Abstract
An extensive repertoire of protein 4.1R isoforms is predominantly generated by alternative pre-mRNA splicing and differential usage of two translation initiation sites. The usage of the most upstream ATG (ATG-1) generates isoforms containing N-terminal extensions of up to 209 aa compared with those translated from the downstream ATG (ATG-2). To characterize nonerythroid 4.1R proteins translated from ATG-1 and analyze their intracellular localization, we cloned 4.1R cDNAs containing this translation initiation site. Six different clones were isolated from the nucleated human MOLT-4 T-cell line by reverse transcriptase–PCR techniques. Transient expression of the six ATG-1-translated 4.1R isoforms tagged with a c-Myc epitope revealed that all of them predominantly distributed to the plasma membrane and the endoplasmic reticulum. Staining of MOLT-4 cell plasma membranes but not nuclei was also observed by immunofluorescence microscopy by using an antibody specific to the N-terminal extension. Consistent with this, the antibody reacted with a major endogenous protein of ≈145 kDa present in nonnuclear but absent from nuclear fractions prepared from MOLT-4 cells. Because these data suggested that ATG-1-translated 4.1R isoforms were predominantly excluded from the nucleus, we fused the 209-aa domain to nuclear 4.1R isoforms encoded from ATG-2 and observed that this domain inhibited their nuclear targeting. All these results indicate that the N-terminal domain of ATG-1-translated 4.1R isoforms plays a pivotal role in differential targeting of proteins 4.1R.
Many important structural proteins are expressed not as simple polypeptides but rather as families of closely related isoforms. The structural protein 4.1R was originally identified as a protein of 78–80 kDa of the erythroid membrane skeleton, where it acts as a linking molecule, attaching the spectrin/actin cytoskeletal scaffold to the lipid bilayer via interactions with cytoplasmic domains of transmembrane proteins (reviewed in ref. 1). Subsequent studies showed that multiple immunoreactive proteins 4.1R, varying considerably in size from 30 to 210 kDa, were also detected in nucleated cells, including avian erythroid cells and nonerythroid cells of many mammal species (2, 3). Immunological studies indicated that, in nucleated cells, 4.1 epitopes are not strictly confined to the cell membrane and have been detected at intracellular sites such as stress fibers (4), perinuclear regions (5), centrosomes (6), and the nucleus (7–11). The nuclear localization of 4.1R has been confirmed by transfection experiments of 4.1R cDNAs isolated from a human T-cell line (11) and from human erythroblasts (12).
Posttranslational modification accounts partly for the impressive diversity of 4.1R proteins; however, alternative splicing of the 4.1R premRNA is the major cause of the generation of the extensive repertoire of 4.1R isoforms. The prototypical erythroid protein 4.1R (80 kDa) is produced when 17 nucleotides 5′ upstream from exon 2 are spliced out, and translation is initiated at the downstream start site present in exon 4 (ATG-2). Synthesis of isoforms larger than 80 kDa occurs when the 17-nt sequence, which contains the upstream ATG (ATG-1) translation initiation codon, is included, thus converting part of the untranslated 5′ extremity of the mRNA into a translatable reading frame contiguous with that of the known 80-kDa erythroid protein 4.1R. This event originates 4.1 isoforms with an extended N-terminal region of up to 209 aa (see Fig. 1a). Thus, two different 4.1R isoform groups can be generated depending on whether ATG-2 or ATG-1 is used as the translation initiation site. These two groups are denominated low molecular-weight (LMW) and high molecular-weight (HMW) 4.1R isoforms, respectively (reviewed in ref. 1). The number, the intracellular location, and the function of the HMW 4.1R isoforms, mainly abundant in nonerythroid tissues, have not been well characterized. Besides the predicted isoforms exhibiting different N-terminal structures, diverse variants of the domains involved in the binding to spectrin-actin networks and to glycophorin can also be generated by combinatorial splicing among the alternative exons identified to date. Protein 4.1R represents, therefore, an extreme variation on the theme of isoform multiplicity. This scenario has become even more complicated because of the recent discovery of the existence of a family of distinct protein 4.1 genes (13, 14).
Figure 1.
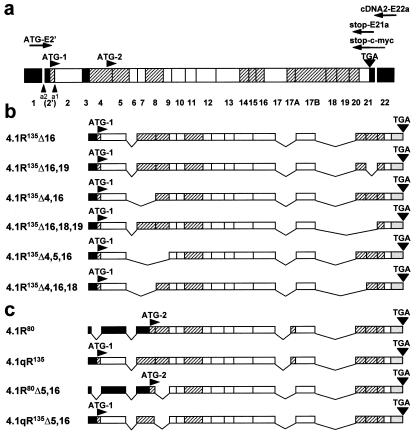
Cloning of lymphoid HMW protein 4.1R cDNAs. (a) Schematic representation of the exon map of the protein 4.1R. Exons are coded as follows: shaded, alternative; white, constitutive; black, noncoding region. The number of each individual exon is indicated at the bottom. Two translation initiation sites at exons 2′ (ATG-1) and 4 (ATG-2) are indicated, as well as the stop codon (TGA) at exon 21. a1 represents the alternative 3′-splice acceptor site used by LMW 4.1R encoding mRNAs; a2 represents the alternative 3′-splice acceptor site used by HMW 4.1R encoding mRNAs. The primers used for reverse transcription–PCR and for epitope-tagging are indicated: cDNA2-E22a for reverse transcription, ATG-E2′ and stop-E21a for the specific PCR amplification reaction, and ATG-E2′ and stop-c-myc for epitope-tagging. (b) Exon composition of the six cloned HMW 4.1R cDNAs. (c) Exon composition of two nuclear LMW 4.1R protein cDNAs (4.1R80 and 4.1R80Δ5, 16) isolated from MOLT-4 and that of the composite 4.1qR135 and 4.1qR135Δ5,16 cDNA constructs used in this study.
In this study, we have approached the molecular characterization of nonerythroid 4.1R proteins translated from the upstream ATG-1 translation initiation site, the analysis of their intracellular location, and the role that the extra N-terminal domain plays in 4.1R distribution. The molecular cloning of 4.1R cDNAs has provided us with the opportunity to analyze the exonic composition of six different clones, to characterize their protein products in vivo and in vitro, and to determine their intracellular distribution. Our results show that these HMW 4.1R isoforms distribute predominantly to nonnuclear sites: the plasma membrane and the endoplasmic reticulum (ER). An anti-4.1R antibody directed against the N-terminal extension of HMW 4.1R proteins recognized endogenous proteins of ≈145 kDa in nonnuclear fractions of MOLT-4 T-cells by immunoblot analysis. This antibody consistently stained the plasma membrane of the cell, leaving the nuclei unstained, whereas an anti-4.1R antibody recognizing both HMW and LMW 4.1R isoforms stained the plasma membrane and the nucleus of the cells. In addition, we demonstrate by constructing and analyzing composite 4.1R cDNAs that the N-terminal 209-aa domain plays an important role in the differential intracellular localization of proteins 4.1R. This region was sufficient to abrogate the nuclear targeting of proteins 4.1R encoded from ATG-2. These results further corroborate the hypothesis that proteins 4.1R containing the N-terminal extension are predominantly excluded from the nucleus.
Materials and Methods
Cell Culture and Transfection.
Human T lymphoid MOLT-4 cells were used as the RNA source for cDNA cloning and cell fractionation experiments, and COS-7 cells for transient cDNA expression, immunofluorescence, and biochemical analyses. Cells were grown as described (11). Transient transfections were performed in COS-7 cells by electroporation by using the Electro Cell Manipulator 600 (BTX, San Diego). Cells were processed 48 h after transfection.
RNA Extraction, cDNA Cloning, and in Vitro Protein Expression.
Cytoplasmic RNA was extracted from MOLT-4 cells, as previously described (15). 4.1R mRNA was specifically reverse transcribed, as reported (11). The 4.1R cDNAs were amplified by PCR under the conditions previously described (16) by using Taq polymerase (Promega), stopE21a (11), and ATG-E2′ (CAGCCCAGCAACATCATGA), a sense primer that includes exon 2′ sequences (Fig. 1a). This allowed the 4.1R cDNA population containing the first translation initiation site (ATG-1) to be obtained. Reaction products were cloned into the pMOSBlue vector (Amersham Pharmacia Biotech). For expression experiments, 4.1R cDNAs were tagged at their 3′ termini by PCR. Reactions were carried out by using ATG-E2′ and stop-c-myc, an antisense primer complementary to the 3′ end of the 4.1R coding sequence (Fig. 1a) and encoding the c-Myc 9E10 epitope (EQKLISEEDL) (17). The cDNAs encoding epitope-tagged 4.1R proteins were finally cloned into the pSRα (18) or pCR3 (Invitrogen) mammalian expression vectors. 4.1R cDNAs were sequenced as reported (11). In vitro protein expression was achieved by coupled in vitro transcription and translation reactions, as described (11).
Composite cDNA Constructs.
4.1qR135 was made by substituting the ScaI–BamHI fragment of pSRα 4.1H (11), renamed here as 4.1R80, by the ScaI–BamHI fragment of pSRα 4.1R135Δ16, thus rendering a chimeric cDNA that encodes a protein containing the 209-aa N-terminal extension present in HMW 4.1R isoforms fused to the LMW nuclear 4.1R80.
4.1qR135Δ5,16 was constructed by using the overlap extension technique (19). PCRs were performed by using pSRα 4.1R135Δ16 and pSRα 4.1R80Δ5,16 (a LMW nuclear isoform, similar to 4.1R80 but lacking exons 5 and 16; see Fig. 1c) as templates, and ATG-E2′, stop-c-myc, E2s (a sense oligonucleotide complementary to exon 2 sequences, GCAGCTCCTGAACCGGAACTC), and E4a (an antisense oligonucleotide complementary to exon 4 sequences, GAAACCTTGCAGTGCATGT) as primers. The product from these reactions was inserted into the pCR3 mammalian expression vector. This yielded a cDNA encoding a 4.1R isoform containing the complete 209-aa N-terminal domain fused to 4.1R80Δ5,16.
Antibodies.
Anti-c-Myc monoclonal antibody 9E10 (17) was obtained from the American Type Culture Collection. Anti-4.1R (10b) antibody has been previously characterized (20) and recognizes a sequence encoded by exon 17. Anti-4.1R (762) antibody was raised against a synthetic peptide (TKNKERTSESRGC) whose sequence is encoded by exon 2 and anti-4.1R (766) antibody against a synthetic peptide (AAQTDDNSGDLC) whose sequence is encoded by the joining of exons 18 and 19. Each peptide was coupled to keyhole lymphet hemocyanine and used to immunize rabbits, as previously described (20). Specificity of antibody 762 was demonstrated by immunofluorescence microscopy and by Western blotting of COS-7 cells expressing HMW 4.1R or LMW 4.1R isoforms (see Fig. 4). Similarly, specificity of antibody 766 was also checked in COS-7 cells expressing HMW 4.1R isoforms containing or lacking exon 18- and 19-encoded sequences (data not shown). Anti-CD4 mouse monoclonal antibody was a generous gift from B. Alarcón (Centro de Biología Molecular, Madrid). Antiprotein disulfide isomerase (PDI) rabbit polyclonal antibody was a gift from J. G. Castaño (Instituto de Investigaciones Biomédicas, Madrid). Polyclonal antinuclear lamin A, B, and C antibodies were generous gifts from S. D. Georgatos (University of Crete, Greece). Antitubulin antibody (YL1/2) was purchased from Sera-Lab (Crawley Down, Sussex, U.K.). Goat anti-mouse or anti-rabbit Ig secondary antibodies, conjugated with horseradish peroxidase, fluorescein isothiocyanate, or Texas red isothiocyanate were obtained from Southern Biotechnology Associates.
Figure 4.
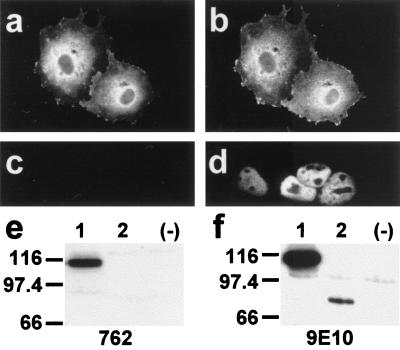
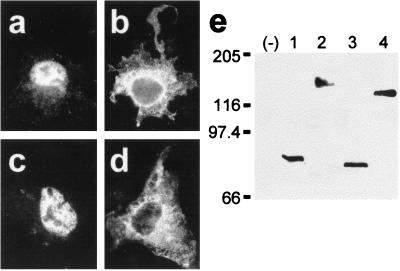
Characterization of antibody 762 directed against the N-terminal extension of HMW 4.1R proteins. COS-7 cells transfected with the HMW 4.1R135Δ16,18,19 cDNA (a and b) or with the LMW 4.1R80Δ5,16 cDNA (c and d) were double labeled with antibodies 762 and 9E10 (a and c; b and d, respectively) 48 h after transfection and examined by epifluorescence microscopy. Antibody 9E10 recognized both epitope-tagged 4.1R proteins (b and d), whereas antibody 762 recognized the HMW 4.1R protein (a) but not the LMW 4.1R protein (c). (e and f) Western blot analysis of total extract proteins from untransfected COS-7 cells (−) or from cells transfected with the HMW 4.1R135Δ16,18,19 cDNA (lanes 1) or with the LMW 4.1R80Δ5,16 cDNA (lanes 2) as revealed with antibodies 762 (e) or 9E10 (f). Antibody 9E10 recognized both epitope-tagged 4.1R proteins (f, lanes 1 and 2), whereas antibody 762 recognized the HMW 4.1R protein (Fig. 4e, lane 1) but not the LMW 4.1R protein (Fig. 4e, lane 2).
Immunofluorescence Microscopy.
Cells grown on glass coverslips were fixed, permeabilized, and blocked as described (11). Cells were incubated with the appropriate antibodies and processed as reported (8). Preparations were examined with a Zeiss epifluorescence microscope. Controls to assess the specificity and the lack of crosslabeling included incubations with irrelevant monoclonal antibodies and nonimmune rabbit sera or omission of either of the primary antibodies.
Protein Extractions, Subcellular Fractionation, and Western Blotting.
Cells grown on Petri dishes were rinsed twice in PBS and lysed in 100 μl of solubilizing Laemmli buffer (21). Nuclear and nonnuclear fractions were prepared as described (22). Protein fractions were separated by NaDodSO4/PAGE and transferred to Immobilon (Millipore)-poly(vinylidene difluoride) in 3-(cyclohexylamino)-1-propanesulfonic acid buffer, pH 11. Membranes were processed and developed as reported (8).
Results and Discussion
Cloning and Exon Composition of Lymphoid HMW 4.1R Protein cDNAs.
To carry out a systematic analysis of the number, structure, and intracellular distribution of HMW 4.1R isoforms present in lymphoid cells, we took the approach of directly cloning the coding sequences corresponding to this subset of 4.1R isoforms in a single DNA fragment. We adopted a reverse transcription–PCR approach by using RNA from the MOLT-4 human T-cell line as the starting template and 4.1R-specific oligonucleotide primers. The different clones obtained were analyzed by Southern blot by using exon-specific oligonucleotide probes. Six different groups of 4.1R cDNAs were identified on the basis of the presence or absence of specific exons. Representative clones from each group were sequenced to confirm their exon composition, which is shown schematically in Fig. 1b. The clones have been named according to the nomenclature of Gascard et al. (12) of 4.1R cDNAs isolated from human erythroblasts. The 4.1R135Δ4,5,16 cDNA used in this work has been reconstituted from the previous 4.1E cDNA (23) with the same exon composition but bearing a nucleotide deletion in exon 7 that causes premature termination of translation. The largest and most abundant 4.1R cDNA that we have cloned corresponds to 4.1R135Δ16, which codes for a protein of 810 aa; the smallest corresponds to 4.1R135Δ4,16,18, which codes for a protein of 696 aa. Interestingly, 4.1R135Δ16, 4.1R135Δ16,18,19, and 4.1R135Δ16,19 cDNAs contained all three exons 2′, 2, and 4, the complete coding region for the N-terminal 209-aa domain. By contrast, 4.1R135Δ4,16,18, 4.1R135Δ4,5,16 and 4.1R135Δ4,16 cDNAs lacked exon 4 sequences, therefore their protein products contained shorter N-terminal extensions.
Notable features regarding the exon composition of the cloned 4.1R cDNAs are that all of them contain the alternative exons 8 and 20, none contains alternative exon 16, and, as expected, all of them lack exon 3. Thus, the diversity of the HMW 4.1R isoforms that we have isolated from MOLT-4 T cells arises from the presence/absence of exons 4-, 5-, 18-, and/or 19-encoded sequences. Most of these results are consistent with results from experiments in which small 4.1R cDNA pieces containing specific alternative sequences were amplified by using RNA from diverse tissues as starting templates (16, 24, 25). Indeed, it has been reported that deletion of exon 8 sequences appears to be a rare event detected only by sensitive PCR techniques and that, therefore, its physiological significance is uncertain (16). It is also possible that expression of exon 8 is tissue-specifically regulated and spliced in all MOLT-4 4.1R RNAs. Consistent with our results, the presence of exon 8-encoded sequences has also been detected in all of the 4.1R isoforms isolated from human erythroblasts (12). 4.1R cDNAs lacking both exon 18 and 19 have not been reported previously. In this study, we have isolated a 4.1R cDNA designated 4.1R135Δ16,18,19 that lacks both exons 18 and 19. Thus, the discovery of a 4.1R cDNA with a combination of previously unidentified exonic sequences adds more variety to the already complex map of alternative splicing events described for the 4.1R premRNA. All of the 4.1R isoforms isolated in this study lack exon 16-encoded sequences. This sequence and that encoded by exon 17 constitute the spectrin-actin binding domain of protein 4.1R (26). It is notable that most of the major isoforms isolated from human erythroblasts contain both exon 16- and 17-encoded sequences (12), thus confirming a previously reported (16, 27, 28) distinctive feature between erythroid and nonerythroid cells.
In Vivo and in Vitro Characterization of the Cloned HMW 4.1R Isoforms.
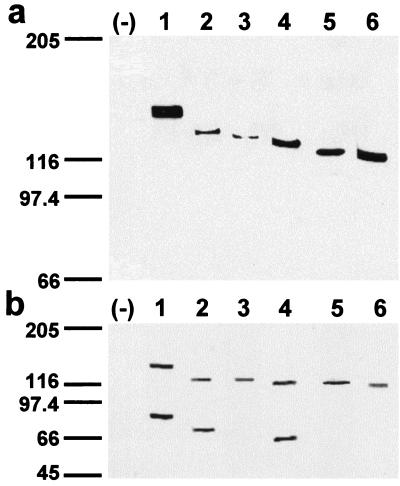
The predicted sizes of the protein 4.1R isoforms that we have cloned are: ≈91 kDa (4.1R135Δ16); ≈87 kDa (4.1R135Δ16, 19); ≈82 kDa (4.1R135Δ16, 18, 19, and 4.1R135Δ4, 16) and ≈78 kDa (4.1R135Δ4, 16, 18, and 4.1R135Δ4, 5, 16). The apparent molecular masses of the in vivo- and in vitro-expressed proteins were determined by NaDodSO4/PAGE. To distinguish ectopic proteins from endogenous 4.1 proteins, the different 4.1R proteins were tagged with the c-Myc 9E10 epitope at their C terminus. Fig. 2a shows the sizes of the epitope-tagged 4.1R isoforms isolated from extracts prepared from transiently transfected COS-7 cells and immunoblotted with antitag antibodies. Proteins of apparent molecular masses ranging from approximately 115-kDa (4.1R135Δ4, 16, 18) to 145-kDa (4.1R135Δ16) were detected. To analyze the in vitro expression of the six 4.1R cDNAs, they were independently transcribed in vitro with T7 RNA polymerase and translated in vitro by using a rabbit reticulocyte lysate. The in vitro-translated products (Fig. 2b) have apparent molecular masses similar to those observed for the in vivo-expressed proteins (Fig. 2a). Additional protein bands were observed in the in vitro translation experiments of the three ATG-2-containing 4.1R cDNAs (Fig. 2b, lanes 1, 2, and 4). They might correspond to 4.1R proteolytic fragments or, most likely, they may arise from the usage of the ATG-2 translation initiation site, because they correspond in size to products translated from this site. Furthermore, no additional protein bands were observed in the in vitro translation assays of the three ATG-2-lacking 4.1R cDNAs (Fig. 2b, lanes 3, 5, and 6). The absence of additional protein bands in the in vivo experiments (Fig. 2a) indicates that their synthesis is probably a rare event occurring within the cell.
Figure 2.
In vivo and in vitro expression of the HMW 4.1R isoforms. (a) Western blot analysis with anti-c-Myc antibody of total extract proteins from untransfected COS-7 cells (−) or from cells transfected with the following cDNAs: 4.1R135Δ16 (lane 1); 4.1R135Δ16,19 (lane 2); 4.1R135Δ4,16 (lane 3); 4.1R135Δ16,18,19 (lane 4); 4.1R135Δ4,5,16 (lane 5); and 4.1R135Δ4,16,18 (lane 6). (b) (1–6) In vitro coupled transcription and translation products of the cloned 4.1R cDNAs indicated in a, labeled with [35S]methionine and autoradiographed. (−), a control containing all components of the mixture except template cDNA.
Intracellular Localization of Exogenous Epitope-Tagged HMW 4.1R Isoforms.
To analyze the intracellular distribution of each 4.1R isoform, COS-7 cells were transiently transfected with each specific 4.1R cDNA, and the expressed tagged 4.1R protein was analyzed 48 hr after transfection by immunofluorescence microscopy by using 9E10 antibody. The six 4.1R isoforms distributed predominantly to the plasma membrane and the ER, and none of them localized predominantly to the nucleus. Fig. 3 a–c shows representative images of the predominant distribution patterns observed. To confirm the presence of HMW 4.1R proteins in the ER and the plasma membrane, we performed double-labeling immunofluorescence microscopy using antibodies recognizing protein markers of these cellular sites together with an antibody recognizing the exogenous tagged-HMW 4.1R proteins. Anti-PDI antibody was used to stain the ER and anti-CD4 antibody to label the plasma membrane of COS-7 cells cotransfected with CD4 cDNA. Fig. 3 b and d shows the immunostaining patterns observed with 9E10 and α-PDI antibodies, respectively and Fig. 3 c and e those observed with 10b and anti-CD4 antibodies, respectively. Colocalization of protein 4.1R with PDI and with CD4 indicates that HMW 4.1R isoforms are indeed distributed to the ER and the plasma membrane. Our finding that HMW 4.1R proteins localize to the ER supports the notion that erythroid cytoskeletal proteins may also be playing roles at intracellular sites other than the membrane skeleton. In this regard, specific isoforms of other erythroid cytoskeletal proteins, such as spectrin and ankyrin, have been identified in the ER/Golgi apparatus, where they may form a vesicular Golgi-associated membrane skeleton (29–31). Whether 4.1R, spectrin, and ankyrin are components that interact in the ER as they do in the erythroid membrane skeleton remains to be established.
Figure 3.
Subcellular localization of HMW 4.1R isoforms. COS-7 cells transfected with each of the cloned HMW 4.1R cDNAs were labeled with different antibodies and examined by epifluorescence microscopy 48 h after transfection. (a) Cells stained with anti-c-Myc antibody; (b and d) cells double stained with anti-c-Myc antibody (b) and anti-PDI antibody (d), a marker of the ER; (c and e) cells double stained with anti-4.1R (10b) antibody (c) or anti-CD4 mouse monoclonal antibody (e), a marker of the plasma membrane. Representative images are shown: (a) predominant expression at the plasma membrane and the ER; (b) at the ER; (c) at the plasma membrane. These expression patterns were observed for the six HMW 4.1R isoforms.
Intracellular Localization of Endogenous HMW 4.1R Proteins.
None of the six HMW 4.1R isoforms analyzed above was distributed predominantly to the nucleus, but all were found at nonnuclear sites. Therefore, we investigated whether the endogenous HMW 4.1R proteins followed the same distribution patterns. To distinguish specifically HMW 4.1R immunoreactive proteins from LMW 4.1R isoforms, we prepared antibody 762, which recognizes a region of the N-terminal extension encoded by exon 2 (see Materials and Methods). Specificity of the antibody was established by showing that it reacted with HMW 4.1R isoforms but not with LMW 4.1R isoforms both in immunofluorescence (Fig. 4 a and c, respectively) and Western blot experiments (Fig. 4e, lanes 1 and 2, respectively) of COS-7 cells transfected with the appropriate cDNAs. In these experiments, antibody 9E10 recognized both types of 4.1R isoforms (Fig. 4 b and d and Fig. 4f, lanes 1 and 2). The preimmune serum did not recognize any of the expressed 4.1R proteins (data not shown).
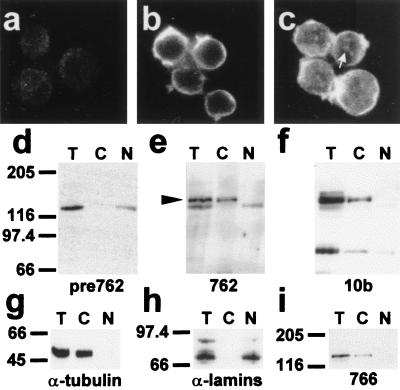
The intracellular distribution of endogenous HMW 4.1R immunoreactive proteins present in MOLT-4 cells was analyzed by immunofluorescence microscopy by using antibody 762. As shown in Fig. 5b, the antibody exclusively stained the plasma membrane of the cells suggesting that 4.1R proteins containing the N-terminal epitope recognized by antibody 762 are, indeed, localized to nonnuclear sites. Antibody 10b, which recognizes a sequence present in both HMW and LMW 4.1R isoforms (20), stained the plasma membrane in addition to the nucleus of the cells, in which a dot-like pattern could be distinguished (Fig. 5c). This latter result suggests that 4.1R proteins lacking the N-terminal epitope recognized by antibody 762 are present in the nucleus.
Figure 5.
Intracellular distribution of endogenous immunoreactive HMW 4.1R proteins in MOLT-4 T-cells. Immunofluorescence microscopy of MOLT-4 T-cells probed with preimmune serum for antibody 762 (a), antibody 762 (b) and antibody 10b (c). The white arrow indicates the dot-like staining pattern detected with 10b antibody within the nucleus. (d–i) Western blot analysis of MOLT-4 subcellular fractions isolated as described in Materials and Methods (equivalent cell volumes were loaded in each lane; T, total cell extracts; C, cytoplasm or nonnuclear fractions; N, nuclear fractions). The antibodies used were: preimmune serum for antibody 762 (d), antibody 762 (e), antibody 10b (f), antitubulin antibody (g), antilamins (h), and antibody 766. The black arrowhead indicates the specific ≈145-kDa band fractionating to the nonnuclear fraction that is recognized by antibody 762. This band is also recognized by antibodies 10b and 766.
To characterize further the endogenous 4.1R immunoreactive proteins containing the N-terminal extension, we performed cell fractionation analysis of MOLT-4 T-cells. Antilamin (Fig. 5h) and antitubulin (Fig. 5g) antibodies were used as controls for the correct fractionation of the nucleus and that of the cytoplasm, respectively. The lamins were detected in the nuclear fraction (Fig. 5h, lane N) but not in the nonnuclear fraction (Fig. 5h, lane C), whereas tubulin was observed in the nonnuclear one (Fig. 5g, lane C). Antibody 762 specifically recognized a band of ≈145 kDa that fractionated to the nonnuclear fraction (compare lanes T, C, and N in Fig. 5e with those of the preimmune serum in Fig. 5d). This ≈145-kDa band was also recognized by antibodies 10b (Fig. 5f, lanes T and C) and 766 (Fig. 5i, lanes T and C). It may be that this ≈145-kDa band is the product of the largest and most abundant cDNA (4.1R135Δ16) cloned by us from these cells.
The results concerning the distribution of endogenous HMW 4.1R isoforms obtained by both immunofluorescence and subcellular fractionation of MOLT-4 T-cells further support the notion that HMW 4.1R proteins are predominantly excluded from the nucleus.
Consistent with the results obtained in the immunofluorescence assays, antibody 10b also recognized intensely a band of ≈80 kDa that fractionated to the nuclear (Fig. 5f, lane N) and the nonnuclear fractions (Fig. 5f, lane C). Minor bands of larger sizes than ≈80 kDa were also detected with antibody 10b. These bands were not observed with either antibody 762 (Fig. 5e) or 766 (Fig. 5i).
The 209-aa Domain of HMW 4.1R Isoforms Affects 4.1R Intracellular Distribution.
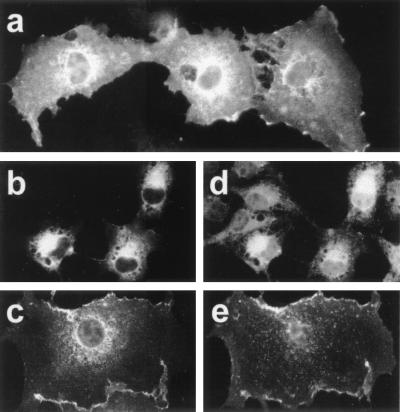
To investigate further whether the 209-aa domain plays a role in the differential intracellular distribution of proteins 4.1R, we constructed composite 4.1R cDNAs and analyzed the distribution of their products by transfection experiments. Protein 4.1H, renamed here as 4.1R80, is a LMW 4.1R isoform encoded from ATG-2 that is distributed in the nucleus (11). This protein contains a basic sequence motif (KKKR), generated by the joining of exon 13 and 16 sequences, which is necessary but not sufficient for 4.1R80 nuclear import (11). A composite cDNA designated 4.1qR135 was constructed by adding the 209-aa sequence to the N terminus of protein 4.1R80 (see Fig. 1c). The intracellular distributions of the composite protein 4.1qR135 and that of 4.1R80 were compared by immunofluorescence microscopy by using the anti-c-Myc antibody. Although 4.1R80 was predominantly distributed in the nucleus (Fig. 6a), protein 4.1qR135 was not (Fig. 6b). Indeed, the percentage of transfected cells analyzed (≈300 cells from 6 independent transfection experiments) presenting only nuclear staining was 75% for 4.1R80 compared with 4% for 4.1qR135. The remaining 25% (4.1R80) compared with 96% (4.1qR135) of the cells displayed intense nonnuclear and faint nuclear stainings.
Figure 6.
Fusion of the 209-aa domain of HMW 4.1R isoforms to nuclear LMW 4.1R isoforms inhibits their targeting to the nucleus. COS-7 cells were transfected with the indicated plasmids, subjected to immunofluorescence analysis with anti-c-Myc antibody 48 h after transfection, and examined by epifluorescence microscopy (a–d). The intracellular localization of (a) 4.1R80 and (c) 4.1R80Δ5,16 was compared with that of the (b) 4.1qR135 and (d) 4.1qR135Δ5,16 chimeras. Western blot analysis with anti-c-Myc antibody of total extract proteins from untransfected COS-7 cells (−) or from cells transfected with the following cDNAs: 4.1R80 (lane 1); 4.1qR135 (lane 2); 4.1R80Δ5,16 (lane 3); and 4.1qR135Δ5,16 (lane 4).
Protein 4.1R80Δ5,16 is another nuclear LMW 4.1R isoform that, unlike LMW 4.1R80, lacks exon 16- and exon 5-encoded sequences (see Fig. 1c). Because protein 4.1R80Δ5,16 does not contain exon 16-encoded sequences, regions still uncharacterized and different from the basic tetrapeptide KKKR must be involved in 4.1R80Δ5,16 nuclear entry. To investigate further whether the extra N-terminal region of HMW 4.1R isoforms could also eclipse nuclear entry of the LMW 4.1R80Δ5,16 isoform lacking exon 16-encoded sequences, we constructed a composite cDNA (designated 4.1qR135Δ5, 16) by fusing the 209-aa sequence to the N terminus of protein 4.1R80Δ5,16 (see Fig. 1c). The intracellular distribution of the composite protein 4.1qR135Δ5,16 and that of 4.1R80Δ5,16 was subsequently analyzed by immunofluorescence microscopy. Fig. 6 c and d shows representative images of the predominant staining patterns observed. The LMW 4.1R isoform, 4.1R80Δ5,16, distributed only in the nucleus of most of the transfected cells (83%). This percentage was dramatically reduced to less than 1% when the HMW construct, 4.1qR135Δ5,16, was assayed. The remaining 17% (4.1R80Δ5, 16) compared with 99% (4.1qR135Δ5, 16) of the cells analyzed (≈275 cells from 5 transfection experiments) expressed the proteins strongly at nonnuclear sites and poorly in the nucleus, a result that is consistent with that obtained for 4.1R80 and 4.1qR135. Therefore, the 209-aa domain present in HMW 4.1R isoforms, when added to the N terminus of nuclear LMW 4.1R isoforms 4.1R80 and 4.1R80Δ5,16, eclipses the effect of their different nuclear targeting sequences. The addition of the 209-aa domain to the nuclear localization signal of SV40 large T-antigen fused to bacterial β-galactosidase also resulted in a reduction in the number of cells presenting only nuclear expression (from 89% when transfection was carried out with the construct lacking the 209 aa to 69% when transfection was carried out with the construct containing the 209 aa; data not shown). Consistent with our findings, it has been observed that the nuclear expression of a specific HMW 4.1R isoform (4.1R135) was lower than that of its LMW 4.1R counterpart (4.1R80) (12), and that a HMW 4.1R protein (4.1R135) fused to the green fluorescent protein was located in nonnuclear regions of most of the transfected cells (32).
In summary, the intracellular localization of the exogenous-tagged HMW 4.1R proteins, as well as that of the endogenous immunoreactive HMW 4.1 proteins present in MOLT-4 T-cells, supports the hypothesis that HMW 4.1R proteins are mainly nonnuclear proteins. This finding was confirmed further, because the nuclear entry of two different LMW 4.1R isoforms was inhibited when the 209-aa domain was fused to the N terminus of their sequences. These results are consistent with the hypothesis that the effect of an N-terminal targeting sequence eclipses the effect of an internal or C-terminal targeting determinant (reviewed in ref. 33). Taken together, all these results indicate that the 209-aa domain of HMW 4.1R isoforms plays an important role in 4.1R distribution, counterbalancing nuclear targeting determinants present in proteins 4.1R.
Acknowledgments
We thank Dr. T. K. Tang for generous help. We are also very grateful to Dr. B. Alarcón (Centro de Biología Molecular “Severo Ochoa,” Madrid, Spain) for providing us with the pSRα plasmid containing the CD4 cDNA and the polyclonal antibody against CD4, and to Dr. J. G. Castaño (Instituto de Investigaciones Biomédicas, Madrid, Spain) for the anti-PDI antibody. C.M.L. and C.M.P.-F. are recipients of fellowships from the Comunidad de Madrid and the Ministerio de Educación y Cultura, respectively. This work was supported by grants nos. PM95-0030 and PM98-0002 from the Ministerio de Educación y Cultura. The authors also thank the Fundación Ramón Areces for institutional financial support.
Abbreviations
- HMW
high molecular weight
- LMW
low molecular weight
- ER
endoplasmic reticulum
- PDI
protein disulfide isomerase
- ATG-1
upstream ATG
- ATG-2
downstream ATG
References
- 1.Conboy J G. Semin Hematol. 1993;30:58–73. [PubMed] [Google Scholar]
- 2.Granger B L, Lazarides E. Cell. 1984;37:595–607. doi: 10.1016/0092-8674(84)90390-8. [DOI] [PubMed] [Google Scholar]
- 3.Anderson R A, Correas I, Mazzucco C, Castle J D, Marchesi V T. J Cell Biochem. 1988;37:269–284. doi: 10.1002/jcb.240370303. [DOI] [PubMed] [Google Scholar]
- 4.Cohen C M, Foley S F, Korsgren C. Nature (London) 1982;299:648–650. doi: 10.1038/299648a0. [DOI] [PubMed] [Google Scholar]
- 5.Leto T L, Pratt B M, Madri J A. J Cell Physiol. 1986;127:423–431. doi: 10.1002/jcp.1041270311. [DOI] [PubMed] [Google Scholar]
- 6.Krauss S W, Chasis J A, Rogers C, Mohandas N, Krockmalnic G, Penman S. Proc Natl Acad Sci USA. 1997;94:7297–7302. doi: 10.1073/pnas.94.14.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correas I. Biochem J. 1991;279:581–585. doi: 10.1042/bj2790581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Cárcer G, Lallena M J, Correas I. Biochem J. 1995;312:871–877. doi: 10.1042/bj3120871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krauss S W, Larabell C A, Lockett S, Gascard P, Penman S, Mohandas N, Chasis J A. J Cell Biol. 1997;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lallena M J, Correas I. J Cell Sci. 1997;110:239–247. doi: 10.1242/jcs.110.2.239. [DOI] [PubMed] [Google Scholar]
- 11.Luque C M, Lallena M J, Alonso M A, Correas I. J Biol Chem. 1998;273:11643–11649. doi: 10.1074/jbc.273.19.11643. [DOI] [PubMed] [Google Scholar]
- 12.Gascard P, Lee G, Coulombel L, Auffray I, Lum M, Parra M, Conboy J G, Mohandas N, Chasis J A. Blood. 1998;92:4404–4414. [PubMed] [Google Scholar]
- 13.Parra M, Gascard P, Walensky L D, Snyder S H, Mohandas N, Conboy J G. Genomics. 1998;49:298–306. doi: 10.1006/geno.1998.5265. [DOI] [PubMed] [Google Scholar]
- 14.Peters L L, Weier H U, Walensky L D, Snyder S H, Parra M, Mohandas N, Conboy J G. Genomics. 1998;54:348–350. doi: 10.1006/geno.1998.5537. [DOI] [PubMed] [Google Scholar]
- 15.Favaloro J, Treisman R, Kamen R. Methods Enzymol. 1980;65:718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- 16.Conboy J G, Chan J Y, Chasis J A, Kan Y W, Mohandas N. J Biol Chem. 1991;266:8273–8280. [PubMed] [Google Scholar]
- 17.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 20.Correas I, Speicher D W, Marchesi V T. J Biol Chem. 1986;261:13362–13366. [PubMed] [Google Scholar]
- 21.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.De Cárcer G, Cerdido A, Medina F J. Planta. 1997;201:487–495. doi: 10.1007/s004250050093. [DOI] [PubMed] [Google Scholar]
- 23.Lallena M J, Martínez C, Valcárcel J, Correas I. J Cell Sci. 1998;111:1963–1971. doi: 10.1242/jcs.111.14.1963. [DOI] [PubMed] [Google Scholar]
- 24.Conboy J G, Chan J, Mohandas N, Kan Y W. Proc Natl Acad Sci USA. 1988;85:9062–9065. doi: 10.1073/pnas.85.23.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baklouti F, Huang S C, Vulliamy T J, Delaunay J, Benz E J., Jr Genomics. 1997;39:289–302. doi: 10.1006/geno.1996.4512. [DOI] [PubMed] [Google Scholar]
- 26.Correas I, Leto T L, Speicher D W, Marchesi V T. J Biol Chem. 1986;261:3310–3315. [PubMed] [Google Scholar]
- 27.Tang T K, Leto T L, Correas I, Alonso M A, Marchesi V T, Benz E J., Jr Proc Natl Acad Sci USA. 1988;85:3713–3717. doi: 10.1073/pnas.85.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang T K, Qin Z, Leto T, Marchesi V T, Benz E J., Jr J Cell Biol. 1990;110:617–624. doi: 10.1083/jcb.110.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beck K A, Buchanan J A, Malhotra V, Nelson W J. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck K A, Buchanan J A, Nelson W J. J Cell Sci. 1997;110:1239–1249. doi: 10.1242/jcs.110.10.1239. [DOI] [PubMed] [Google Scholar]
- 31.Devarajan P, Stabach P R, Mann A S, Ardito T, Kashgarian M, Morrow J S. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattagajasingh S N, Huang S C, Hartenstein J S, Snyder M, Marchesi V T, Benz E J. J Cell Biol. 1999;145:29–43. doi: 10.1083/jcb.145.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danpure C J. Trends Cell Biol. 1995;5:230–238. doi: 10.1016/s0962-8924(00)89016-9. [DOI] [PubMed] [Google Scholar]