Abstract
ICER is a member of the CREM family of basic leucine zipper transcription factors that acts as a dominant negative regulator of gene transcription. Four different isoforms of ICER (I, Iγ, II and IIγ) are transcribed from the P2 promoter of the Crem gene. We previously found that each of the ICER isoforms is induced by parathyroid hormone in osteoblasts. The goal of the present study was to assess the function of ICER in bone by overexpressing ICER in osteoblasts of transgenic mice. ICER I and ICER II cDNAs, each containing an N-terminal FLAG epitope tag, were cloned downstream of a fragment containing 3.6 kb of the rat Col1a1 promoter and most of the rat Col1a1 first intron to produce pOBCol3.6-ICER I and pOBCol3.6-ICER II transgenes, respectively. Multiple lines of mice were generated bearing the ICER I and ICER II transgenes. At 8 weeks of age, ICER I and ICER II transgenic mice had lower body weights and decreased bone mineral density of femurs and vertebrae. Further studies were done with ICER I transgenic mice, which had had greatly reduced trabecular bone volume and a markedly decreased bone formation rate in femurs. Osteoblast differentiation and osteocalcin expression were reduced in ex vivo bone marrow cultures from ICER I transgenic mice. ICER I antagonized the activity of ATF4 at its consensus DNA binding site in the osteocalcin promoter in vitro. Thus, transgenic mice with osteoblast-targeted overexpression of ICER resulted in osteopenia caused primarily by reduced bone formation. We speculate that ICER regulates the activity and/or expression of ATF/CREB factors required for normal bone formation.
Keywords: Bone, osteoblast, CREM, ICER, transgenic, bone morphometry, ATF4
Introduction
The cAMP responsive element modulator (CREM) is a member of the CREB/ATF family of basic leucine zipper (bZIP) transcription factors [1]. Crem encodes multiple isoforms that give rise to both activators and inhibitors of gene expression. Crem expression is regulated at multiple levels, including transcription from four different promoters [1–4], mRNA splicing [4, 5] and the use of alternative polyadenylation [4] and translational initiation sites [6, 7].
The inducible cAMP early repressor (ICER) is transcribed from the P2 promoter of the Crem gene [8, 9]. The P2 promoter is located within the 10 kb intron between exons G and γ. The P2 promoter contains two contiguous pairs of cAMP response element (CRE) sites termed cAMP autoregulatory response elements (CAREs) that are strongly inducible by cAMP [10]. Four ICER isoforms (I, Iγ, II, and IIγ) can be generated by alternative splicing of the γ domain and DNA binding domain I. The transcripts for ICER I and Iγ contain the contiguous DNA binding domains I and II sequences. However, a stop codon at the end of the DNA binding domain I prevents translation of DNA binding domain II. Due to alternative splicing, the transcripts for ICER II and IIγ contain only DNA binding domain II. DNA binding domains I and II are very similar and thus, all ICER proteins, which consist almost exclusively of the bZip domain of CREM, are thought to have similar activity as transcriptional repressors [1].
ICER was first discovered in pineal gland and plays a role in the regulation of circadian rhythms [11]. ICER was subsequently shown to regulate a variety of other cellular functions including interleukin-2 [12, 13] and interleukin-4 [14] production in T cells, cyclin A expression and cell proliferation in AtT20 cells [15] and Fas ligand expression in T and natural killer lymphocytes [16]. Rat and human prostate tumor cells engineered to overexpress ICER are unable to form tumors in nude mice [17, 18]. The sustained induction of ICER leads to cardiac myocyte apoptosis [19]. An important aspect of ICER biology is its ability to repress its own production. ICER homodimers inhibit transactivation of the P2 promoter by binding to the CAREs [1, 20]. This represents an autoregulatory feedback loop that allows the resetting of ICER-inhibited gene expression. Thus, ICER may be responsible for shaping the transient induction of gene expression in response to cAMP.
We previously reported that each of the four ICER isoforms is rapidly and transiently induced by PTH in osteoblasts via the cAMP-PKA signaling pathway [21, 22]. Moreover, we showed that induction of a transfected Pghs2 promoter-reporter construct with forskolin, a direct adenylase cyclase stimulator, is inhibited by transfection of an ICER IIγ expression construct in MC3T3-E1 cells. This led us to speculate that ICER induction in osteoblasts might represent a mechanism for regulating gene expression in response to PTH and other agonists that increase cAMP levels. To gain insight into the actions of ICER in vivo, we developed transgenic mice that overexpress ICER I and ICER II broadly in cells of the osteoblast lineage. Osteoblast-targeted ICER transgenic mice showed reduced body size, trabecular bone volume and bone formation. Bone marrow cultures from ICER transgenic mice displayed reduced osteoblast differentiation.
Materials and Methods
Animals
All animal care procedures were reviewed and approved by the University of Connecticut Health Center Animal Care Committee. To produce ICER transgenic mice, FLAG-ICER I and FLAG-ICER II cDNAs were amplified by PCR from pCR3.1-F-ICER with an Xba I-built-in 5’ primer and a 3’ primer corresponding to the 3’ end of ICER I. The PCR products were cloned directly to a pCR2.1 vector (Invitrogen Company, Carlsbad, CA). After verifying the orientation and the sequence of the inserts, the Xba I fragment was released and cloned into a ClaPa polylinker, which is flanked by Cla sites and contains a the bovine growth hormone polyadenylation (bGH poly A) sequence [23]. The FLAG-ICER-bGH poly A cassette was released by digestion with Cla1 and cloned into pBC-SK+, which contains the rat Col1a1 gene fragment from –3518 to +1594 bp including part of the first exon in which the AUG initiator has been mutated and most of the first intron [23]. This step produced the pOBCol3.6-ICER transgenes (Figure 1). The downstream Xho I site of the pOBCol3.6-ICER transgenes were mutated to an Sst II site with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Sst II fragments of pOBCol3.6-ICER I and II were gel purified and microinjected into CD-1 embryos to generate transgenic lines. The transgenes were detected as a 700 nt product by PCR of tail DNA using primers corresponding to the rat Col1a1 first intron (5’-ACCCTCCTCCATTTTAGCC) and the FLAG sequence (5’-CATCGTCGTCCTTGTAGTC) with denaturation at 94 C for 30 sec, annealing at 65 C for 30 sec and extension at 72 C for 2 min for 32 cycles [24]. To generate experimental mice, mating units typically consisted of wild type females and hemizygous transgenic males.
Figure 1.
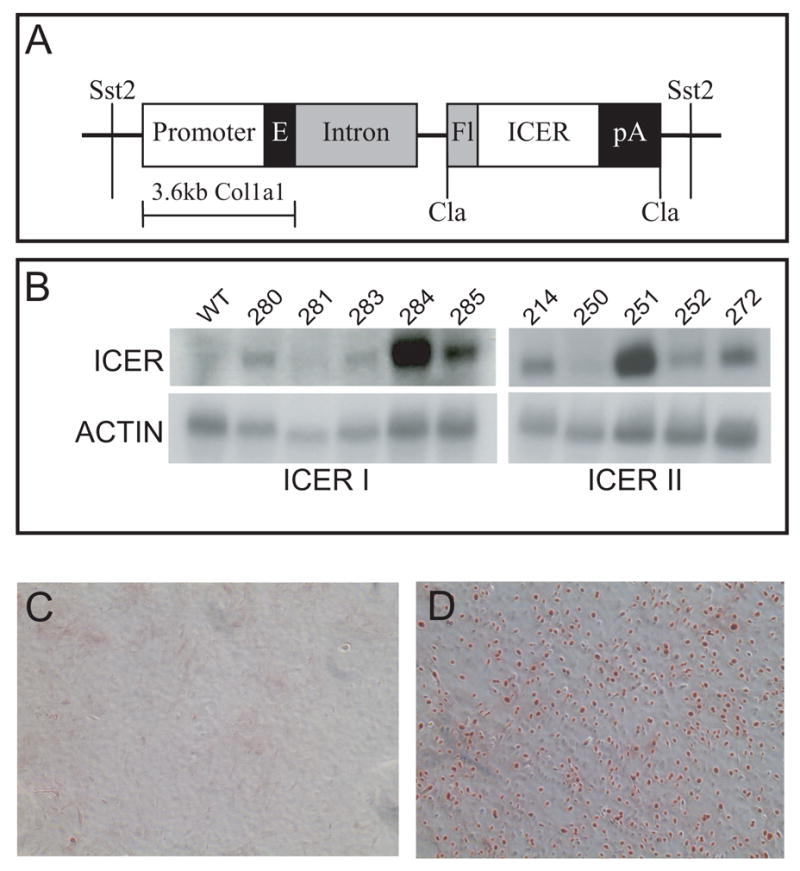
Generation of ICER transgenic mice. (A) The pOBCol3.6-ICER transgenes consist of the ICER I or II cDNA, a 3.6-kb fragment of the rat Col1a1 gene including 3.5 kb of promoter and 115 bp of first exon, 1.6 kb of the rat Col1a1 first intron and the bovine growth hormone polyadenylation site. (B) Northern blot analysis of ICER mRNA in long bones expressing either the ICER I or ICER II transgenes. (C, D) Immunostaining of primary calvarial osteoblast cultures established from wild type (WT) and ICER I (line 284) transgenic (TG) mice. Primary cultures were prepared from neonatal calvaria as described in Materials and Methods. Cultures were grown almost to confluence and stained with the M2 antibody that recognizes the FLAG epitope in the ICER I transgene.
In some experiments, pOBCol3.6-ICER I mice (line 284) were bred with mice containing a green fluorescent protein (GFP) transgene driven by a 2.3-kb fragment of the rat Col1a1 promoter and part of the first Col1a1 intron (pOBCol2.3-GFP) (kindly provided by Dr. David Rowe), which is a real time, fluorescent marker of mature osteoblasts [25]. Homozygous pOBCol2.3-GFP mice were bred with hemizygous pOBCol3.6-ICER I mice. This yielded progeny that were all hemizygous for the pOBCol2.3-GFP transgene and either wild type or hemizygous for the pOBCol3.6-ICER I transgene.
Immunostaining
Primary calvarial osteoblasts from 7-day-old wild type and transgenic mice were prepared as previously described [26]. Immunostaining for the FLAG-tagged ICER I transgene was performed as previously described with some modifications [24]. Primary neonatal calvarial osteoblasts were plated in αMEM medium with 10% heat-inactivated fetal calf serum at 8600 cells/well in a 4-well chamber slide. Cells were grown to 60–80% confluence. The medium was removed and cells were washed, fixed and stained using the M2 monoclonal primary antibody that recognizes the FLAG epitope (Sigma-Aldrich, St. Louis, MO), a biotinylated secondary anti-mouse IgG (1:200) and horseradish perioxidase streptavidin (1:100).
Transient transfection
MC3T3-E1 and HEK293 cells were transfected with the expression constructs pCR3.1-ICER I and pCMV-ATF4 and a promoter-reporter luciferase construct: one containing 4 tandem OSE1 sites driving luciferase (p4OSE1-luc) and one containing 6 tandem OSE2 sites driving luciferase (p6OSE2-luc) [27]. Transfections were performed using the Lipofectamine PLUSTM kit (Invitrogen). The amount of DNA in each transfection reaction was equalized using empty expression vectors, (pCR3.1-MT or pCMV-MT). Luciferase constructs were added at 0.25 μg/reaction for all reactions. Luciferase activity was measured 24 hours after transfection using a Promega Assay kit. Cells were lysed with 1X Cell Culture Lysis Reagent and supernatants harvested by centrifugation. Ten μl of each lysate was analyzed using a Berthold LB 9501/16 luminometer (Wallac, Inc.) and normalized to protein content [28].
Bone mineral density and morphometry
Femurs and the third lumbar vertebra (L3) were dissected from 8-week-old mice and stored in 70% ethanol. Bone mineral density (BMD) of the femurs and L3 vertebrae were determined by dual energy X-ray absorptiometry (DEXA) using PIXImus mouse densitometer (Lunar Corporation, Madison, WI). Cortical and trabecular morphometry of femurs was quantified at mid-diaphysis and within the distal metaphysis, respectively, using conebeam X-ray computed microtomography (microCT) using a μCT40 instrument (Scanco Medical AG, Bassersdorf, Switzerland) as previously described [26]. Segmentation of bone from marrow and soft tissue was performed in conjunction with a constrained Gaussian filter (to reduce noise) at density thresholds of 712 mg/cm3 and 474 mg/cm3 for cortical and trabecular compartments, respectively. Volumetric regions for trabecular analysis were selected within the endosteal borders to include the secondary spongiosa of femoral metaphyses located 960 μm from the growth plate and extending 1 mm proximally. Trabecular morphometry was assessed by measuring bone volume fraction (BV/TV), trabecular thickness (TbTh) and trabecular number (TbN). Cortical morphometry was quantified and averaged for 50 serial cross-sections (600 μm) extending distally from the diaphyseal midpoint between proximal and distal growth plates. Cortical measurements included average cortical thickness, cross-sectional area of cortical bone, and sub-periosteal cross-sectional area (bone plus marrow, an index of cross-sectional dimensions).
Bone histomorphometry
Seven-week-old wild type and ICER I (line 284) female mice were injected intraperitoneally with calcein (10 mg/kg body weight) and 7 days later with xylenol orange (90 mg/kg body weight). Mice were euthanized 2 days after the second injection. Femurs were dissected free of tissue and fixed in 4% paraformaldehyde. The undecalcified femurs were dehydrated in increasing concentrations of ethanol, cleared in xylene, and embedded in methyl methacrylate. Five-micron-thick longitudinal serial sections were taken from the middle of the femur as defined by the location of a central vein. Measurements were made blinded as to the treatment group using an OsteoMeasure computerized image analysis system (OsteoMetrics, Inc. Atlanta, GA) interfaced with an Optiphot Nikon microscope (Nikon Inc., Melville, NY) at a magnification of 20x. All measurements were confined to an area of the secondary spongiosa between 400 and 2000 microns distal to the growth plate-metaphyseal junction of the distal femur. We used terminology and units recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research [29].
Osteoblast differentiation
Bone marrow was flushed from the long bones of 8-week-old male mice into αMEM and sieved through a 40-μm cell strainer (Becton Dickinson, Franklin Lakes, NJ). Cells were plated at 1.7 x 106/ml in 35 mm wells in αMEM supplemented with 10% non-heat-inactivated FCS, 100 U/ml penicillin and 50 μg/ml streptomycin. On day 3, half of the medium was removed and replenished with fresh medium. On day 7, the medium was completely replenished and cells were cultured in osteogenic differentiation medium containing α-MEM with 10% non heat-inactivated FCS, 100 U/ml penicillin, 50 μg/ml streptomycin and 8 mM β-glycerophosphate and 50 μg/ml ascorbic acid. Medium was changed every 2 days until day 28.
Cultures were analyzed for alkaline phosphatase and mineral staining on days 14, 21 and 28 while RNA was isolated on days 14 and 21 for gene expression studies. Alkaline phosphatase staining was performed using a commercial kit (Sigma Diagnostics, Inc., St. Louis, MO). To assess mineralization, plates stained with alkaline phosphatase were further stained with von Kossa reagent. Plates were rinsed briefly with water and treated with LiCO3 for 20 min. After a brief rinse with water, cells were covered with 5% AgNO3 for 1 h under bright light. To assess GFP fluorescence, cultures were analyzed using a Fluorimager SI instrument (Molecular Dynamics, Sunnyvale, CA). A 515nm long pass emission filter was with a photomultiplier tube voltage (PMT) of 800 was utilized for optimal detection. Digital images of excitation spectra were processed using ImageQuaNT software (Molecular Dynamics). To assess gene expression, total RNA was extracted using TRIzol Reagent (GibcoBRL) as previously described [30]. RNA was redissolved in GTC buffer and precipitated in isopropanol. RNA was washed in 80% ethanol, dissolved in diethylpyrocarbonate (DEPC) water and stored at -80 C until use. For Northern blot analysis, total RNA was denatured and fractionated on an agarose gel and transferred to a nitrocellulose membrane by capillary transfer. Transferred RNA was hybridized to the membrane, which was then exposed to existing 32P-labelled cDNA probes for Col1a1, bone sialoprotein (BSP), osteocalcin (OC) and β-actin. Signals were quantified as described previously [30].
Osteoclast formation
Marrow was flushed from the long bones of 8-week-old female with □MEM and filtered with a 40-μm cell strainer (Becton Dickinson, Franklin Lakes, NJ). The harvested marrow cells were plated at 25,000/well in a 48-well dishes in αMEM supplemented with 10% heat-inactivated FCS and 30 μg/ml each of M-CSF and RANK ligand (both from R&D Systems, Inc., Minneapolis, MN). The culture medium was changed on day 3. Tartrate resistant acid phosphatase (TRAP) staining was performed on days 3, 4 and 5 using a leukocyte acid phosphatase staining kit (Sigma Diagnostics Inc., St. Louis, MO) according to the manufacturer’s protocol. TRAP-positive cells containing 3 or more nuclei per cell were counted under a microscope.
Serum markers
Serum osteocalcin was measured by radioimmunoassay using a goat anti-mouse osteocalcin antibody as previously described [31]. Serum C-terminal telopeptide of α1(I) collagen (CTX) was measured by an enzyme-linked immunosorbent assay (ELISA) using a RatLaps kit (Nordic Bioscience Diagnostics, A/S).
Results
Transgenic lines overexpressing pOBCol3.6-ICER I and pOBCol3.6-ICER II in osteoblasts were generated by embryo microinjection. Transgene mRNA was highly expressed in calvaria, femur and vertebra, with lower expression in skin and tendon and virtually no expression in other tissues (not shown). Transgene mRNA expression varied among the lines. Of the ICER I transgenic lines, expression was highest in lines 284 and 285, which were chosen for further examination (Figure 1B). Of the ICER II lines, expression was highest in line 251, but this line was lost. Therefore, line 214 was chosen for examination. Primary calvarial osteoblasts from wild type and ICER I mice (line 284) were cultured and stained for FLAG-tagged ICER protein using the M2 antibody, which recognizes the FLAG epitope. Staining was not seen in wild type cells (Figure 1C), while strong immunostaining was seen in most transgenic cells (Figure 1D). These data indicated that the ICER transgenes were highly expressed in osteoblasts at both the mRNA and protein level.
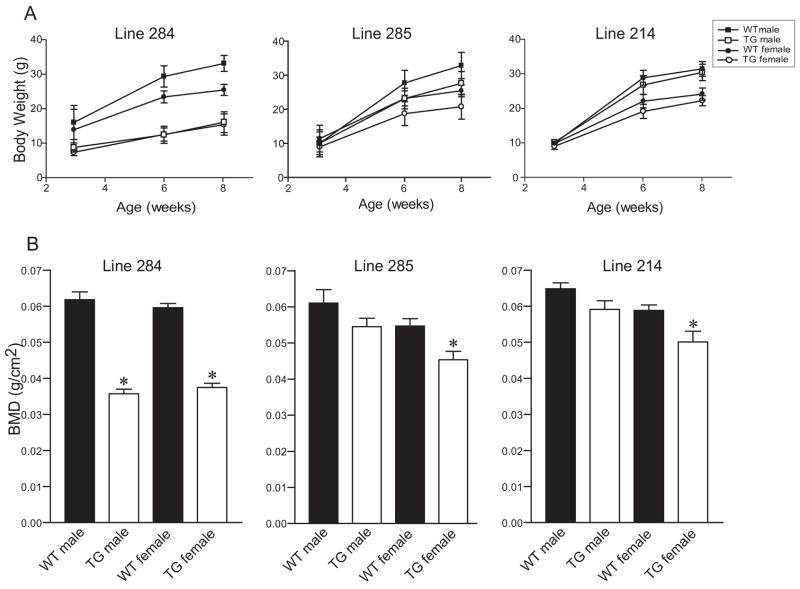
Six- and eight-week-old line 284 mice (ICER I) had reduced body weight compared to wild-type littermates (Figure 2A). Lines 285 and 214 (ICER I and ICER II, respectively), which expressed lower levels of the ICER transgene, also showed a weight difference, although not as marked as line 284 (Figure 2A). The BMD of dissected femurs was greatly reduced in males and females mice of line 284. (Figure 2B). There was also a reduction in BMD in femurs of lines 285 and 214, although females were more affected than males (Figure 2B and C). The BMD differences for males of lines 285 and 214 did not reach statistical significance.
Figure 2.
Phenotypic analysis of ICER transgenic (TG) mice. (A) Body weights were measured at 3, 6 and 8 weeks of age. Each value represents the mean ± SD of 7–22 mice for line 284 (ICER I), 2–5 mice for line 285 (ICER I) and 8–21 mice for line 214 (ICER II) mice except for the line 285 TG male group at 3 weeks, which is one mouse. Body weights for TG mice were significantly different (p<0.05) from the respective sex-matched wild type (WT) groups for all age points for line 284 and for the 6- and 8-week age points for line 214. (B) BMD measurements assessed by DEXA in 6- to 8-week-old mice in these lines. Each value is the mean ± SEM for 5–13 mice. *Different from the sex-matched WT group, p<0.01.
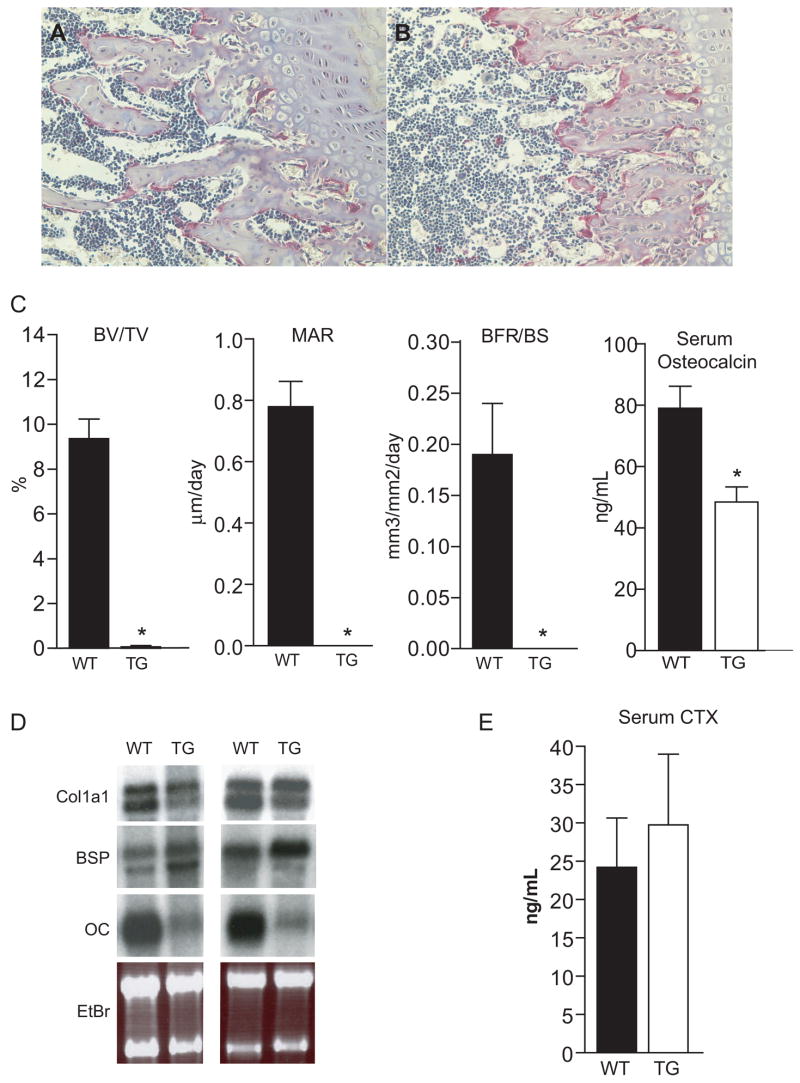
Adult line 284 mice (ICER I) had a scarcity of trabecular bone in the primary spongiosa of distal femurs compared to wild type littermates (Figure 3A and B). Histomorphometric analysis of line 284 femurs showed a significant reduction in trabecular bone volume fraction (BV/TV%) (Figure 3C). The mineral apposition rate (MAR) and bone formation rate per bone surface (BFR/BS) were profoundly decreased in transgenic mice (Figure 3C). Neonatal calvaria from ICER I mice (line 284) showed a marked reduction in expression of osteocalcin mRNA, a marker of mature osteoblasts; however, Col1a1 was not greatly affected while bone sialoprotein (BSP) was increased (Figure 3D). Serum osteocalcin, a marker for bone formation was reduced in sera of line 284 mice (ICER I) compared to wild type littermates (Figure 3C). Together, these data indicated that bone formation was reduced in ICER I mice.
Figure 3.
In vivo analyses of wild type and ICER I (line 284) transgenic mice. Hematoxylin and eosion staining of longitudinal sections of a wild type (A) and transgenic (B) femur counterstained with TRAP at 20X magnification. (C) Histomorphometric and serum osteocalcin measurements. Bone volume normalized to total volume (BV/TV) was measured by static histomorphometric analysis. Each value represents mean ± SEM for 14 wild type (WT) and 4 transgenic (TG) femurs. Mineral apposition rate (MAR) and bone formation rate normalized to bone surface (BFR/BS) were determined by dynamic histomorphometric analysis of double-labeled surface of active bone formation. Each value represents the mean ± SEM of 10 WT and 5 TG mice. Serum osteocalcin protein level measured by ELISA was utilized as a biochemical marker of bone formation from sera of 6- to 8-week-old mice. Each value represents the mean ± SEM of 8 WT and 9 TG mice. Values for males and females were pooled. *Different from WT, p<0.01. (D) Northern blot analysis of a1(I) collagen (Col1a1), bone sialoprotein (BSP) and osteocalcin (OC) mRNA levels in calvaria of neonatal WT and ICER I (line 284) TG mice. EtBr, ethdidium bromide. (E) Serum levels of the C-terminal telopeptide of α1(I) collagen (CTX) links in WT and TG mice. Each value is the mean ± SEM of 9 WT and 9 TG mice. Values for males and females were pooled.
TRAP staining was seen in the primary spongiosa of both wild type and line 284 mice (ICER I, Figures 3A and B). Osteoclast number per bone surface was not significantly altered in transgenic femurs (not shown), although this may have been due to lack of a sufficient amount of trabecular template for an adequate assessment. Serum CTX was unchanged in line 284 (ICER I, Figure 3E). These data indicate that osteoclast formation and bone resorption were not greatly affected in ICER I mice.
Bone morphometry of line 284 mice (ICER I) was evaluated by microCT (Figure 4). Three-dimensional images showed dramatic reduction in the trabecular bone compartment in line 284 (Figure 4B) compared to wild type femurs (Figure 4C). Trabecular bone volume, number and thickness were greatly reduced in line 284 femurs Figure 4C). Cortical bone morphometry at the mid-diaphyseal region of femurs was affected in line 284 mice (Figure 4D and E). The subperiosteal area of male line 284 femurs was significantly reduced, although this parameter was not altered in female line 284 femurs (Figure 4F). Cortical bone area and thickness were decreased in both male and female line 284 femurs (Figure 4F).
Figure 4.
Microcomputed tomographic (MicroCT) analysis of femurs from 7.5-week-old ICER I mice (line 284). Three-dimensional images corresponding to the trabecular bone compartment of (A) wild type (WT) and (B) transgenic (TG) femurs. (C) BV/TV, trabecular number (Tb.N) and trabecular thickness (Tb.Th) measurements in male and female WT and TG mice. Cross-sectional images displaying the cortical bone integrity of WT (D) and TG (E) femurs. (F) Cortical bone measurements extrapolated from MicroCT analysis including cross-sectional area, cortical bone area and cortical thickness in male and female WT and TG mice. Each value is the mean ± SD for 12 WT males, 8 TG males, 6 WT females and 6 TG females.
To determine the effect of the ICER transgene on osteoblast differentiation, bone marrow cultures were established from ICER I (line 284) and wild type mice that were bred to incorporate a pOBCol2.3-GFP transgene, which is a real time, fluorescent marker of mature osteoblasts [25]. Homozygous pOBCol2.3-GFP mice were bred with hemizygous pOBCol3.6-ICER I transgenic mice. This yielded progeny that were all hemizygous for the pOBCol2.3-GFP transgene and either wild type or hemizygous for the pOBCol3.6-ICER I transgene (line 284). In bone marrow stromal cultures, von Kossa staining was greatly reduced in line 284 (ICER I) cultures at day 21 (Figure 5A). GFP fluorescence at day 21 (Figure 5B), and BSP and OC mRNA levels at days 14 and 21 (Figure 5C), were reduced in line 284 (ICER I) cultures.
Figure 5.
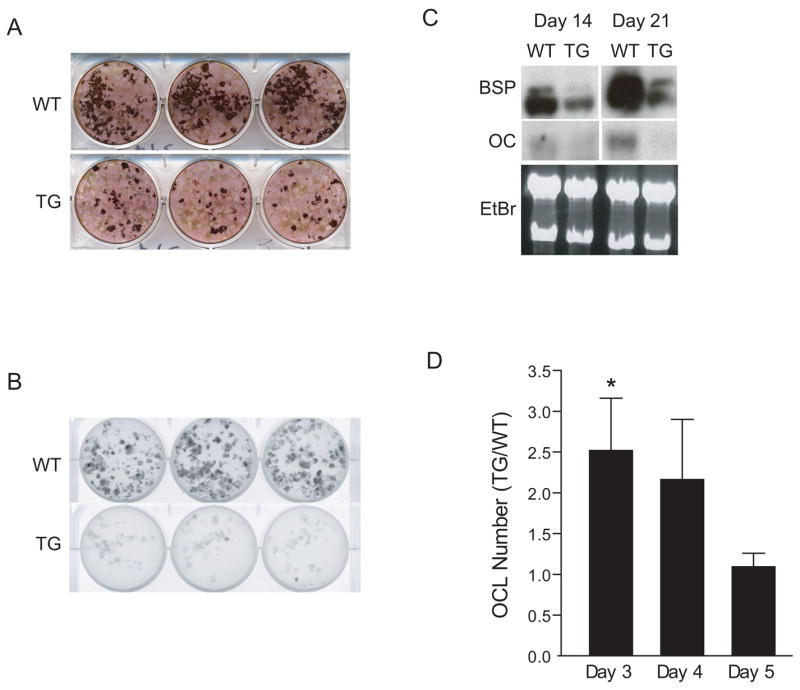
Ex vivo culture of bone cells from ICER I mice (line 284). (A) Bone marrow stromal cells from 6- to 8-week-old wild type (WT) and transgenic (TG) mice were cultured in osteogenic medium until day 21 and stained with alkaline phosphatase and von Kossa stain. Three replicate wells are shown. (B) Heterozygous pOBCol3.6-ICER I mice (line 284) were bred homozygous pOBCol2.3-GFP mice. All progeny were heterozygous for the pOBCol2.3-GFP transgene and either wild type (WT) or hemizygous for the pOBCol3.6-ICER I (line 284) transgene (TG). Bone marrow stromal cultures were prepared from WT and TG mice and examined for GFP fluorescence with a fluorimager. GFP fluorescence is shown in WT and TG cultures at day 21. Three replicate wells are shown. (C) Analysis of mRNA markers of osteoblast differentiation. Bone sialoprotein (BSP) and osteocalcin (OC) expression was analyzed by Northern blot analysis of cultures at day 14 and 21 in WT and TG mice. Ethidium bromide (EtBr) staining was used as a loading control. These data are representative of three experiments. (D) Osteoclastogenesis was assessed in ex vivo bone marrow cells cultured with M-CSF (30 ng/ml) and RANKL (30 ng/ml) as described. For each day of culture, osteoclast number (OCL) in TG cultures was normalized to WT values. Each value represents the mean ± SEM of three experiments. *Significant effect of the transgene, p<0.05.
Bone marrow cells from 8-week-old line 284 (ICER I) and wild type female mice were treated with M-CSF and RANKL to induce osteoclast differentiation. The number of TRAP-positive multinucleated cells in transgenic cultures was increased by 2-fold on day 3 of culture but was not changed at days 4 and 5 (Figure 5D). Taken together, the ex vivo cellular differentiation studies indicated that the ICER I transgene primarily affected osteoblast differentiation.
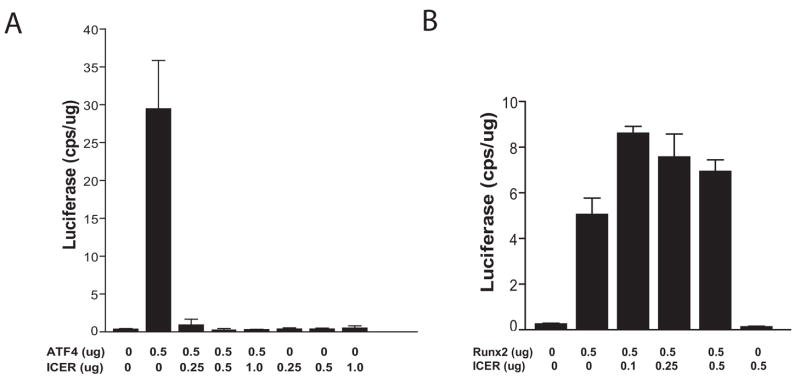
ICER I (line 284) mice showed a dramatic loss of femoral trabecular bone in vivo and a marked reduction in osteocalcin expression but Col1a1 expression was not dramatically altered. This phenotype was reminiscent of ATF4 knockout mice and raised the possibility that ICER antagonizes the activity of ATF4 in transgenic mice [32]. We therefore examined ICER I function at the OSE1 site that binds ATF4 in the mouse osteocalcin promoter. We utilized an OSE1 construct, containing four OSE1 sequences in tandem driving luciferase expression (p4OSE1-Luc). ATF4 induced p4OSE1-Luc activity (Figure 6A). However, when cells were co-transfected with increasing amounts of an ICER I expression plasmid (pCR3.1-ICER I), the induction of the OSE1 construct was potently repressed. Thus, ICER I repressed ATF4 activity in at the OSE1 site. To determine the specificity of this transcriptional repression, we examined the effect of ICER I at the OSE2 site of the osteocalcin promoter. This site was chosen because Runx2 is the primary transcriptional regulator of osteocalcin expression and because it cooperates with ATF4 at the OSE2 site. Runx2 induced a multimerized OSE2 construct driving luciferase (p6OSE2-Luc) in HEK293 cells (Figure 6B). At low levels (0.1 μg), ICER I appeared to augment OSE2 activity in response to Runx2. As the concentration of ICER I plasmid increased, p6OSE2-luc activity decreased, although not below the level induced by Runx2 alone. This was in sharp contrast to the repression of ATF4 activity by ICER I at the OSE1 site.
Figure 6.
(A) ICER dose response to ATF4 at the OSE1 site. MC3T3-E1 cells (MC-4 clone) were transfected with pCMV-ATF4 and 4OSE1-Luc and co-transfected with increasing concentrations of pCR3.1-ICER I. Luciferase activity was normalized to total protein using the BCA assay. (B) ICER does not modulate Runx2 activity at the OSE2 site. Increasing concentrations of ICER expression plasmid were co-transfected with Runx2 vector and 6OSE2-luc in HEK293 cells. Luciferase activity was normalized to total protein.
Discussion
We previously showed that all ICER isoforms are induced by PTH in cultured osteoblasts and in bone in vivo [21]. To explore the effects of ICER in vivo, we generated transgenic mice in which a 3.6-kb fragment of the rat Col1a1 promoter and most of the first intron of Col1a1 directed ICER I and II expression to osteoblasts. This promoter fragment has been well characterized with respect to its temporal and spatial pattern of expression in mouse tissues [25, 33, 34]. We found that the bone mineral density was reduced in transgenic femurs. This change was seen in two independent lines of ICER I mice with varying levels of transgene expression and one line of ICER II mice. In ICER I transgenic lines, the strength of the phenotype was associated with the level of transgene expression indicating that the bone phenotype was independent of the transgene integration site. The phenotypes of the ICER I and ICER II transgenic lines were similar. Due to alternative splicing of the CREM gene, ICER I and ICER II proteins differ in their DNA binding domain. The alternative DNA binding domains I and II encoded by the Crem gene are very similar. As a result, the dominant negative function of the ICER I and ICER II proteins are thought to be equivalent, although this has not been studied systematically. Each of the four ICER proteins repressed the inhibin α-subunit promoter in transfected mouse granulosa cells [35] and the Fas ligand promoter in transfected T lymphocytes [16]. In primary cultures of human endometrial stromal cells, ICER I and ICER II equally repressed PKA-induced expression of transfected prolactin promoter-reporter constructs [7].
Transgenic distal femurs of ICER I mice (line 284) had a striking paucity of trabecular bone. Osteoblast differentiation was markedly impaired in bone marrow cultures and serum osteocalcin levels were reduced in these animals. By contrast, osteoclast number per bone surface in distal femurs and serum CTX were unchanged, and osteoclast formation was only transiently increased in bone marrow cultures from line 284. Taken together, these data suggest that the decrease in bone mass in ICER I mice was largely due to decreased bone formation. Using a similar in vivo strategy, tissue selective promoters have been used to generate transgenic mice with ICER expression targeted to pancreatic β-cells [36] and thymocytes [37].
bZIP factors are grouped into thirteen families based on their dimerization properties [38]. ICER is classified as a member of the CREB/CREM/ATF1 family within the broader ATF/CREB superfamily. Members of the CREB/CREM/ATF1 family form both homo- and heterodimers within the family that bind to palindromic CRE sites [38]. ICER consists almost exclusively of the bZIP domain of CREM and is devoid of transactivation domains rendering it a potential dominant negative transcription factor. ICER homodimers can function as transcriptional repressors by binding with high affinity to CRE sites [1] precluding the recruitment of other factors required for necessary for transcriptional initiation [39]. For example, ICER prevents α-inhibin transcription by in ovarian granulosa cells by preventing CREB binding to a CRE site [35]. Alternatively, ICER can form inactive heterodimers with transcription factors such as CREB and CREM [3] and NFAT [13, 14].
Of paramount interest are the observations that Atf4 knockout mice have low bone mass characterized by reduced trabecular bone volume and that Atf4 knockout osteoblasts show impaired differentiation ex vivo [32, 40]. ATF4 induces osteocalcin expression and a distinct binding site for ATF4 was discovered as OSE1, a cis-acting element in the osteocalcin promoter [32]. In addition, although Col1a1 mRNA expression is maintained in osteoblasts from Atf4 knockout mice, type I collagen protein synthesis is greatly decreased, suggesting a role for ATF4 in post-translational modification of type I collagen. ATF4 and Runx2 cooperate in the activation of the osteocalcin gene [27]. Due to noteworthy parallels in the phenotype we have observed in ICER I transgenic mice, specifically with regard to dramatic effects on the trabecular bone compartment and osteocalcin expression in ex vivo osteoblast cultures as well as in vivo expression (data not shown), we believe it is possible that ICER interacts with ATF4 to disrupt its ability to activate genes necessary for physiologic bone remodeling. To address the role of ICER in modulating ATF4 at its OSE1 site, we utilized a promoter-reporter construct using a multimerized OSE1 promoter driving luciferase (p4OSE1-Luc). Consistent with previous studies [27, 32], ATF4 potently activated the OSE1 construct driving high expression of the luciferase reporter in both MC3T3-E1 (clone MC-4) and HEK293 cells. However, when cells were co-transfected with both ATF4 and ICER, there was a profound suppression of ATF4-mediated activation to baseline, showing that ICER dramatically disrupted ATF4 function at its OSE1 binding site. However, ICER did not repress Runx2 activity at the OSE2 site, showing specificity of the ICER effect. In preliminary studies, we found that ICER does not bind strongly if at all to the OSE1 site; rather it may decrease the binding of ATF4 to the OSE1 site (data not shown). Further studies will be necessary to support these results. Recently, the factor inhibiting ATF4-mediated transcription (FIAT) was identified in osteoblasts [41]. FIAT, a bZIP factor inhibits ATF4-mediated transcription by forming an inactive heterodimer with ATF4. Transgenic expression of FIAT with a 2.3-kb fragment of the mouse Col1a1 promoter resulted in greatly decreased trabecular bone volume and osteocalcin expression [41]. This model has interesting phenotypic similarities with ICER transgenic mice, thus strengthening our hypothesis that ATF4 is a target of ICER.
Although we hypothesize that ATF4 is a target of ICER, it is very likely that ICER interacts with and regulates the activity of other transcription factors in osteoblasts. Components of the dimeric AP-1 complex, which include FOS and JUN proteins, may also be direct targets of ICER. Although the c-FOS is best known for its role in osteoclast formation [42, 43], the anabolic effect of PTH on bone acquisition is in part dependent on the c-Fos gene [44]. Fra-1 and deltaFosB, a splice variant of FosB, are stimulators of osteoblast differentiation and function. Mice with a conditional knockout of Fra-1 develop osteopenia [45], while Fra-1 [46, 47] and deltaFosB [48, 49] transgenic mice display osteosclerosis. C/EBPβ is another intriguing candidate for interacting with ICER since C/EBPβ synergizes with Runx2 to induce osteocalcin gene expression during osteoblast differentiation [50, 51]. Moreover, C/EBP factors can heterodimerize with selected ATF/CREB proteins [52]. Therefore, it is conceivable that transgenic ICER protein could have prevented C/EBPβ from interacting with Runx2, thus blocking osteoblast differentiation. ICER transgenic mice have phenotypic similarities to mice with an osteoblast-targeted C/EBPβ p20 transgene including a marked decrease in femoral trabecular bone volume. Since p20 can act as a dominant negative molecule, ICER and p20 transgenes may converge on a similar pathway [53]. In our model, ICER is expressed constitutively at supraphysiological levels. This raises the possibility that transgenic ICER may interact with transcription factors that are not normally engaged by physiological levels of ICER.
The low bone mass in ICER transgenic mice was due largely by an inhibition of bone formation. Interestingly, we have shown that Crem knockout mice, which are devoid of ICER and other CREM isoforms, have slightly increased bone mass, consistent with the loss of a repressor such as ICER [54]. The ICER transgenic models raise the possibility that ICER is a mechanism to regulate the strength and timing of the activity of ATF/CREB factors that participate in osteoblast differentiation and function (i.e. a fine tuning mechanism). Our past work has shown that ICER is induced by PTH and other agonists that increase cAMP [21, 22]. We reason that by antagonizing the activity of multiple factors such as CREB and ATF4, ICER modulates pathway that control bone formation. Downstream targets of ICER may include hormones such as insulin-like growth factor-I (IGF-I), which is known to be regulated by cAMP [55] and mediates in part the anabolic effects of PTH on bone mass accrual [56] and the Wnt pathway, which is stimulated by PTH-induced cAMP [57]. Further studies in our laboratory are designed to identify the molecular targets of ICER that regulate bone formation.
Acknowledgments
This project described was made possible by grant number R01 AR46542 to BEK from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). YFH and TKC received support on the institutional Skeletal, Craniofacial and Oral Biology training grant T32 DE007302 from the National Institute of Dental and Craniofacial Research. We acknowledge support from the Core Center for Musculoskeletal Disorders grant (P30 AR046026, NIAMS) and the University of Connecticut Health Center Microcomputed Tomography Facility. We thank the Yale University Core Center for Musculoskeletal Disorders program for assistance with measuring serum turnover markers. We thank Dr. Renny Franceschi for providing the OSE1 and OSE2 reporter plasmids and the ATF4 expression plasmid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Taranpreet K. Chandhoke, Email: Chandhoke@student.uchc.edu.
Yu-Feng Huang, Email: whuang@csmu.edu.tw.
Fei Liu, Email: feiliu@umich.edu.
Gloria A. Gronowicz, Email: gronowicz@nso1.uchc.edu.
Douglas J. Adams, Email: dadams@nso.uchc.edu.
John R. Harrison, Email: harrison@nso1.uchc.edu.
Barbara E. Kream, Email: kream@nso1.uchc.edu.
References
- 1.Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–86. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 2.Daniel PB, Rohrbach L, Habener JF. Novel cyclic adenosine 3',5'-monophosphate (cAMP) response element modulator theta isoforms expressed by two newly identified cAMP-responsive promoters active in the testis. Endocrinology. 2000;141:3923–30. doi: 10.1210/endo.141.11.7758. [DOI] [PubMed] [Google Scholar]
- 3.Stehle JH, Foulkes NS, Molina CA, Simonneaux V, Pevet P, Sassone-Corsi P. Adrenergic signals direct rhythmic expression of transcriptional repressor CREM in the pineal gland. Nature. 1993;365:314–20. doi: 10.1038/365314a0. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes NS, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–49. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 5.Foulkes NS, Mellstrom B, Benusiglio E, Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature. 1992;355:80–4. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- 6.Delmas V, Laoide BM, Masquilier D, de Groot RP, Foulkes NS, Sassone-Corsi P. Alternative usage of initiation codons in mRNA encoding the cAMP-responsive-element modulator generates regulators with opposite functions. Proc Natl Acad Sci U S A. 1992;89:4226–30. doi: 10.1073/pnas.89.10.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gellersen B, Kempf R, Telgmann R. Human endometrial stromal cells express novel isoforms of the transcriptional modulator CREM and up-regulate ICER in the course of decidualization. Mol Endocrinol. 1997;11:97–113. doi: 10.1210/mend.11.1.9875. [DOI] [PubMed] [Google Scholar]
- 8.Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 9.Stehle JH, Foulkes NS, Molina CA, Simonneaux V, Pevet P, Sassone-Corsi P. Adrenergic signals direct rythmic expression of transcriptional repressor CREM in the pineal gland. Nature. 1993;365:314–320. doi: 10.1038/365314a0. [DOI] [PubMed] [Google Scholar]
- 10.Behr R, Weinbauer GF. cAMP response element modulator (CREM): an essential factor for spermatogenesis in primates? Int J Androl. 2001;24:126–35. doi: 10.1046/j.1365-2605.2001.00277.x. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi JS. Circadian rhythms. ICER is nicer at night (sir!) Curr Biol. 1994;4:165–8. doi: 10.1016/s0960-9822(94)00040-0. [DOI] [PubMed] [Google Scholar]
- 12.Bodor J, Spetz AL, Strominger JL, Habener JF. cAMP inducibility of transcriptional repressor ICER in developing and mature human T lymphocytes. Proc Natl Acad Sci U S A. 1996;93:3536–41. doi: 10.1073/pnas.93.8.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodor J, Bodorova J, Gress RE. Suppression of T cell function: a potential role for transcriptional repressor ICER. J Leukoc Biol. 2000;67:774–9. doi: 10.1002/jlb.67.6.774. [DOI] [PubMed] [Google Scholar]
- 14.Bodor J, Habener JF. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J Biol Chem. 1998;273:9544–51. doi: 10.1074/jbc.273.16.9544. [DOI] [PubMed] [Google Scholar]
- 15.Lamas M, Molina C, Foulkes NS, Jansen E, Sassone-Corsi P. Ectopic ICER expression in pituitary corticotroph AtT20 cells: effects on morphology, cell cycle, and hormonal production. Mol Endocrinol. 1997;11:1425–34. doi: 10.1210/mend.11.10.9987. [DOI] [PubMed] [Google Scholar]
- 16.Bodor J, Bodorova J, Bare C, Hodge DL, Young HA, Gress RE. Differential inducibility of the transcriptional repressor ICER and its role in modulation of Fas ligand expression in T and NK lymphocytes. Eur J Immunol. 2002;32:203–12. doi: 10.1002/1521-4141(200201)32:1<203::AID-IMMU203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 17.Memin E, Yehia G, Razavi R, Molina CA. ICER reverses tumorigenesis of rat prostate tumor cells without affecting cell growth. Prostate. 2002;53:225–31. doi: 10.1002/pros.10149. [DOI] [PubMed] [Google Scholar]
- 18.Yehia G, Razavi R, Memin E, Schlotter F, Molina CA. The expression of inducible cAMP early repressor (ICER) is altered in prostate cancer cells and reverses the transformed phenotype of the LNCaP prostate tumor cell line. Cancer Res. 2001;61:6055–9. [PubMed] [Google Scholar]
- 19.Yan C, Miller CL, Abe J. Regulation of phosphodiesterase 3 and inducible cAMP early repressor in the heart. Circ Res. 2007;100:489–501. doi: 10.1161/01.RES.0000258451.44949.d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thonberg H, Lindgren EM, Nedergaard J, Cannon B. As the proliferation promoter noradrenaline induces expression of ICER (induced cAMP early repressor) in proliferative brown adipocytes, ICER may not be a universal tumour suppressor. Biochem J. 2001;354:169–77. doi: 10.1042/0264-6021:3540169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tetradis S, Nervina JM, Nemoto K, Kream BE. Parathyroid hormone induces expression of the inducible cAMP early repressor in osteoblastic MC3T3-E1 cells and mouse calvariae. J Bone Miner Res. 1998;13:1846–51. doi: 10.1359/jbmr.1998.13.12.1846. [DOI] [PubMed] [Google Scholar]
- 22.Nervina JM, Tetradis S, Huang YF, Harrison D, Molina C, Kream BE. Expression of inducible cAMP early repressor is coupled to the cAMP-protein kinase A signaling pathway in osteoblasts. Bone. 2003;32:483–90. doi: 10.1016/s8756-3282(03)00056-5. [DOI] [PubMed] [Google Scholar]
- 23.Kalajzic Z, Liu P, Kalajzic I, Du Z, Braut A, Mina M, Canalis E, Rowe DW. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–60. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 24.Jiang J, Lichtler AC, Gronowicz GA, Adams DJ, Clark SH, Rosen CJ, Kream BE. Transgenic mice with osteoblast-targeted insulin-like growth factor-I show increased bone remodeling. Bone. 2006;39:494–504. doi: 10.1016/j.bone.2006.02.068. [DOI] [PubMed] [Google Scholar]
- 25.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 26.Sher LB, Harrison JR, Adams DJ, Kream BE. Impaired cortical bone acquisition and osteoblast differentiation in mice with osteoblast-targeted disruption of glucocorticoid signaling. Calcif Tissue Int. 2006;79:118–25. doi: 10.1007/s00223-005-0297-z. [DOI] [PubMed] [Google Scholar]
- 27.Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280:30689–96. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 28.Smith PK, Krohn RI, Hermanson GT, Mailia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Oslson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Analytic Biochemistry. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 29.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 30.Sher LB, Woitge HW, Adams DJ, Gronowicz GA, Krozowski Z, Harrison JR, Kream BE. Transgenic expression of 11beta-hydroxysteroid dehydrogenase type 2 in osteoblasts reveals an anabolic role for endogenous glucocorticoids in bone. Endocrinology. 2004;145:922–9. doi: 10.1210/en.2003-0655. [DOI] [PubMed] [Google Scholar]
- 31.Gundberg CM, Clough ME, Carpenter TO. Development and validation of a radioimmunoassay for mouse osteocalcin: paradoxical response in the Hyp mouse. Endocrinology. 1992;130:1909–15. doi: 10.1210/endo.130.4.1547718. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–98. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 33.Bogdanovic Z, Bedalov A, Krebsbach PH, Pavlin D, Woody CO, Clark SH, Thomas HF, Rowe DW, Kream BE, Lichtler AC. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J Bone Miner Res. 1994;9:285–92. doi: 10.1002/jbmr.5650090218. [DOI] [PubMed] [Google Scholar]
- 34.Pavlin D, Lichtler AC, Bedalov A, Kream BE, Harrison JR, Thomas HF, Gronowicz GA, Clark SH, Woody CO, Rowe DW. Differential utilization of regulatory domains within the alpha 1(I) collagen promoter in osseous and fibroblastic cells. J Cell Biol. 1992;116:227–36. doi: 10.1083/jcb.116.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burkart AD, Mukherjee A, Mayo KE. Mechanism of repression of the inhibin alpha-subunit gene by inducible 3',5'–cyclic adenosine monophosphate early repressor. Mol Endocrinol. 2006;20:584–97. doi: 10.1210/me.2005-0204. [DOI] [PubMed] [Google Scholar]
- 36.Inada A, Hamamoto Y, Tsuura Y, Miyazaki J, Toyokuni S, Ihara Y, Nagai K, Yamada Y, Bonner-Weir S, Seino Y. Overexpression of inducible cyclic AMP early repressor inhibits transactivation of genes and cell proliferation in pancreatic beta cells. Mol Cell Biol. 2004;24:2831–41. doi: 10.1128/MCB.24.7.2831-2841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodor J, Feigenbaum L, Bodorova J, Bare C, Reitz MS, Jr, Gress RE. Suppression of T-cell responsiveness by inducible cAMP early repressor (ICER) J Leukoc Biol. 2001;69:1053–9. [PubMed] [Google Scholar]
- 38.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–35. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–22. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 40.Yang X, Karsenty G. ATF4, whose osteoblast accumulation is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J Biol Chem. 2004 doi: 10.1074/jbc.M410010200. [DOI] [PubMed] [Google Scholar]
- 41.Yu VW, Ambartsoumian G, Verlinden L, Moir JM, Prud'homme J, Gauthier C, Roughley PJ, St-Arnaud R. FIAT represses ATF4-mediated transcription to regulate bone mass in transgenic mice. J Cell Biol. 2005;169:591–601. doi: 10.1083/jcb.200412139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grigoriadis AE, Wang ZQ, Cecchini MG, Hofstetter W, Felix R, Fleisch HA, Wagner EF. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–8. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 43.Grigoriadis AE, Wang ZQ, Wagner EF. Fos and bone cell development: lessons from a nuclear oncogene. Trends Genet. 1995;11:436–41. doi: 10.1016/s0168-9525(00)89142-8. [DOI] [PubMed] [Google Scholar]
- 44.Demiralp B, Chen HL, Koh AJ, Keller ET, McCauley LK. Anabolic actions of parathyroid hormone during bone growth are dependent on c-fos. Endocrinology. 2002;143:4038–47. doi: 10.1210/en.2002-220221. [DOI] [PubMed] [Google Scholar]
- 45.Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L, Amling M, Wagner EF. The Fos-related antigen Fra-1 is an activator of bone matrix formation. Embo J. 2004;23:2789–99. doi: 10.1038/sj.emboj.7600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jochum W, David JP, Elliott C, Wutz A, Plenk H, Matsuo K, Wagner EF. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Natural Medicine. 2000;6:980–984. doi: 10.1038/79676. [DOI] [PubMed] [Google Scholar]
- 47.Roschger P, Matsuo K, Misof BM, Tesch W, Jochum W, Wagner EF, Fratzl P, Klaushofer K. Normal mineralization and nanostructure of sclerotic bone in mice overexpressing Fra-1. Bone. 2004;34:776–82. doi: 10.1016/j.bone.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 48.Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med. 2000;6:985–90. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- 49.Kveiborg M, Sabatakos G, Chiusaroli R, Wu M, Philbrick WM, Horne WC, Baron R. DeltaFosB induces osteosclerosis and decreases adipogenesis by two independent cell-autonomous mechanisms. Mol Cell Biol. 2004;24:2820–30. doi: 10.1128/MCB.24.7.2820-2830.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutierrez S, Javed A, Tennant DK, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB. CCAAT/enhancer-binding proteins (C/EBP) beta and delta activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem. 2002;277:1316–23. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 51.McCarthy TL, Ji C, Chen Y, Kim KK, Imagawa M, Ito Y, Centrella M. Runt domain factor (Runx)-dependent effects on CCAAT/enhancer-binding protein delta expression and activity in osteoblasts. J Biol Chem. 2000;275:21746–53. doi: 10.1074/jbc.M002291200. [DOI] [PubMed] [Google Scholar]
- 52.Vallejo M, Ron D, Miller CP, Habener JF. C/ATF, a member of the activating transcription factor family of DNA-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to cAMP response elements. Proc Natl Acad Sci U S A. 1993;90:4679–83. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison JR, Huang YF, Wilson KA, Kelly PL, Adams DJ, Gronowicz GA, Clark SH. Col1a1 promoter-targeted expression of p20 CCAAT enhancer-binding protein beta (C/EBPbeta), a truncated C/EBPbeta isoform, causes osteopenia in transgenic mice. J Biol Chem. 2005;280:8117–24. doi: 10.1074/jbc.M410076200. [DOI] [PubMed] [Google Scholar]
- 54.Liu F, Lee SK, Adams DJ, Gronowicz GA, Kream BE. CREM deficiency in mice alters the response of bone to intermittent parathyroid hormone treatment. Bone. 2007;40:1135–43. doi: 10.1016/j.bone.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarthy TL, Centrella M, Canalis E. Cyclic AMP induces insulin-like growth factor I synthesis in osteoblast-enriched cultures. J Biol Chem. 1990;265:15353–6. [PubMed] [Google Scholar]
- 56.Bikle DD, Sakata T, Leary C, Elalieh H, Ginzinger D, Rosen CJ, Beamer W, Majumdar S, Halloran BP. Insulin-like growth factor I is required for the anabolic actions of parathyroid hormone on mouse bone. J Bone Miner Res. 2002;17:1570–8. doi: 10.1359/jbmr.2002.17.9.1570. [DOI] [PubMed] [Google Scholar]
- 57.Kulkarni NH, Halladay DL, Miles RR, Gilbert LM, Frolik CA, Galvin RJ, Martin TJ, Gillespie MT, Onyia JE. Effects of parathyroid hormone on Wnt signaling pathway in bone. J Cell Biochem. 2005;95:1178–90. doi: 10.1002/jcb.20506. [DOI] [PubMed] [Google Scholar]