Abstract
Background/Aim
The pathophysiology of irritable bowel syndrome (IBS) remains enigmatic; abnormalities in serotonin metabolism have been implicated. Two proteins that influence the function of serotonin and serotonergic receptors are serotonin transporter protein (SERT or soluble carrier protein SLC6A4) and p11 (S-100A10, or calpactin I light chain). Both proteins are reported to be associated with depression-like states, a frequent co-morbid condition in IBS. We explored the hypothesis that expression of these two proteins in colonic and rectal mucosa is abnormal in patients with IBS as compared to healthy controls.
Methods
mRNA expression of SLC6A4 and p11 was measured in sigmoid and rectal mucosal biopsies. Genotype of the promoter for SLC6A4 was also assessed in all participants. Validation studies explored reproducibility of two biopsies taken from the same region, and biopsies taken an average of ~3 months apart.
Results
We found normal colonic mucosal expression of SLC6A4 in diarrhea (IBS-D) or constipation predominant IBS (IBS-C). On the other hand, p11 expression was increased in IBS. No significant effect on p11 mRNA expression in sigmoid colon or rectum was noted from antidepressant treatment in any of the analyzed subgroups.
Conclusion
Colonic mucosal expression of SLC6A4 in IBS is normal. Given that overexpression of p11 can increase serotonergic receptor functions (e.g. 5-HT1B receptors), these data support the need for further study of the interaction between p11 expression in health and disease and its role in the therapeutic response to serotonergic agents, including antidepressants.
Keywords: Genotype, mucosal gene expression, IBS phenotype, SERT, SLC6A4, p11
INTRODUCTION
There is increasing evidence for disordered enteric serotoninergic signaling in IBS. Serotonin is produced in enteroendocrine cells in the gastrointestinal mucosa, and the enteric nervous system. After food ingestion or passage of a bolus through the intestine, there is release of serotonin and increased levels are observed in IBS-D (1,2). This biogenic amine is inactivated by the serotonin transporter protein (SERT), which belongs to the family of SLC 6A4 transporters (3). In one study, mRNA for SERT and tryptophan hydroxylase protein levels in rectal mucosa were found to be markedly decreased in rectal mucosa from patients with IBS, whether associated with diarrhea or constipation (4). This led to two intriguing hypotheses: first, that diarrhea resulted from the failure to inactivate serotonin and activation of receptors for serotonin that accelerate transit (e.g. 5-HT4 receptors); second, that constipation in IBS resulted from down-regulation of the receptor secondary to failure to inactivate the serotonin (4).Serotoninergic 5-HT3 receptor antagonists are used to treat diarrhea-predominant IBS while 5-HT4 receptor agonists are used to treat constipation-predominant IBS. However, it is unclear why only a subgroup of patients responds to these agents. It is conceivable that the genotype influences the response to these agents (5).
Most attention has focused on the role of 5-HT3 and 5-HT4 receptor subtypes in the enteric serotoninergic signaling. Serotoninergic 5-HT1B receptors also play a crucial role in regulating serotonin neurotransmission (6,7) The 5-HT1B/1D receptor agonist sumatriptan retarded gastric emptying in healthy subjects (8). Sumatriptan also increased gastric accommodation (9) and reduced perception of gastric distension in patients with functional dyspepsia and hypersensitivity (10). A recent study found that a protein (i.e., p11), which is a member of the S100 EF-hand protein family, increased the localization of 5-HT1B receptors to the cell surface (11). Moreover, p11 expression was reduced in humans and in animal models of depression and p11 knockout mice had features of depression (11). Conversely, mice with overexpression of p11 had increased 5-HT1B receptor function and acted as is they were treated with an antidepressant. p 11 also facilitates the translocation of annexin II to the cell surface and the functional expression of ion channels (NaV1.8, TASK-1, TRPV5/6) was shown earlier (12–15).
pH protein belongs to a family of calcium-binding proteins. Among these, S100A1, S100A4, S100A6 and S100A10 (also known as p11), but not S100B, are expressed in guinea-pig smooth muscle, and could be potentially involved in the regulation of cytoplasmic Ca(2+)-concentration and/or in signal transduction in smooth muscle (16). S100-mediated signal transduction pathways also play an important role in nervous system function, and studies implicate S100A1 in the neuronal cell dysfunction or death that occurs in Alzheimer's disease (17). Given these functions and the association between depression and IBS, it is conceivable that this protein may be relevant to functional gastrointestinal and motility disorders.
Therefore, our aim was to assess the mRNA expression of SLC6A4 and p11 in mucosal biopsies from IBS patients. A better understanding of the expression of SLC6A4 and p11 may provide mechanistic clues to IBS and improve our understanding of the variation in the efficacy of serotonergic agents in this disorder.
PATIENTS AND METHODS
Patients with Irritable Bowel Syndrome and Healthy Participants
Sixty-five participants (19–73 years old) completed a validated bowel disease questionnaire [BDQ (18) including questions that corresponded to Rome II criteria (19)]. Forty IBS participants were selected from an administrative database of 752 patients with IBS residing within 150 miles radius of Rochester, Minnesota, U.S.A, and were recruited by mailing. All IBS patients had already been evaluated by a staff gastroenterologist by clinically indicated tests including endoscopy, biopsies and tests of rectal evacuation. Patients were selected based on their predominant bowel dysfunction which was confirmed at the time of study by means of a standard questionnaire (18). Healthy volunteers were recruited by public advertisement in Rochester, MN. All participants signed informed consent for the study, which was approved by the Mayo Clinic Institutional Review Board.
Collection and RNA Isolation from Rectal and Sigmoid Colon Mucosal Biopsies
Participants attended the Mayo General Clinical Research Center to undergo a flexible sigmoidoscopy and biopsies in order to isolate mRNA from the biopsies. Due to the risk of bleeding from the biopsy procedure, participants taking aspirin, platelet inhibitors or anticoagulants were excluded if these medications could not be stopped at least one week prior to the endoscopy. Two phosphosoda enemas (Fleet® enema, C.B. Fleet, Lynchburg, VA) were administered one hour prior to sigmoidoscopy. One rectal and two sigmoid colon mucosal biopsies were collected from the 14 IBS-C, 26 IBS-D patients and 25 controls using standard large size biopsy forceps.
Ten of these subjects (5 controls and 5 IBS patients) were randomly selected to have a second, IRB-approved sigmoidoscopy two to three months after the first collection in order to assess data reproducibility. At the second examination, one rectal and one sigmoid biopsy were collected. Biopsies were taken from normal appearing mucosa only, i.e., areas with edema due to endoscope pressure were avoided. All sigmoidoscopies were performed without sedation, as is the clinical standard at Mayo Clinic, and the subjects were monitored for 60 minutes after the biopsies to ensure that they were stable for dismissal from the research center without signs of any bleeding or other complications. Immediately after collection, biopsies were submerged in 5 volumes of RNA Later Solution (Ambion Inc., Austin, TX) and stored at −20°C until analyzed. Tissue was homogenized in a mixer mill 501 (Qiagen, Venlo, The Netherlands) in RLT cell lysis buffer (Qiagen), followed by RNA extraction from the disrupted cells using the RNeasy Kit (Qiagen) with DNase treatment on the column.
Measurement of the Expression of SERT-P and p11 mRNA in Mucosal Biopsies
cDNA synthesis was performed using 500 nanograms of total RNA as template, random hexamer primers and Superscript III reverse transcriptase in a volume of 20µl during 1 hour at 50°C. This was followed by inactivation of the enzyme at 70°C for 10 minutes, according to the recommendations of the manufacturer (Invitrogen, Carlsbad, CA). Finally, real-time quantitative PCR (RTQ-PCR) was performed using the ABI Prism® 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) and the qPCR Core Kit (Eurogentec, Seraing, Belgium), with primers and probes from validated fluorogenic TaqMan gene expression assays-on-demand (Applied Biosystems). The thermal cycling conditions were 10 minutes at 95°C, followed by 45 cycles of 15 seconds at 95°C and 1 minute at 60°C. The following assays-on-demand were applied: Hs00169010_m1 (SERT, SLC6A4), Hs00741221_m1 (p11, S100A10, exon 1–2), Hs00237010_m1 (p11, S100A10, exon 2–3), and assays for control genes Hs99999903_m1 (β-actin, ACTB), Hs00193002_m1 (SART1, encoding the squamous cell carcinoma antigen recognized by T cells), and Hs99999907_m1 (β2-microglobulin, B2M). Threshold cycle (Ct) values representing the PCR cycle at which an increase in reporter fluorescence above a baseline signal was first detected were automatically calculated by the Sequence Detector Software SDS 2.1 (Applied Biosystems). Transcript quantification was performed in triplicate for every sample.
The relative quantity of our genes of interest (SLC6A4 and S100A10) was then determined by the comparative cycle threshold method for relative quantification, as described in a user bulletin of Applied Biosystems (20). Briefly, relative gene expression within one sample was expressed as ΔCt = Ctgene-of-interest − Ctcontrol-gene. Using a sigmoid colon mucosal sample from a healthy subject as the calibrator, relative gene expression was then calculated as ΔΔCt (=ΔCtsample − ΔCtcalibrator) to allow for comparison of gene expression levels of all samples in the study. Finally, the relative gene expression levels were converted and expressed as fold difference (= 2−ΔΔCt).
In order to ensure the validity of our analyses, we expressed each of the genes of interest relative to a control gene with a similar level of expression. Therefore, SLC6A4 was expressed relative to SART1, and S100A10 relative to β-2 microglobin (B2M). Because SLC6A4 was expressed relative to ACTB (β actin) in the original paper of Coates et al. (4), we also performed this analysis, despite the large difference in expression level between SLC6A4 and ACTB. A batch correction of the data was performed to avoid bias caused by technical variations during sample processing at different time points.
Genotyping SLC6A4-Promoter
Venous blood drawn from a forearm vein was stored as de-identified samples for genetic analysis. Genomic DNA was isolated from whole blood using the Purgene DNA purification performed on the Autopure LS (Gentra Systems, Minneapolis, MN). Molecular assays to genotype the SERT-P (SLC6A4) promoter were performed as described earlier (21).
Histological Assessment of Sigmoid and Rectal Biopsies
A single expert pathologist (T.S.) did a conventional histological assessment on formalin-fixed rectal and sigmoid biopsy tissue from each subject using hematoxylin and eosin–stained sections and standardized, published criteria (22).
Data and Statistical Analysis
Because the RTQ-PCR analyses were performed independently for both of the two collected biopsy samples of sigmoid mucosa per subject, the average fold difference value was calculated for each subject and used for the statistical analyses. The Kruskal-Wallis, or non-parametric ANOVA test, was used for comparing the gene expression in the three groups: controls, IBS-C, and IBS-D. When the p-value for the three-group comparison was 0.05 or less, we compared the content in each IBS group versus the control group separately using the Mann-Whitney test. All comparisons were two-tailed.
RESULTS
Patient demographics are listed in Table 1. All participants completed studies without complications.
Table 1.
Phenotypic Characteristics and Relevant Medication Use of IBS Patients and Controls
| Controls | IBS-C | IBS-D | |
|---|---|---|---|
| No. of participants | 25 | 16 | 24 |
| Caucasian (n, %) | 24 (96) | 16 (100) | 24 (100) |
| Gender (n, % female) | 23 (92) | 16 (100) | 21 (88) |
| Age (mean ± SEM) (range) | 39 ± 2 (19 – 60) | 47 ± 3 (27 – 73) | 40 ± 3 (22 – 64) |
| BMI (mean ± SEM) (range) | 26.1 ± 1.2 (18.3 – 40.2) | 25.7 ± 1.4 (20.0 – 42.6) | 29.2 ± 1.3 (20.9 – 43.6) |
| Medication Type | |||
| Laxatives/Fiber (%) | 0 | 9 (56) | 1 (4) |
| Antidiarrheals/antispasmodics (%) | 0 | 1 (6) | 4 (14) |
| Clonidine (%) | 0 | 0 | 2 (8) |
| SSRI (%) | 4 (16) | 4 (25) | 8 (33) |
| 5-HT3 Antagonists (alosetron) (%) | 0 | 0 | 2 (8) |
| 5-HT4 Agonists (tegaserod) (%) | 0 | 2 (13) | 0 |
| Other Antidepressants (%) | 2 (8) (buproprion) | 4 (25) (3 buproprion, 1 amitriptyline) | 2 (8) (buproprion, amitriptyline) |
Histological Assessment of Mucosal Biopsies
Using established criteria, hematoxylin- and eosin-stained sections of rectal and sigmoid biopsies were normal in most healthy subjects and patients with C-IBS and D-IBS. One healthy subject and 2 patients with D-IBS had focal acute colitis affecting one of two sites (i.e., either sigmoid colon or rectum) in each subject. Melanosis coli were observed in 3 patients with C-IBS and 1 patient with D-IBS. The thickness of the subepithelial collagen layer was at the upper limit of normal (i.e., 10 µm) in 1 healthy subject, 1 patient with C-IBS, and 2 patients with D-IBS. Differences among groups were not statistically significant.
Expression of SLC6A4 mRNA
The expression of SLC6A4 mRNA was not significantly different in the three groups (healthy control, IBS-C, IBS-D) in either sigmoid or rectal mucosa, whether expressed as the ratio relative to SART1 or ACTB (Figure 1A and 1B). As this was a surprising result, we performed validation studies, assessing reproducibility of SLC6A4 mRNA measurements in the two sigmoid mucosal biopsies obtained in the same biopsy session and in biopsy session several weeks apart. Thus, Figure 2 shows Bland-Altman plots of SLC6A4 mRNA expression level in two sigmoid biopsies (taken at the same sigmoidoscopy) in controls and IBS, using SART1 or ACTB as control gene. Overall Pearson correlation coefficients were 0.75 (p<0.0001) and 0.71 (p<0.0001) for SLC6A4/SART1 and SLC6A4/ACTB, respectively.
Figure 1.
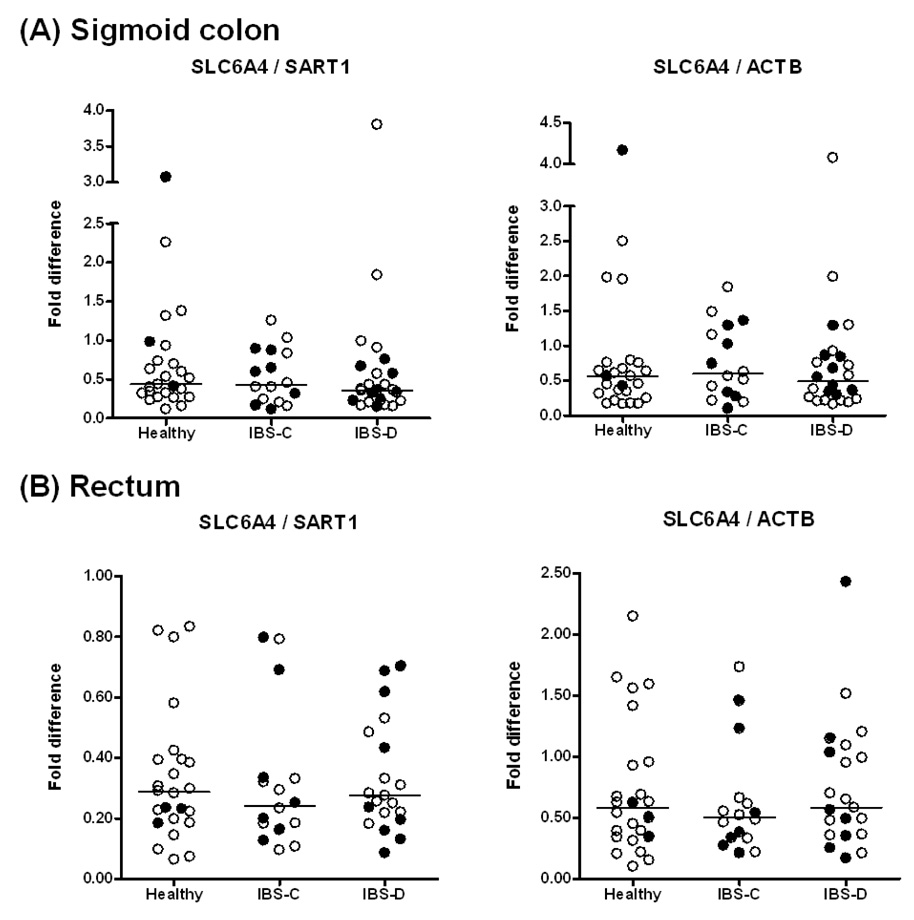
Relative SLC6A4 mRNA expression in mucosal biopsies from (A) sigmoid colon and (B) rectum in healthy controls, IBS-D and IBS-C patients. Filled circles refer to participants receiving antidepressant medications. Data are expressed relative to the control genes SART1 (left) and ACTB (right). For sigmoid colon, data points represent the average value of two samples per subject. Median values per group are shown as horizontal lines.
Figure 2.
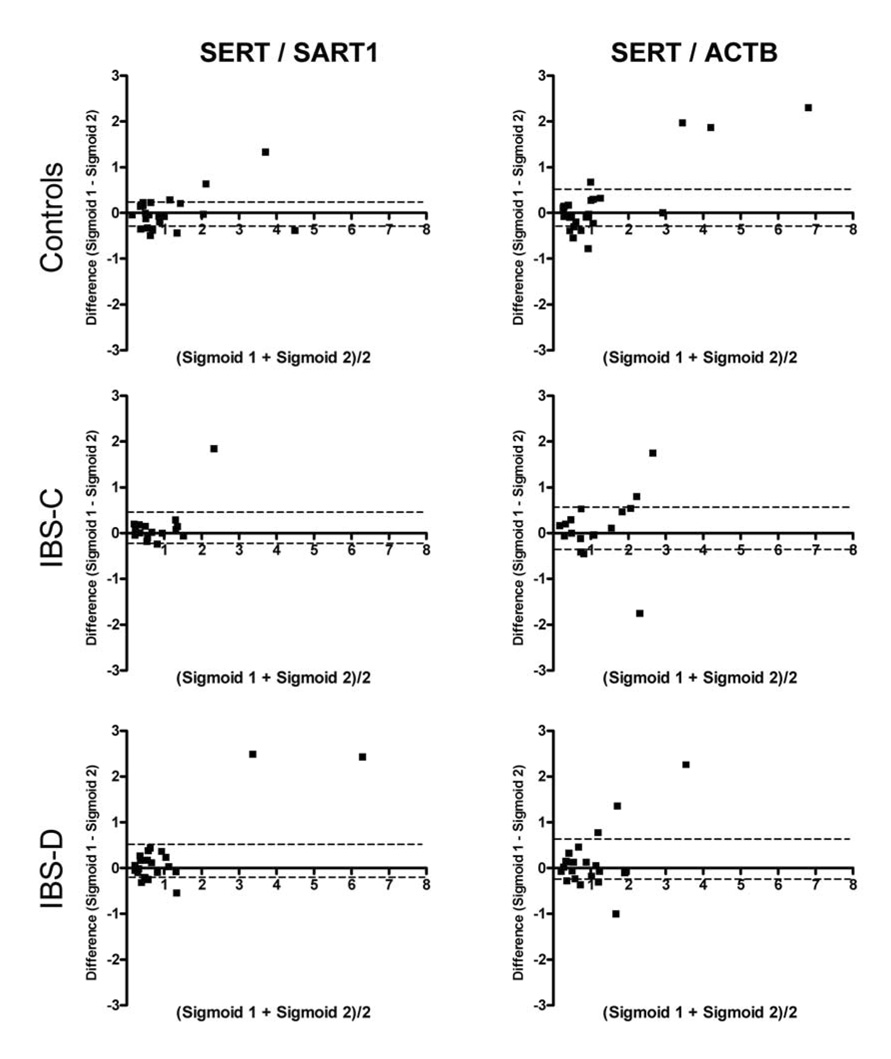
Bland-Altman plots of the relative SLC6A4 mRNA expression level in two sigmoid biopsies taken at the same sigmoidoscopy in controls and IBS. Data are expressed relative to the control genes SART1 (left) and ACTB (right). Units of both axes are fold difference of SLC6A4 over control gene. Dashed lines represent the 95% confidence intervals for the mean difference between the biopsies.
Second, we determined variation in mucosal SLC6A4 mRNA content in rectal and sigmoid colon biopsies over time in 10 randomly selected subjects (5 healthy controls and 5 IBS). Median time between the two endoscopies was 82 days (IQR 71–97, range 57–105). The Spearman correlation (rs) analysis, demonstrated also in Bland-Altman plots in Figure 3, shows that SLC6A4 expression is stable over time in sigmoid colon samples (rs > 0.7, p<0.02). This reproducibility was not observed in rectal mucosal samples (rs <0.15, p= NS). Together, these analyses demonstrate that the lack of a difference in mRNA expression of SLC6A4 in IBS patients versus healthy controls cannot be attributed to sampling error or day-to-day variability.
Figure 3.
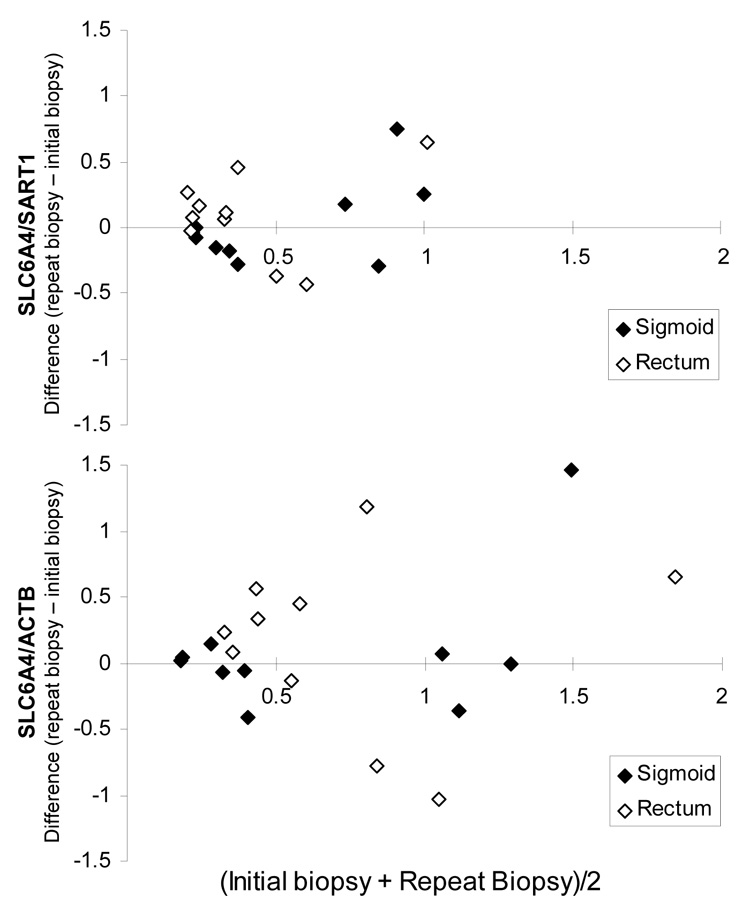
Bland-Altman plots of the relative SLC6A4 mRNA expression level within 10 subjects (5 controls and 5 IBS) in rectal and sigmoid colon mucosal biopsies over time. Median time between endoscopies was 82 days. Data are expressed relative to the control genes SART1 (top row) and ACTB (bottom row). Units of both axes are fold difference of SLC6A4 over control gene. The averaged values from the initial two sigmoid biopsies were used for this analysis.
Association of SLC6A4 Promoter Genotype and Mucosal SLC6A4 mRNA in IBS
No overall significant association was found between SLC6A4 promoter genotype and SLC6A4 mRNA expression in the entire study population (Figure 4) or in any of the phenotype groups (data not shown).
Figure 4.
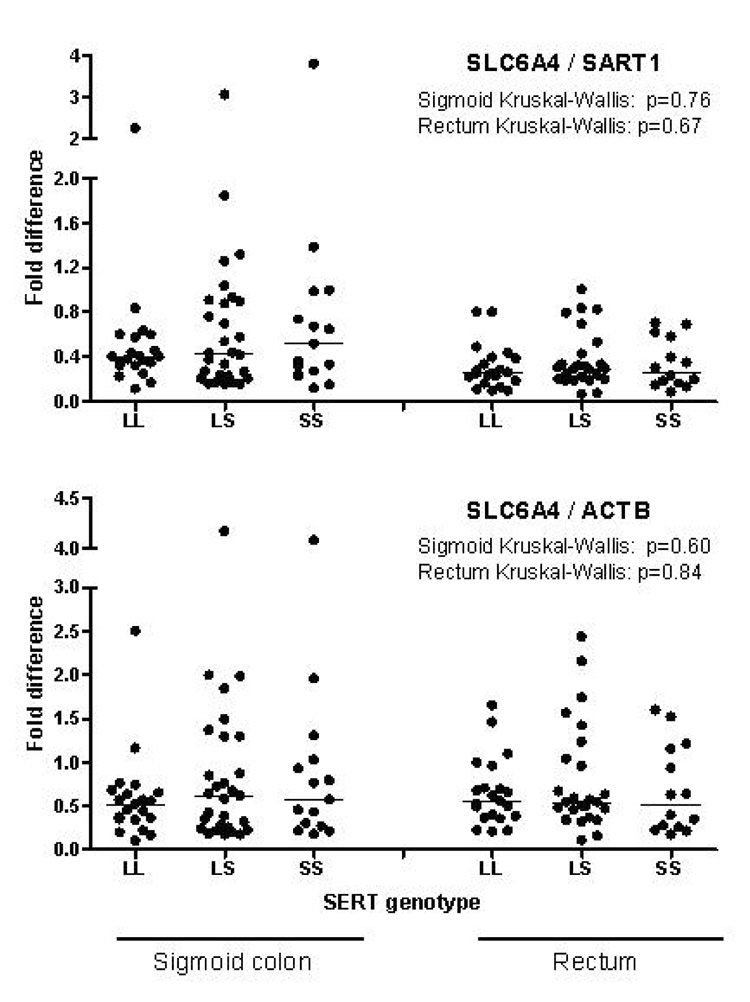
Comparison of mucosal SLC6A4 mRNA expression between SLC6A4 promoter genotypes. Data of SLC6A4 mRNA are expressed in relation to the control genes SART1 (upper panel) and ACTB (lower panel). Results from sigmoid and rectal biopsies are shown on the left and right, respectively. Horizontal lines represent medians.
p11 mRNA Expression
In contrast to the lack of difference in expression of SLC6A4, a significant increase was observed in the mRNA expression of p11 in the sigmoid mucosa of IBS patients. Figure 5 shows the mRNA expression of p11 (S100A10) relative to β2 microglobulin (B2M) as measured by two different RTQ-PCR assays. According to both assays, Hs00741221_m1, spanning p11 exons 1 and 2 and Hs00237010_m1, spanning p11 exons 2 and 3, a significant increase was found in IBS-C patients compared to controls. It should be noted that several IBS-D individuals also had higher p11 mRNA expression on both assays, though statistical significance was not achieved when comparing median group values. However, when IBS-D and IBS-C patients were pooled into one group of IBS patients and compared to the group of healthy controls, p11 mRNA was significantly increased relative to controls for both RTQ-PCR assays ( p<0.016 and p<0.013 respectively for p11 exon 1–2 and p11 exon 2–3).
Figure 5.
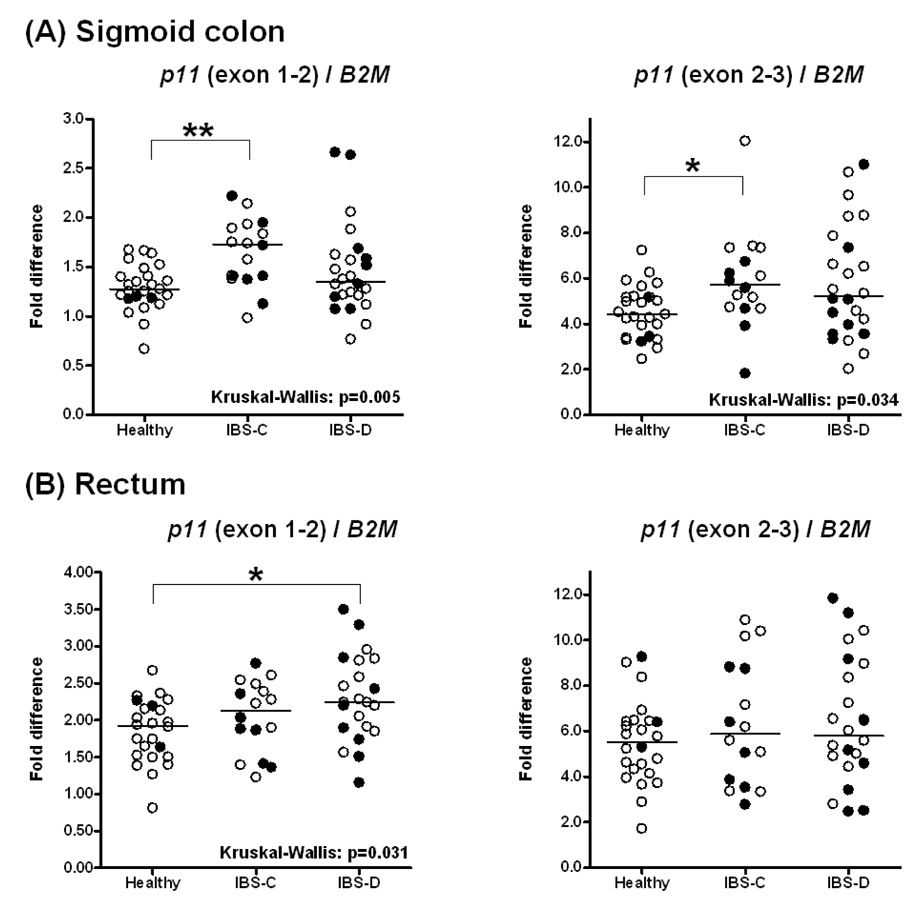
S100A10 (p11) mRNA expression in mucosal biopsies from (A) sigmoid colon and (B) rectum in healthy controls and IBS-D and IBS-C patients based on two different RTQ-PCR TaqMan assays (p11 exon 1–2 and p11 exon 2–3). Filled circles refer to participants receiving antidepressant medications. Data are expressed relative to the control gene B2M. For sigmoid colon, data points represent the average value of two samples per subject. Median values per group are shown as horizontal lines. Asterisks represent p-values <0.05 (*) or <0.01 (**) in two-group comparisons following significant Kruskal-Wallis tests.
In rectal mucosal samples, a significantly higher p11 expression compared to the control group was observed in the exon 1–2 assay for the IBS-D subgroup, but no statistical significance was reached for the IBS-C group‥ When all IBS patients were pooled into one group, a significantly (p<0.018) higher expression was found relative to controls. No significant effect on p11 mRNA expression in sigmoid colon or rectum was noted from antidepressant treatment in any of the analyzed subgroups [healthy, IBS-C, IBS-D (Figure 5)].
DISCUSSION
This is the first demonstration that the sigmoid mucosal expression of p11, a protein critical to 5-HT1B receptor functions, was increased in IBS. However, in contrast to a previous study (4), mRNA expression of SLC6A4 in the sigmoid and rectal mucosa, assessed on two separate occasions in a subset of subjects, was not significantly different in patients with IBS-D or IBS-C relative to controls. Moreover, there was no statistically significant relationship between the germ line DNA (SLC6A4 promoter genotype) and sigmoid or rectal mucosal SLC6A4 mRNA expression.
We considered several potential explanations for these differences between this and the previous study in the published literature (4).
First, we considered whether the control gene’s expression may have influenced the relative expression of the mRNA for SLC6A4. In our study, the SLC6A4 mRNA expression data were analyzed using two control genes, SART1 and ACTB. We noted somewhat greater deviation from the line of identity in the ratio of SLC6A4 to ACTB than the ratio to SART1 in these mucosal samples, as seen in Figure 2. The higher correlations that are found when using SART1 as the control gene may be explained by the large difference in the expression level between SLC6A4 and ACTB. This is also demonstrated by Coates et al, who measured a 105–106 difference in rectal mucosal concentrations of SLC6A4 and ACTB mRNA. In contrast, this difference in mucosal concentration between SCL6A4 and SART1 is much smaller (~102). However, irrespective of the control gene (ACTB or SART1), mRNA for SCL6A4 was not different between IBS and controls.
Second, we considered the possibility that, in contrast to the observation of Coates et al (4), our controls may have had low levels of expression of SCL6A4 mRNA, and this low level may have resulted from low grade inflammation as a result of coincidental prior gastroenteritis or intake of nonsteroidal agents. Thus, we carefully examined the rectal and sigmoid mucosal biopsies for evidence of inflammation. The histopathological examination, performed by a single histopathologist who was blinded to the clinical diagnosis, suggests that there were no features to suggest low grade inflammation in the IBS or control participants. Hence, the normal level of expression of SCL6A4 mRNA in IBS patients relative to healthy controls cannot be attributed to subclinical inflammation reducing expression of SCL6A4 in the controls.
Third, we considered the possibility that SLC6A4 expression fluctuates over time. Therefore we repeated biopsies in a subset of subjects. Taken together, the results demonstrate that the results for SLC6A4 expression from the two initial sigmoid biopsies were highly correlated. Moreover, SLC6A4 expression within subjects was highly correlated between the initial and subsequent biopsies taken almost three months later.
Fourth, we considered the possibility that concomitant medications influenced expression of SLC6A4. Thus, SSRIs affect SLC6A4 activity; however, in our study, an equal proportion of participants in each group was taking an SSRI. Moreover, Coates et al (4) observed that the use of an SSRI, a 5-HT4 receptor agonist, or a non-serotonin-specific antidepressant had no effect on any of the elements of 5-HT signaling evaluated in their study.
It is unclear why mucosal SLC6A4 mRNA expression differed between the sigmoid and rectal mucosa in our study. Participants did receive a sodium phosphate enema two hours prior to the procedure in our study. Details on bowel preparation of participants were not provided in the paper by Coates et al (4). Overall, we observed that data from biopsies of the sigmoid mucosa are more reproducible, and these data suggest that this region may represent a favorable site for monitoring changes in SLC6A4 expression levels, for example in response to drug treatment. Further studies of the expression of key functional proteins in sigmoid and rectal mucosa are awaited to determine whether there are systematic differences in mucosal expression of these proteins.
We observed over-expression of p11 mRNA in sigmoid mucosa from patients with IBS compared to controls. This is in contrast to the deficit in p11 expression in depression (11,23) and of SLC6A4 expression or genetics in anxiety, life event stress responses and depression (3,24–26). Though the effects of increased p11 mRNA expression on gastrointestinal sensorimotor functions are unknown, p11 is co-localized with 5-HT1B receptors, stimulation of which is known to relax the gastric fundus and delay stomach emptying in humans. Thus, it is certainly conceivable that increased p11 might retard colonic transit through activation of the serotonergic receptors by serotonin released in response to luminal chemical or mechanical stimuli. Also, the physiological significance of increased p11 mRNA expression in at least a subset of IBS-D patients is, unclear and awaits further study of the potential role of p11 in mediating response to stimulation of other serotonergic receptors.
p11 is known to be involved in trafficking members of the voltage-gated sodium and potassium channel families as well as transient receptor potential and chloride channels. It plays a selective role in enhancing functional expression of ASIC1a (27). ASICs are involved in visceral nociception and, hence, p11 signaling may also be relevant to the pain component of IBS (28). Thus, over-expression of p11 may conceivably contribute to visceral hypersensitivity or pain experienced in IBS.
Antidepressants regulate p11 expression to restore normal functions. In view of the normal or elevated expression of p11 in individual IBS patients, this variation may also explain the relative variation in efficacy of antidepressants in the treatment of IBS (29) whose function in other neural tissues is altered by the expression of p11 mRNA. Further studies are needed to explore whether concomitant anxiety or depression influence p11 expression and the response to antidepressants. However, our preliminary analysis on the effect of treatment with antidepressants in this observational study did not reveal any significant effect on mucosal p11 mRNA expression. We also perceive that these data should stimulate further studies of the p11 expression at the protein level, the physiological correlates of this expression, the effects of serotonergic agents and antidepressants on p11 expression, and its effects on colonic motor and sensory functions.
In summary, over-expression of p11 in IBS has the potential to modulate the function of serotonergic receptors, including 5-HT1B receptors, to induce symptoms. The normal mRNA expression of SLC6A4 in rectal and sigmoid mucosa of well characterized patients with IBS raises questions about the association reported in another study.
Acknowledgment
This study was supported in part by General Clinical Research Center grant RR00585. Dr. Camilleri is supported by grants R01 DK54681 and K24 DK02638 from National Institutes of Health. This work was also supported by a Research grant from Johnson & Johnson Pharmaceutical Research & Development. We thank Mrs. Cindy Stanislav for excellent secretarial assistance and Mr. Thomas Ford for assistance with questionnaire analysis.
Conflict of Interest Disclosure
The research was supported by a grant from Johnson & Johnson Pharmaceutical Research & Development. The following authors are employees of Johnson & Johnson Pharmaceutical Research & Development: Jeroen Aerssens, Leen Thielemans, Hinrich Göhlmann, Ilse Van Den Wyngaert, and Bernard Coulie.
Abbreviations used
- IBS
irritable bowel syndrome
- IBS-C
irritable bowel syndrome with predominant constipation
- IBS-D
irritable bowel syndrome with predominant diarrhea
- NS
not significant
- SERT
serotonin transporter
- SERT-P
serotonin transporter promoter
- SLC6A4
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael Camilleri, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Christopher N. Andrews, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Adil E. Bharucha, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Paula J. Carlson, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Irene Ferber, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Debra Stephens, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Thomas C. Smyrk, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Raul Urrutia, Clinical Enteric Neuroscience Translational and Epidemiological Research, (C.E.N.T.E.R.) Group, and Gastroenterology Research Unit, Mayo Clinic College of Medicine, Rochester, Minnesota
Jeroen Aerssens, Johnson & Johnson Pharmaceutical Research & Development, Beerse, Belgium
Leen Thielemans, Johnson & Johnson Pharmaceutical Research & Development, Beerse, Belgium
Hinrich Göhlmann, Johnson & Johnson Pharmaceutical Research & Development, Beerse, Belgium
Ilse Van Den Wyngaert, Johnson & Johnson Pharmaceutical Research & Development, Beerse, Belgium
Bernard Coulie, Johnson & Johnson Pharmaceutical Research & Development, Beerse, Belgium
REFERENCES
- 1.Dunlop SP, Coleman NS, Blackshaw E, Perkins AC, Singh G, Marsden CA, Spiller RC. Abnormalities of 5-hydroxytryptamine metabolism in irritable bowel syndrome. Clin Gastroenterol Hepatol. 2005;3:349–357. doi: 10.1016/s1542-3565(04)00726-8. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson W, Lockhart S, Whorwell PJ, Keevil B, Houghton LA. Altered 5-hydroxytryptamine signaling in patients with constipation- and diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2006;130:34–43. doi: 10.1053/j.gastro.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 3.Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, Benjamin J, Muller CR, Hamer DH, Murphy DL. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science. 1996;274:1527–1531. doi: 10.1126/science.274.5292.1527. [DOI] [PubMed] [Google Scholar]
- 4.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, Moses PL. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrome. Gastroenterology. 2004;126:1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Camilleri M, Atanasova E, Carlson PJ, Ahmad U, Kim HJ, Viramontes BE, McKinzie S, Urrutia R. Serotonin-transporter polymorphism pharmacogenetics in diarrhea-predominant irritable bowel syndrome. Gastroenterology. 2002;123:425–432. doi: 10.1053/gast.2002.34780. [DOI] [PubMed] [Google Scholar]
- 6.Maroteaux L, Saudou F, Amlaiky N, Boschert U, Plassat JL, Hen R. Mouse 5HT1B serotonin receptor: cloning, functional expression, and localization in motor control centers. Proc Natl Acad Sci USA. 1992;89:3020–3024. doi: 10.1073/pnas.89.7.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moret C, Briley M. The possible role of 5-HT(1B/D) receptors in psychiatric disorders and their potential as a target for therapy. Eur J Pharmacol. 2000;404:1–12. doi: 10.1016/s0014-2999(00)00581-1. [DOI] [PubMed] [Google Scholar]
- 8.Coulie B, Tack J, Maes B, Geypens B, De Roo M, Janssens J. Sumatriptan, a selective 5-HT1 receptor agonist, induces a lag phase for gastric emptying of liquids in humans. Am J Physiol. 1997;272:G902–G908. doi: 10.1152/ajpgi.1997.272.4.G902. [DOI] [PubMed] [Google Scholar]
- 9.Boeckxstaens GE, Hirsch DP, Kuiken SD, Heisterkamp SH, Tytgat GN. The proximal stomach and postprandial symptoms in functional dyspeptics. Am J Gastroenterol. 2002;97:40–48. doi: 10.1111/j.1572-0241.2002.05421.x. [DOI] [PubMed] [Google Scholar]
- 10.Tack J, Caenepeel P, Corsetti M, Janssens J. Role of tension receptors in dyspeptic patients with hypersensitivity to gastric distention. Gastroenterology. 2004;127:1058–1066. doi: 10.1053/j.gastro.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Svenningsson P, Chergui K, Rachleff I, Flajolet M, Zhang X, El Yacoubi M, Vaugeois JM, Nomikos GG, Greengard P. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- 12.Deora AB, Kreitzer G, Jacovina AT, Hajjar KA. An annexin 2 phosphorylation switch mediates p11-dependent translocation of annexin 2 to the cell surface. J Biol Chem. 2004;279:43411–43418. doi: 10.1074/jbc.M408078200. [DOI] [PubMed] [Google Scholar]
- 13.Okuse K, Malik-Hall M, Baker MD, Poon WY, Kong H, Chao MV, Wood JN. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature. 2002;417:653–656. doi: 10.1038/nature00781. [DOI] [PubMed] [Google Scholar]
- 14.van de Graaf SF, Hoenderop JG, Gkika D, Lamers D, Prenen J, Rescher U, Gerke V, Staub O, Nilius B, Bindels RJ. Functional expression of the epithelial Ca(2+) channels (TRPV5 and TRPV6) requires association of the S100A10-annexin 2 complex. EMBO J. 2003;22:1478–1487. doi: 10.1093/emboj/cdg162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard C, Tinel N, Terrenoire C, Romey G, Lazdunski M, Borsotto M. p11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. EMBO J. 2002;21:4439–4448. doi: 10.1093/emboj/cdf469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daub B, Schroeter M, Pfitzer G, Ganitkevich V. Expression of members of the S100 Ca2+-binding protein family in guinea-pig smooth muscle. Cell Calcium. 2003;33:1–10. doi: 10.1016/s0143-4160(02)00167-7. [DOI] [PubMed] [Google Scholar]
- 17.Zimmer DB, Chaplin J, Baldwin A, Rast M. S100-mediated signal transduction in the nervous system and neurological diseases. Cell Mol Biol. 2005;51:201–214. [PubMed] [Google Scholar]
- 18.Talley NJ, Phillips SF, Wiltgen CM, Zinsmeister AR, Melton LJ., 3rd Assessment of functional gastrointestinal disease: the bowel disease questionnaire. Mayo Clin Proc. 1990;65:1456–1479. doi: 10.1016/s0025-6196(12)62169-7. [DOI] [PubMed] [Google Scholar]
- 19.Thompson WG, Longstreth GF, Drossman DA, Heaton KW, Irvine EJ, Muller-Lissner SA. Functional bowel disorders and functional abdominal pain. Gut. 1999;45 Suppl 2:II43–II47. doi: 10.1136/gut.45.2008.ii43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ. ABI Prism 7700 Sequence Detection System, User Bulletin 2, PE Applied Biosystems. 1997 [Google Scholar]
- 21.Kim HJ, Camilleri M, Carlson PJ, Cremonini F, Ferber I, Stephens D, McKinzie S, Zinsmeister AR, Urrutia R. Association of distinct alpha(2) adrenoceptor and serotonin transporter polymorphisms with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53:829–837. doi: 10.1136/gut.2003.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins D, Balsitis M, Gallivan S, et al. Guidelines for the initial biopsy diagnosis of suspected chronic idiopathic inflammatory bowel disease. The British Society of Gastroenterology Initiative. J Clin Pathol. 1997;50:93–105. doi: 10.1136/jcp.50.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp T. Neuroscience. A new molecule to brighten the mood. Science. 2006;311:45–46. doi: 10.1126/science.1122819. [DOI] [PubMed] [Google Scholar]
- 24.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 25.Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, Egan MF, Weinberger DR. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 26.Sibille E, Lewis DA. SERT-ainly involved in depression, but when? Am J Psychiatry. 2006;163:8–11. doi: 10.1176/appi.ajp.163.1.8. [DOI] [PubMed] [Google Scholar]
- 27.Donier E, Rugiero F, Okuse K, Wood JN. Annexin II light chain p11 promotes functional expression of acid-sensing ion channel ASIC1a. J Biol Chem. 2005;280:38666–38672. doi: 10.1074/jbc.M505981200. [DOI] [PubMed] [Google Scholar]
- 28.Cervero F, Laird JM. Understanding the signaling and transmission of visceral nociceptive events. J Neurobiol. 2004;61:45–54. doi: 10.1002/neu.20084. [DOI] [PubMed] [Google Scholar]
- 29.Quartero AO, Meineche-Schmidt V, Muris J, Rubin G, de Wit N. Bulking agents, antispasmodic and antidepressant medication for the treatment of irritable bowel syndrome. Cochrane Database Syst Rev. 2005;2:CD003460. doi: 10.1002/14651858.CD003460.pub2. [DOI] [PubMed] [Google Scholar]


