxic stress and can trigger the onset of DNA repair or apoptosis. Indeed, the ability of p53 to induce apoptosis is unequivocally accepted. The intricacies of p53 in DNA repair are, however, less well known. Both NER and BER pathways can involve p53 through its ability to interact with components of the repair machinery, for example, APE and PolA (29, 30). p53 is a short-lived protein with a half-life of approximately 5 to 20 minutes in most types of cells studied. It is regulated by posttranslational modifications (phosphorylation and acetylation) and is modulated by intracellular redox state (60). Protein levels of p53 can rapidly increase severalfold after DNA damage, mainly by posttranslational mechanisms. The elevation in p53 protein levels occurs through stabilization stimulated by phosphorylation. Phosphorylation of serine15 is a key response to DNA damage. Phosphorylation also regulates the interactions of p53 monomers. p53 can form homotetramers and must be in tetrameric form for sequence-specific DNA binding and transcriptional activation (60). Tetramerization is stimulated by phosphorylation of serine392. This phosphorylation increases 10-fold the association constant for tetramer formation. Recent work has revealed an interesting connection between DNA/RNA helicases and p53-mediated apoptosis. p68, a prototypic member of the DEAD box family of helicase proteins, functions as a coactivator of p53 (99). Suppression of p68 can block apoptosis in response to DNA damage (99).
p53 is degraded rapidly in an ubiquitination-dependent proteasomal pathway (97) that is known to be compromised in several neurodegenerative disorders. Murine double minute 2 has a crucial role in this degradation pathway (97). Murine double minute 2 seems to function in a feedback loop to limit the duration or magnitude of the p53 response to DNA damage. Expression of the Mdm-2 gene is controlled by p53 (97). Murine double minute 2 binds to the N-terminal transcriptional activation domain of p53 and regulates the transactivator activity and the stability of p53 by direct association. Murine double minute 2 seems to possess ubiquitin ligase activity for p53 through the ubiquitin-conjugating enzyme E2. Stabilization of p53 is achieved through phosphorylation of serine15, resulting in inhibition of formation of Mdm-2–p53 complexes. Ataxia telangiectasia mutated also regulates the stabilization of p53 through Mdm-2 phosphorylation, thus preventing Mdm-2-dependent p53 degradation.
DNA DAMAGE AND DNA REPAIR IN NEURONS: FUTURE DIRECTIONS
Future work needs to be done on the identification of how DNA damage can lead to the death of neurons in culture models with confirmed DNA lesions. Such studies will then lay the foundation for future work on the mechanisms in human neurodegenerative diseases and their animal models. Key issues that need to be addressed are: 1) the propensities of postmitotic neurons to accumulate specific forms of DNA damage; 2) the chromosomal and gene distributions of DNA damage; 3) the efficiency at which neurons repair these DNA lesion as a function of age; 4) the threshold levels of different types of DNA damage required to cause neuronal death; 5) the molecular sensors that engage repair pathways and death pathways and the molecular switches that determine cell fate in response to DNA damage; and 6) the possibility that mechanisms of genotoxicity depend on degree of neuron differentiation. The reinvigorated concept that envisions neuron vulnerability to degeneration as dictated by proneness to progressive accumulation of unrepaired DNA lesions is very exciting and can be very relevant to the formulation of novel molecular mechanism-based therapies for neuroprotection in progressive age-related neurodegenerative disorders.
Acknowledgments
This article is dedicated to the memory of my father Joseph G. Martin (June 10, 1926–December 21, 2007). The author thanks several individuals in his laboratory, including Dr. Zhiping Liu for developing the comet assay, Dr. Karen McClendon for developing the DNA-AP site assay, Dr. Margaret Wong for developing the recombinant adenoviral DNA repair gene technology, and Yan Pan and Ann Price for technical assistance.
This work was supported by the US Public Health Service Grant Nos. NS034100 and NS052098 from the National Institutes of Health-National Institute of Neurological Disorders and Stroke, the US Public Health Service Grant No. AG016282 from the National Institutes of Health-National Institute on Aging
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Cleaver JE. Defective DNA repair replication in xeroderma pigmentosum. Nature. 1968;218:652–56. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 3.Ronen A, Glickman BE. Human DNA repair genes. Environ Mol Mutagen. 2001;37:241–83. doi: 10.1002/em.1033. [DOI] [PubMed] [Google Scholar]
- 4.Kraemer KH, Patronas NJ, Schiffmann R, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–96. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks PJ. DNA repair in neural cells: Basic science and clinical implications. Mutat Res. 2002;509:93–108. doi: 10.1016/s0027-5107(02)00222-1. [DOI] [PubMed] [Google Scholar]
- 6.Date H, Onodera O, Tanaka H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–88. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 7.Moreira MC, Klur S, Watanabe M, et al. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet. 2004;36:225–27. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- 8.Takashima H, Boerkoel CF, John J, et al. Mutation of TDP1, encoding a topoisomerase I–dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–72. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 9.Rao KS. Genomic damage and its repair in young and aging brain. Mol Neurobiol. 1993;7:23–48. doi: 10.1007/BF02780607. [DOI] [PubMed] [Google Scholar]
- 10.Atamna H, Cheung I, Ames BH. A method for detecting abasic sites in living cells: Age-dependent changes in base excision repair. Proc Natl Acad Sci U S A. 2000;97:686–91. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kingma PS, Osheroff N. Spontaneous DNA damage stimulates topoisomerase II–mediated DNA cleavage. J Biol Chem. 1997;272:7488–93. doi: 10.1074/jbc.272.11.7488. [DOI] [PubMed] [Google Scholar]
- 12.Toyokuni S, Iwasa Y, Kondo S, et al. Intranuclear distribution of 8-hydroxy-2′-deoxyguanosine: An immunocytochemical study. J Histochem Cytochem. 1999;47:833–35. doi: 10.1177/002215549904700613. [DOI] [PubMed] [Google Scholar]
- 13.Lu T, Pan Y, Kao S-Y, et al. Gene regulation and DNA damage in the aging human brain. Nature. 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 14.Martin LJ, Price AC, McClendon K, et al. Early events in target deprivation/axotomy induced neuronal apoptosis in vivo: Oxidative stress, DNA damage, p53 phosphorylation, and subcellular redistribution of death proteins. J Neurochem. 2003;85:234–47. doi: 10.1046/j.1471-4159.2003.01659.x. [DOI] [PubMed] [Google Scholar]
- 15.Martin LJ, Chen K, Liu Z. Adult motor neuron apoptosis is mediated by nitric oxide and Fas death receptor linked to DNA damage and p53 activation. J Neurosci. 2005;25:6449–59. doi: 10.1523/JNEUROSCI.0911-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin LJ, Brambrink AM, Price AC, et al. Neuronal death in newborn striatum after hypoxia-ischemia is necrosis and evolves with oxidative stress. Neurobiol Dis. 2000;7:169–91. doi: 10.1006/nbdi.2000.0282. [DOI] [PubMed] [Google Scholar]
- 17.Endres M, Biniszkiewicz D, Sobol RW, et al. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J Clin Invest. 2004;113:1711–21. doi: 10.1172/JCI20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tice RR, Agurell E, Anderson D, et al. Single cell gel/comet assay: Guidelines for in vitro and an in vivo genetic toxicological testing. Environ Mol Mutagen. 2000;35:206–21. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Martin LJ. Motor neurons rapidly accumulate DNA single-strand breaks after in vitro exposure to nitric oxide and peroxynitrite and in vivo axotomy. J Comp Neurol. 2001;432:35–60. doi: 10.1002/cne.1087. [DOI] [PubMed] [Google Scholar]
- 20.Martin LJ, Liu Z. Injury-induced spinal motor neuron apoptosis is preceded by DNA single-strand breaks and is p53- and Bax-dependent. J Neurobiol. 2002;50:181–97. doi: 10.1002/neu.10026. [DOI] [PubMed] [Google Scholar]
- 21.Martin LJ, Liu Z. DNA damage profiling in motor neurons: A single-cell analysis by comet assay. Neurochem Res. 2002;27:1093–1104. doi: 10.1023/a:1020961006216. [DOI] [PubMed] [Google Scholar]
- 22.Martin LJ, Pan Y, Price AC, et al. Parkinson’s disease α-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J Neurosci. 2006;26:41–50. doi: 10.1523/JNEUROSCI.4308-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin LJ, Liu Z, Chen K, et al. Motor neuron degeneration in amyotrophic lateral sclerosis mutant superoxide dismutase–1 transgenic mice: Mechanisms of mitochondriopathy and cell death. J Comp Neurol. 2007;500:20–46. doi: 10.1002/cne.21160. [DOI] [PubMed] [Google Scholar]
- 24.Martin LJ, Liu Z, Pipino J, et al. Molecular regulation of DNA damage–induced apoptosis of neurons in cerebral cortex. Cerebral Cortex. doi: 10.1093/cercor/bhn167. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duker NJ, Sperling J, Soprano KJ, et al. β-Amyloid protein induces the formation of purine dimers in cellular DNA. J Cell Biochem. 2001;81:393–400. doi: 10.1002/1097-4644(20010601)81:3<393::aid-jcb1053>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci U S A. 1996;93:8919–23. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klugland A, Rosewell I, Hollenbach S, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin LJ. p53 is abnormally elevated and active in the CNS of patients with amyotrophic lateral sclerosis. Neurobiol Dis. 2000;7:613–22. doi: 10.1006/nbdi.2000.0314. [DOI] [PubMed] [Google Scholar]
- 29.Hazra TK, Das A, Das S, et al. Oxidative DNA damage repair in mammalian cells: A new perspective. DNA Repair. 2007;6:470–80. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hedge ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu Y, Tornaletti S, Pfeifer GP. DNA repair domains within a human gene: Selective repair of sequences near the transcription initiation site. EMBO J. 1996;15:675–83. [PMC free article] [PubMed] [Google Scholar]
- 32.Boiteux S, Radicella JP. The human OGG1 gene: Structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- 33.Hashiguchi K, Stuart JA, de Souza-Pinto NC, Bohr VA. The C-terminal αO helix of human Ogg1 is essential for 8-oxoguanine DNA glycosylase activity: The mitochondrial β-Ogg1 lacks this domain and does not have glycosylase activity. Nucleic Acids Res. 2004;32:5596–5608. doi: 10.1093/nar/gkh863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szczesny B, Hazra TK, Papaconstantinou J, et al. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc Natl Acad Sci U S A. 2003;100:10670–75. doi: 10.1073/pnas.1932854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ocampo MTA, Chuang W, Marenstein DR, et al. Targeted deletion of mNth1 reveals a novel repair enzyme activity. Mol Cell Biol. 2002;22:6111–21. doi: 10.1128/MCB.22.17.6111-6121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Souza-Pinto NC, Eide L, Hogue BA, et al. Repair of 8-oxodeoxyguanosine lesions in mitochondrial DNA depends on oxoguanine DNA glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial DNA of OGG1-defective mice. Cancer Res. 2001;61:5378–81. [PubMed] [Google Scholar]
- 37.Akbari M, Otterlei M, Pena-Diaz J, Krokan HE. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience. 2007;145:1201–12. doi: 10.1016/j.neuroscience.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Nilsen H, Rosewell I, Robins P, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–65. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 39.Parsons JL, Kavli B, Slupphaug G, Dianov GL. NEIL1 is the major DNA glycosylase that processes 5-hydroxyuracil in the proximity of a DNA single-strand break. Biochemistry. 2007;46:4158–63. doi: 10.1021/bi0622569. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, de Souza-Pinto NC, Haraguchi K, et al. Repair of foramidopyridines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–51. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 41.Das A, Wiederhold L, Leppard JB, et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair. 2006;5:1439–48. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vartanian V, Lowell B, Minko IG, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–69. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winters TA, Henner WD, Russel PS, et al. Removal of 3′-phosphoglycolate from DNA strand-break damage in an oligonucleotide substrate by recombinant human apurinic/apyrimidinic endonuclease I. Nucleic Acids Res. 1994;22:18866–73. doi: 10.1093/nar/22.10.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi T, Tada M, Igarashi S, et al. Aprataxin, causative gene product for EAOH/AOA1, repairs DNA single-strand breaks with damaged 3′-phosphate and 3′-phosphoglycolate ends. Nucleic Acids Res. 2007;35:3797–3809. doi: 10.1093/nar/gkm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic enodnuclease: Definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A. 1991;88:11450–54. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jayaraman L, Murthy KGK, Zhu C, et al. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–70. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 47.Meira LB, Devaraj S, Kisby GE, et al. Heterozygosity for the mouse Apex gene result in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–57. [PubMed] [Google Scholar]
- 48.Nouspikel T. DNA repair in differentiated cells: Some new answers to old questions. Neuroscience. 2007;145:1213–21. doi: 10.1016/j.neuroscience.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 49.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription--blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–49. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 50.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–56. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 51.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev. 2006;7:517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 52.Almeida KH, Sobol RW. A unified view of base excision repair: Lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marsin S, Vidal AE, Sossou M, et al. Role of XRCC1 in coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem. 2003;278:44068–74. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 54.Driggers WJ, LeDoux SP, Wilson GL. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem. 1993;268:22042–45. [PubMed] [Google Scholar]
- 55.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: Similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 57.Bradley WG, Krasin F. A new hypothesis of the etiology of amyotrophic lateral sclerosis. The DNA hypothesis. Arch Neurol. 1982;39:677–80. doi: 10.1001/archneur.1982.00510230003001. [DOI] [PubMed] [Google Scholar]
- 58.Chen YZ, Bennett CL, Huynh HM, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–35. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gueven N, Becherel OJ, Kijas AW, et al. Aprataxin, a novel protein that protects against genotoxic stress. Hum Mol Genet. 2004;13:1081–93. doi: 10.1093/hmg/ddh122. [DOI] [PubMed] [Google Scholar]
- 60.Sano Y, Date H, Igarashi S, et al. Aprataxin, the causative protein for EAOH is a nuclear protein with a potential role as a DNA repair protein. Ann Neurol. 2004;55:241–49. doi: 10.1002/ana.10808. [DOI] [PubMed] [Google Scholar]
- 61.Ahel I, Rass U, El-Khamisy SF, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–16. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 62.Chen YZ, Hashemi SH, Anderson SK, et al. Senataxin, the yeast Sen1p orthologue: Characterization of a unique protein in which recessive mutations cause ataxia and dominant mutations cause motor neuron disease. Neurobiol Dis. 2006;23:97–108. doi: 10.1016/j.nbd.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 63.Suraweera A, Becherel OJ, Chen P, et al. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J Cell Biol. 2007;177:969–79. doi: 10.1083/jcb.200701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–40. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 65.El-Khamisy SF, Saifi GM, Weinfeld M, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy–1. Nature. 2005;434:108–13. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 66.Plo I, Liao Z-Y, Barcelo JM, et al. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair. 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 67.Miao Z-H, Agama K, Sordet O, et al. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair. 2006;5:1489–94. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 68.Katyal S, El-Khamisy SF, Russel HR, et al. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–31. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirano R, Interthal H, Huang C, et al. Spinocerebellar ataxia with axonal neuropathy: Consequence of a TDP1 recessive neomorphic mutation? EMBO J. 2007;26:4732–43. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olkowski ZL. Mutant AP endonuclease in patients with amyotrophic lateral sclerosis. Neuroreport. 1998;9:239–42. doi: 10.1097/00001756-199801260-00012. [DOI] [PubMed] [Google Scholar]
- 71.Hayward C, Colville S, Swingler RJ, Brock DJH. Molecular genetic analysis of the APEX nuclease gene in amyotrophic lateral sclerosis. Neurology. 1999;52:1899–1901. doi: 10.1212/wnl.52.9.1899. [DOI] [PubMed] [Google Scholar]
- 72.Shaikh AY, Martin LJ. DNA base-excision repair enzyme apurinic/apyrimidinic endonuclease/redox factor–1 is increased and competent in brain and spinal cord of individuals with amyotrophic lateral sclerosis. NeuroMol Med. 2002;2:47–60. doi: 10.1007/s12017-002-0038-7. [DOI] [PubMed] [Google Scholar]
- 73.Coppede F, Mancuso M, Gerfo AL, et al. Association of the hOGG1 Ser326Cys polymorphism with sporadic amyotrophic lateral sclerosis. Neurosci Lett. 2007;420:163–68. doi: 10.1016/j.neulet.2007.04.067. [DOI] [PubMed] [Google Scholar]
- 74.Vodicka P, Stetina R, Polakova V, et al. Association of DNA repair polymorphisms with DNA repair functional outcomes in healthy human subjects. Carcinogenesis. 2007;28:657–64. doi: 10.1093/carcin/bgl187. [DOI] [PubMed] [Google Scholar]
- 75.Dogru-Abbasoglu S, Inceoglu M, Parildar-Karpuzoglu H, et al. Polymorphisms in the DNA repair genes XPD (ERCC2) and XPF (ERCC4) are not associated with sporadic late-onset Alzheimer’s disease. Neurosci Lett. 2006;404:258–61. doi: 10.1016/j.neulet.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 76.Kovtun IV, Liu Y, Bjoras M, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mullaart E, Boerrigter ME, Ravid R, et al. Increased levels of DNA breaks in cerebral cortex of Alzheimer’s disease patients. Neurobiol Aging. 1990;11:169–73. doi: 10.1016/0197-4580(90)90542-8. [DOI] [PubMed] [Google Scholar]
- 78.Zhang J, Perry G, Smith MA, et al. Parkinson’s disease is associated with oxidative damage to cytoplasmic DNA and RNA in substantia nigra neurons. Am J Pathol. 1999;154:1423–29. doi: 10.1016/S0002-9440(10)65396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fitzmaurice PS, Shaw IC, Kleiner HE, et al. Evidence for DNA damage in amyotrophic lateral sclerosis. Muscle Nerve. 1996;19:797–98. [PubMed] [Google Scholar]
- 80.Martin LJ. Neuronal cell death in nervous system development, disease, and injury. Int J Mol Med. 2001;7:455–78. [PubMed] [Google Scholar]
- 81.Fukae J, Mizuno Y, Hattori N. Mitochondrial dysfunction in Parkinson’s disease. Mitochondrion. 2007;7:58–62. doi: 10.1016/j.mito.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Fang-Kircher SG, Labudova O, Kitzmueller E, et al. Increased steady state mRNA levels of DNA repair genes XRCC1, ERCC2 and ERCC3 in brain of patients with Down syndrome. Life Sci. 1999;64:1689–99. doi: 10.1016/s0024-3205(99)00107-1. [DOI] [PubMed] [Google Scholar]
- 83.Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: Tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 84.Imam SZ, Karahalil B, Hogue BA, et al. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol Aging. 2006;27:1129–36. doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 85.Verjat T, Dhenaut A, Radicella JP, Araneda S. Detection of 8-oxoG DNA glycosylase activity and OGG1 transcripts in the rat CNS. Mutat Res. 2000;460:127–38. doi: 10.1016/s0921-8777(00)00022-7. [DOI] [PubMed] [Google Scholar]
- 86.Duguid JR, Eble JN, Wilson TM, Kelley MR. Differential cellular and subcellular expression of the human multifunctional apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme. Cancer Res. 1995;55:6097–6102. [PubMed] [Google Scholar]
- 87.Waser J, Hubscher U, Kuenzle CC, Spadari S. DNA polymerase beta from brain neurons is a repair enzyme. Eur J Biochem. 1979;97:361–68. doi: 10.1111/j.1432-1033.1979.tb13122.x. [DOI] [PubMed] [Google Scholar]
- 88.Rao KS, Annapurna VV, Raji NS, Harikrishna T. Loss of base excision repair in aging rat neurons and its restoration by DNA polymerase beta. Brain Res Mol Brain Res. 2000:251–59. doi: 10.1016/s0169-328x(00)00266-7. [DOI] [PubMed] [Google Scholar]
- 89.Sugo N, Aratani Y, Nagashima Y, et al. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1379–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Copani A, Hoozemans JJM, Caraci F, et al. DNA polymerase-β is expressed early in neurons of Alzheimer’s disease brain and is loaded into DNA replication forks in neurons challenged with β-amyloid. J Neurosci. 2006;26:10949–57. doi: 10.1523/JNEUROSCI.2793-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu PK, Hsu CY, Dizdaroglu M, et al. Damage, repair, and mutagenesis in nuclear genes after mouse forebrain ischemia-reperfusion. J Neurosci. 1996;16:6795–6806. doi: 10.1523/JNEUROSCI.16-21-06795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gillardon F, Bottiger B, Hossmann KA. Expression of nuclear redox factor ref-1 in the rat hippocampus following global ischemia induced by cardiac arrest. Brain Res Mol Brain Res. 1997;52:194–200. doi: 10.1016/s0169-328x(97)00237-4. [DOI] [PubMed] [Google Scholar]
- 93.Vasko MR, Guo C, Kelly MR. The multifunctional DNA repair/redox enzyme Ape/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair. 2005;4:375–79. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 94.Kastan MB, Lim D-S. The many substrates and functions of ATM. Nat Mol Cell Biol. 2000;1:179–86. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 95.Nakagawa K, Taya Y, Tamai K, Yamaizumi Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol Cell Biol. 1999;19:2828–34. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shangary S, Brown KD, Adamson AW, et al. Regulation of DNA-dependent protein kinase activity by ionizing radiation-activated Abl kinase is an ATM-dependent process. J Biol Chem. 2000;275:301613–68. doi: 10.1074/jbc.M004302200. [DOI] [PubMed] [Google Scholar]
- 97.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 98.Lee Y, Chong MJ, McKinnon PJ. Ataxia telangiectasia mutated–dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J Neurosci. 2001;21:6687–93. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bates GW, Nicol SM, Wilson BJ, et al. The DEAD box protein p68: A novel transcriptional coactivator of the p53 tumour suppressor. EMBO J. 2005;24:543–53. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]


