Abstract
Reassortants between type A and B influenza viruses have not been detected in nature, although both viruses co-circulate in human populations. One explanation for this may be functional incompatibility of RNA transcription and replication between type A and B viruses. To test this possibility, we constructed type A/B mosaic polymerase machinery, containing PB2, PB1, PA and nucleoprotein from each of the two virus types, and assessed their polymerase activities with a type A promoter in a reporter assay. Type B polymerase machinery containing homologous components was functional with the type A promoter albeit to various extents depending on the segments from which the regions downstream of the promoter sequence were derived, indicating functional compatibility between the type A promoter and B polymerase machinery. However, all of the A/B mosaic polymerase machinery, except that containing PA from a type A and the others from a type B virus strain, did not function with the type A promoter, indicating limited compatibility among polymerase components of both types. Taken together, these data suggest that incompatibility among components of the polymerase machinery for RNA transcription and replication alone is not responsible for the lack of heterotypic reassortants.
Keywords: Influenza virus, RNA polymerase
1. Introduction
The genomes of influenza A and B viruses each consist of eight single-stranded RNA segments of negative polarity (Palese and Shaw, 2007). In cells infected with two different type A viruses, homotypic reassortants possessing various combinations of gene segments are readily produced (Wright et al., 2007). However, intertypic reassortants between type A and B viruses have not been detected in nature or in vitro, although both viruses co-circulate in human populations, suggesting the intertypic incompatibility at the RNA level, protein level, or both (Kaverin et al., 1983; Mikheeva and Ghendon, 1982; Tobita et al., 1983).
By contrast, several artificial A/B chimeric viruses have been made by reverse genetics. Muster et al. (1991) produced a mutant type A virus whose non-coding regions (NCRs) of the neuraminidase (NA) segment were replaced with those of the type B NS segment, suggesting that at least the NCRs of type B NS are compatible with type A components with respect to RNA transcription, replication, and packaging. In addition, we have shown that the type A polymerase machinery can express type B HA from the type B HA segment (Horimoto et al., 2003). These observations may be surprising because, although terminal sequences at both ends of the NCRs containing the promoter sequences needed for RNA transcription and replication are conserved among viruses of the same type, they differ between type A and B RNA segments. By contrast, Muster et al. (1991) were unable to generate a mutant virus in which the NCRs for the type A NA segment were replaced with those of the type B NA segment. These data together suggest that functional compatibility between the type A and B NCRs depends on the specific RNA segment.
To gain further insight into heterotypic incompatibility between type A and B viruses at the levels of RNA transcription and replication, here we used a series of reporter constructs containing the NCRs of each type A segment, such that virus RNA (vRNA)-like transcription and replication could be evaluated quantitatively and systemically. Using this series of reporter constructs, we evaluated the activity of A/B mosaic polymerase machinery containing various combinations of polymerase components with respect to their ability to drive the type A promoter.
2. Materials and methods
2.1. Cells
293T human embryonic kidney cells and Madin-Darby canine kidney (MDCK) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS) and in minimal essential medium (MEM) containing 5% newborn calf serum, respectively. The 293T cell line is a derivative of the 293 line into which the gene for the simian virus 40 T antigen was inserted (DuBridge et al., 1987). All cells were maintained at 37°C in 5% CO2.
2.2. Plasmids
A series of plasmids, designated pPolI-χ-SEAP (where χ represents PB2, PB1, PA, HA, NP, NA, M, or NS), each containing the secreted alkaline phosphatase (SEAP) gene flanked by the NCRs of segments from A/WSN/33 (H1N1; A/WSN) between the human RNA polymerase I promoter and the mouse RNA polymerase I terminator were described previously (Maeda et al., 2004). Plasmids constructed to express the PB2, PB1, PA and NP proteins of A/WSN, A/duck/Guangxi/35/2001 (H5N1; A/DKGX) (Li et al., 2005), B/Lee/40 (B/Lee) or B/Panama/45/90 (B/Panama) were inserted into the eukaryotic protein expression vector pCAGGS/MCS (controlled by the chicken β-actin promoter) (Kobasa et al., 1997; Niwa et al., 1991) as described previously (Neumann et al., 1999).
2.3. Expression and measurement of SEAP activity
293T cells in 12-well plates were transfected with 1 μg of each pPolI-χ-SEAP plasmid and with 1 μg each of plasmids designed to express the four influenza viral proteins (PB2, PB1, PA and NP). At 48 hrs post-transfection, SEAP activity in the supernatant was measured by using the Alkaline Phosphatase Yellow (pNPP) liquid substrate system (Sigma Chemical Co., USA) as described previously (Maeda et al., 2004).
2.4. Generation of viruses by plasmid-based reverse genetics
Viruses were generated by transfecting 293T cells with eight plasmids designed to express the vRNA segments, together with the four plasmids designed to express the viral proteins (PB2, PB1, PA and NP) as described previously (Neumann et al., 1999). The viruses in the supernatant of the 293T cells were harvested 48 hrs post-transfection. Virus titers were determined by a plaque assay using MDCK cells.
3. Results
3.1. Activity of type B polymerase machinery with a type A promoter
We constructed a series of plasmids for the synthesis of model vRNAs containing the SEAP open reading frame, instead of the original coding sequences of A/WSN, flanked by the NCRs of one of its segments (Maeda et al., 2004). These model vRNAs are transcribed and replicated only in the presence of functional viral polymerase machinery, composed of three polymerase components (PB2, PB1 and PA) and NP. Upon transfection of 293T cells with one of the model vRNA-synthesizing plasmids together with four plasmids expressing PB2, PB1, PA and NP, levels of transcription and replication for each model vRNA can be evaluated as SEAP activity in cell culture supernatant.
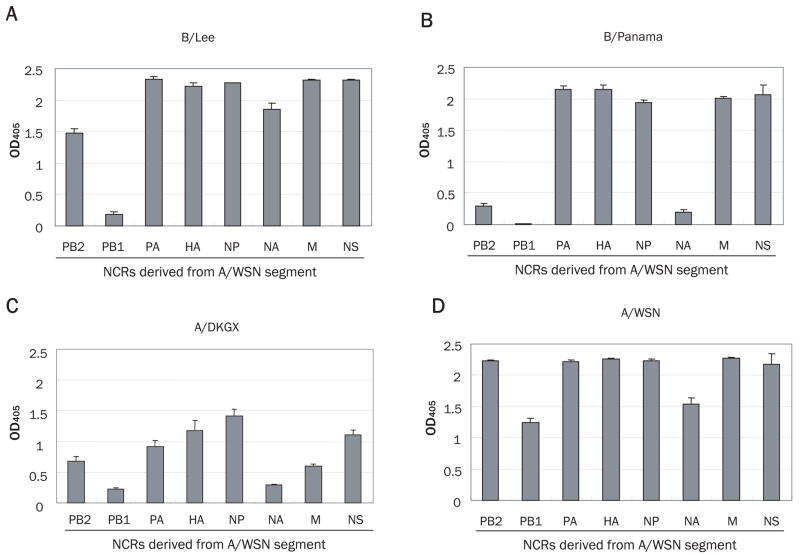
We first measured the SEAP activities of the model vRNAs with A/WSN promoter sequences by the type B polymerase machinery derived from the B/Lee (Fig. 1A) or B/Panama (Fig. 1B) strains, and by type A polymerase machinery derived from A/DKGX (Fig. 1C) or homologous A/WSN as a control (Fig. 1D). The levels of SEAP activity with the B/Lee polymerase machinery were equivalent to those observed with A/WSN (homologous control) in all vRNA-like constructs, except for the PB1 segment. The SEAP value for the PB1 construct with B/Lee polymerase machinery was significantly lower than that for the control A/WSN machinery (p<0.05 by t-test). By contrast, significantly lower levels (p<0.05) of SEAP were detected with the PB2, PB1 and NA segments when the B/Panama polymerase machinery was used. These findings indicate that activity for the type B polymerase machinery with a type A promoter depends on the specific RNA segment and that the levels of activity differ between type B strains. Interestingly, the SEAP activities with all of the A/WSN promoter sequences and the A/DKGX polymerase were significantly lower than for the homologous WSN control, and even for the type B polymerase.
Fig. 1.
Activity of type B polymerase machinery with a type A promoter (non-coding region [NCR] derived from A/WSN segment). Polymerase activities are quantified by use of a SEAP assay of the supernatants of cells transfected with (A) B/Lee-, (B) B/Panama-, or (C) control A/WSN-derived plasmids. The SEAP units are expressed as absorbance values (optical density [OD405]) and are reported as means ± standard deviation (n=3).
To further test the limited activity of the B/Panama polymerase machinery with the type A promoter, we attempted to generate A/WSN virus by plasmid-based reverse genetics using the B/Panama polymerase machinery. The virus was produced in the supernatant of the transfected cells albeit to a lesser extent (4.7x104 PFU/ml) than when using the homologous WSN polymerase machinery (2.1x108 PFU/ml). This finding indicates that type B polymerase machinery is compatible with the type A promoter at the level of RNA transcription and replication.
3.2. Activity of type A/B mosaic polymerase machinery with the type A promoter
To investigate the compatibility among type A and B polymerase components and NP for polymerase activity with the type A promoter, we tested a total of 14 combinations of types A and B polymerase components and NP (Fig. 2A, lanes 2–15), and measured their polymerase activities using a reporter segment that possessed the type A HA segment promoter. Among all combinations tested, only the combination containing PA from A/WSN and NP, PB2 and PB1 from B/Panama (lane 15) showed a level of polymerase activity equivalent to type A (lane 1) or the B homologous control (lane 16); however, the same combination was not functional with B/Lee-derived NP, PB2 and PB1. Similar results were obtained when type A promoters of other segments such as PA, NP, M and NS were used (data not shown). Also, the reverse combination containing PA from B/Panama and NP, PB2 and PB from A/WSN did not show any polymerase activity (lane 5). Thus, the compatibility of PA function between A/WSN and B/Panama is unidirectional. However, the polymerase components and NP of type A and B viruses are largely incompatible with a type A virus promoter.
Fig. 2.
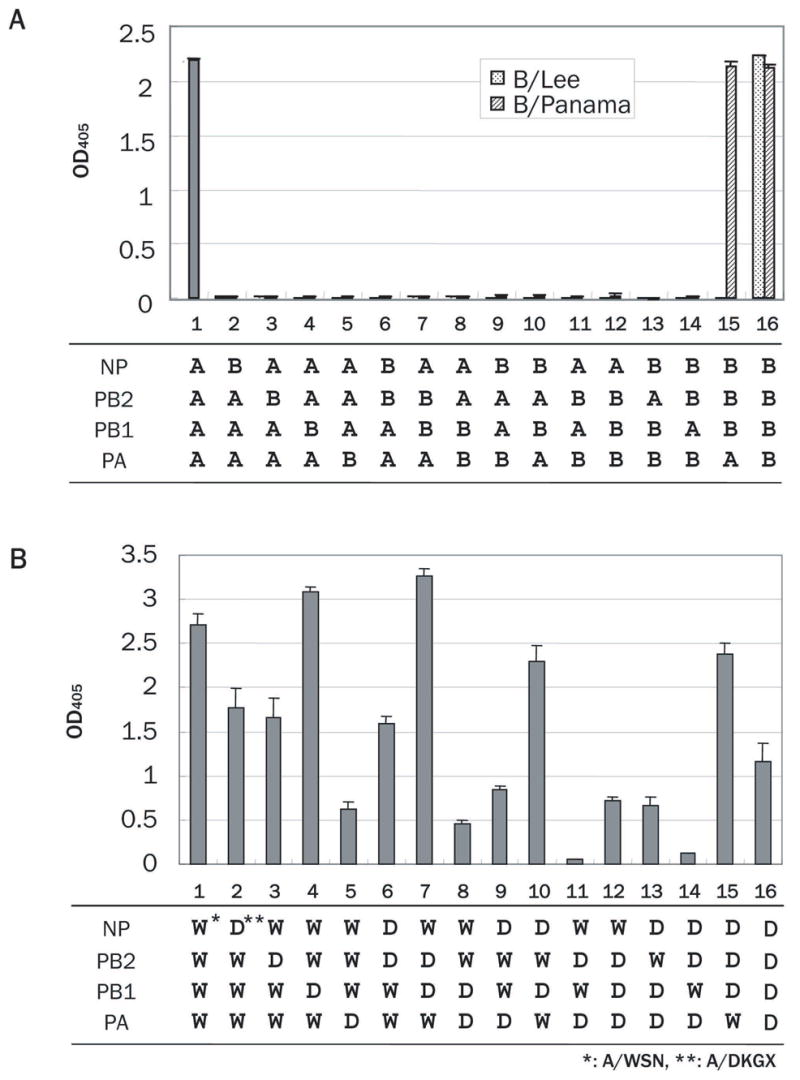
Activity of type A/B mosaic polymerase machinery with a type A HA segment promoter. Combinations of type A and B components are shown at the bottom. Polymerase activities are quantified by use of a SEAP assay with B/Lee-, or B/Panama-derived components as described in the legend to Fig. 1. As controls, SEAP values for homologous type A (lane 1) and type B (lane 16) polymerase machinery are shown.
We, then, tested the activities of various combinations of the polymerase components and NP from the two type B viruses, B/Panama and B/Lee, using a reporter segment possessing the A/WSN HA segment promoter. All 14 combinations of these polymerase components and NP showed similar SEAP activity of more than 2.0, indicating high compatibility between the two type B virus proteins (data not shown). By contrast, the activities of combinations of polymerase components and NP from A/WSN and A/DKGX varied; in particular, all of the combinations containing A/DKGX PA had substantially reduced polymerase activity (Fig. 2B; lanes 5, 8, 9, 11, 12, 13, and 14).
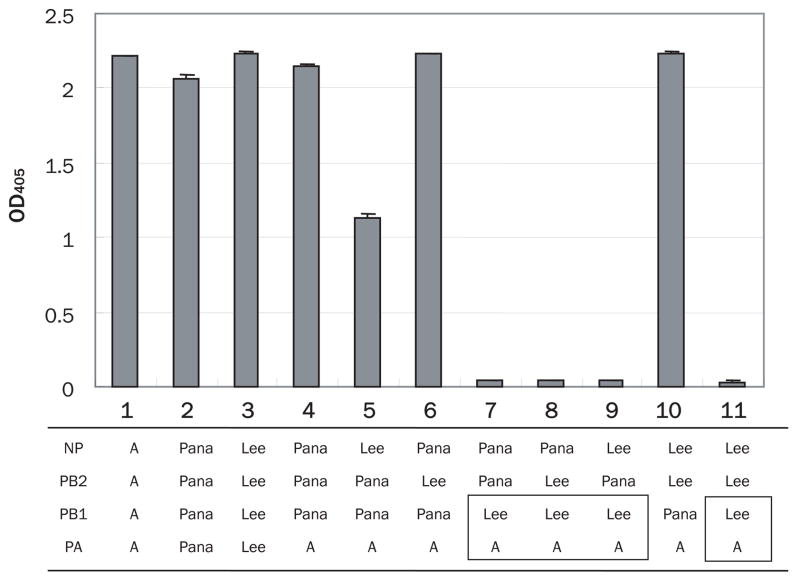
Since the only functional heterotypic polymerase/NP combination possessed PA from type A virus and the rest from B/Panama, we next attempted to determine which polymerase components plus NP of type B viruses are responsible for the formation of functional heterotypic polymerase machinery. To this end, we tested various combinations of type B polymerase components and NP with A/WSN PA (Fig. 3). We found that all of the polymerase machinery containing B/Lee PB1 was non-functional, suggesting that the PA of A/WSN could interact with PB1 from B/Panama but not from B/Lee.
Fig. 3.
Activity of type A/B mosaic polymerase machinery, containing type A PA and the other components from type B, with a type A HA segment promoters Combinations of type B components from B/Panama or B/Lee are shown at the bottom. Polymerase activities are quantified by use of a SEAP assay as described in the legend to Fig. 1. As controls, SEAP values for homologous type A (lane 1) and type B (lane 2 for B/Panama and lane 3 for B/Lee) polymerase machinery are shown.
4. Discussion
Influenza A and B viruses can form homotypic but not heterotypic reassortants. To gain insight into this phenomenon, we assessed the compatibility between type A and B polymerase machinery. Our findings indicate that homologous type B polymerase machinery is functional with the type A promoter albeit to various extents depending upon the segments from which the regions downstream of the promoter sequence were derived. Thus, lack of intertypic reassortment between type A and B viruses cannot be explained by incompatibility at the RNA transcription and replication levels. Although most combinations of polymerase components and NP between type A and B viruses were not functional, one combination did indeed exhibit activity equivalent to either parental polymerase. Previously, we successfully generated type A/B chimeric virus, which contained type B HA in a type A virus background, indicating functional compatibility between type A and B HA glycoproteins (Horimoto et al., 2003). In addition, two studies have reported that type B NA can replace the function of type A NA (Ghate and Air, 1999; Flandorfer et al., 2003). Taken together with our current findings, the lack of intertypic reassortants cannot be explained by incompatibilities at the protein level. Recently, we (K. Fujii et al., 2005; Y. Fujii et al., 2003; Muramoto et al., 2006; Ozawa et al., 2007; Watanabe et al., 2003) and others (De Wit et al., 2006; Dos Santos Afonso et al., 2005; Gog et al., 2007; Liang et al., 2005, 2008; Marsh et al., 2008) have identified regions in the RNA segments of type A viruses that are essential for their incorporation into virions. It may be that incompatibility in these sequences between type A and B viral RNA segments is responsible for the lack of intertypic reassortants.
Interestingly, the activity of the A/DKGX polymerase machinery with A/WSN promoters was significantly lower than that of the homologous WSN control, and even that of the type B polymerase. It may be that the polymerase machinery of this highly pathogenic H5N1 virus may not function optimally in 293T human cells, as used this assay, which would suggest host specificity for the type A polymerase machinery. Our finding that the type A mosaic polymerase machinery containing PA from A/DKGX had reduced polymerase activity suggests that the PA subunit may play a role in the host specificity of type A polymerase activity.
Here, we found that homologous type B polymerase machinery is functional with type A promoters, as reported earlier (Crescenzo-Chaigne et al., 1999), but that the extent of activity depends on the RNA segments from which the NCRs are derived. The NCRs of each segment contain sequences conserved among all eight RNA segments as well as downstream segment-specific sequences (Desselberger et al., 1980). Although promoter activity is mainly determined by the former region (Portela et al., 1999), the latter region could affect the polymerase activity of the type B polymerase machinery with a type A promoter.
We have shown in this study that the PA of A/WSN interacts with PB1 of B/Panama but not with that of B/Lee. Previous studies have reported that direct binding between the carboxyl-terminal region of PA and the amino-terminal region of PB1 is required for functional polymerase machinery (Perez and Donis, 2001). Thus, it is possible that B/Panama PB1 can bind to type A PA, but that B/Lee PB1 cannot. Further studies at the molecular level may shed light on the interplay between PA and PB1 in the polymerase machinery.
Acknowledgments
We thank Sue Watson for editing the manuscript. This work was supported by Grants-in-Aid for Scientific Research and by Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases from the Ministries of Education, Culture, Sports, Science, and Technology, Japan, by CREST (Japan Science and Technology Corporation), by Public Health Service research grants from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Crescenzo-Chaigne B, Naffakh N, van der Werf S. Comparative analysis of the ability of the polymerase complexes of influenza viruses type A, B and C to assemble into functional RNPs that allow expression and replication of heterotypic model RNA templates in vivo. Virology. 1999;265:342–353. doi: 10.1006/viro.1999.0059. [DOI] [PubMed] [Google Scholar]
- De Wit E, Spronken MI, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Evidence for specific packaging of the influenza A virus genome from conditionally defective virus particles lacking a polymerase gene. Vaccine. 2006;24:6647–6650. doi: 10.1016/j.vaccine.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Desselberger U, Racaniello VR, Zazra JJ, Palese P. The 3’ and 5’-terminal sequences of influenza A, B, and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980;8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S, Naffakh N. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5' end of the PB2 segment. Virology. 2005;341:34–46. doi: 10.1016/j.virol.2005.06.040. [DOI] [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandorfer A, García-Sastre A, Basler CF, Palese P. Chimeric influenza A viruses with a functional influenza B virus neuraminidase or hemagglutinin. J Virol. 2003;77:9116–9123. doi: 10.1128/JVI.77.17.9116-9123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. The importance of both the coding and segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J Virol. 2005;79:3766–3774. doi: 10.1128/JVI.79.6.3766-3774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc Natl Acad Sci USA. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghate AA, Air GM. Influenza type B neuraminidase can replace the function of type A neuraminidase. Virology. 1999;26:265–277. doi: 10.1006/viro.1999.9936. [DOI] [PubMed] [Google Scholar]
- Gog JR, Dos Santos Afonso E, Dalton RM, Leclercq I, Tiley L, Elton D, von Kirchbach JC, Naffakh N, Escriou N, Digard P. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Takada A, Iwatsuki-Horimoto K, Hatta M, Goto H, Kawaoka Y. Generation of influenza A virus with chimeric (type A/B) hemagglutinins. J Virol. 2003;77:8031–8038. doi: 10.1128/JVI.77.14.8031-8038.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverin NV, Varich NL, Sklyankaya EI, Amvrosieva TV, Petrik J, Vovk TC. Studies on heterotypic interference between influenza A and B viruses: a differential inhibition of the synthesis of viral proteins and RNAs. J Gen Virol. 1983;64:2139–2146. doi: 10.1099/0022-1317-64-10-2139. [DOI] [PubMed] [Google Scholar]
- Kobasa D, Rodgeres ME, Wells K, Kawaoka Y. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol. 1997;71:6706–6713. doi: 10.1128/jvi.71.9.6706-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J Virol. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Huang T, Ly H, Parslow TG, Liang Y. Mutational analysis of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J Virol. 2008;82:229–236. doi: 10.1128/JVI.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Goto H, Horimoto T, Takada A, Kawaoka Y. Biological significance of the U residue at the -3 position of the mRNA sequences of influenza A viral segments PB1 and NA. Virus Res. 2004;100:153–157. doi: 10.1016/j.virusres.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheeva A, Ghendon YZ. Intrinsic interference between influenza A and B viruses. Arch Virol. 1982;73:287–294. doi: 10.1007/BF01318082. [DOI] [PubMed] [Google Scholar]
- Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among vRNA segments in their role in vRNA incorporation into influenza A virions. J Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Subbarao EK, Enami M, Murphy BR, Palese P. An influenza A virus containing influenza B virus 5’ and 3’ noncoding regions on the neuraminidase gene is attenuated in mice. Proc Natl Acad Sci USA. 1991;88:5177–5181. doi: 10.1073/pnas.88.12.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Fujii K, Muramoto Y, Yamada S, Yamayoshi S, Takada A, Goto H, Horimoto T, Kawaoka Y. Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol. 2007;81:30–41. doi: 10.1128/JVI.01434-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, et al., editors. Fields Virology. 5. Williams & Wilkins; Philadelphia: 2007. pp. 1647–1689. [Google Scholar]
- Perez DR, Donis RO. Functional analysis of PA binding by influenza a virus PB1: effects on polymerase activity and viral infectivity. J Virol. 2001;75:8127–8136. doi: 10.1128/JVI.75.17.8127-8136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Zurcher T, Nieto A, Ortin J. Replication of orthomyxoviruses. Adv Virus Res. 1999;54:319–348. doi: 10.1016/s0065-3527(08)60370-x. [DOI] [PubMed] [Google Scholar]
- Tobita K, Tanaka T, Goto H, Feng SY. Temperature-sensitive influenza A virus clones originated by a cross between A/Aichi/2/68 (H3N2) and B/Yamagata/1/73. Arch Virol. 1983;75:17–27. doi: 10.1007/BF01314124. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J Virol. 2003;77:10575–10583. doi: 10.1128/JVI.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PF, Neumann G, Kawaoka Y. Orthomyxoviruses. In: Knipe DM, Howley PM, et al., editors. Fields Virology. 5. Williams & Wilkins; Philadelphia: 2007. pp. 1691–1740. [Google Scholar]