Introduction
Vitamin C, an essential co-factor for at least eight enzymatic reactions, might also be involved in development or treatment of cancer, cardiovascular diseases, diabetes, and stroke.
Ascorbic acid, the reduced form of vitamin C is transported across epithelial barriers by the sodium dependent vitamin C transporters 1 (SVCT1). SVCT1 is encoded by SLC23A1 and mapped to 5q31.2 [1, 2].
Recently, the pattern of common genetic variants has been characterized for both ascorbic acid transporters, SLC23A1 and SLC23A2, which share common intron/exon borders, are 58% similar in sequence across the coding region, but differ greatly in size and linkage disequilibrium patterns [3].
Here we characterize the genetic variation in the less constrained SLC23A1 gene in more detail and test for functional consequences. SLC23A1 is expressed in tissues critical for absorption and reabsorption of vitamin C (kidney, intestinal, and hepatic tissues) [4]. Therefore functional consequences of variations in SVCT1 would impact on dietary requirements and recommendations.
Materials
Initially genetic variants were identified by re-sequence analysis with coding bias, performed in 48 anonymous controls (24 African American and 24 Caucasian). Later the analysis was expanded to larger sample sizes.
Haplotype structure was estimate by Phase (Version 2.1).
Linkage disequilibrium (LD) was estimated using Haploview Version 2.05.
Genetic conservation was graphed using the online VISTA tools against mus musculus slc23a1.
Site directed mutagenesis was performed with the Promega Gene Editor Kit.
Transport assays in Xenopus laevis oocytes were performed 3 days after injection of 16 ng cRNA representing the sequence validated SVCT1 genotypes and different concentration of radiolabel ascorbic acid as substrate.
Results and conclusions
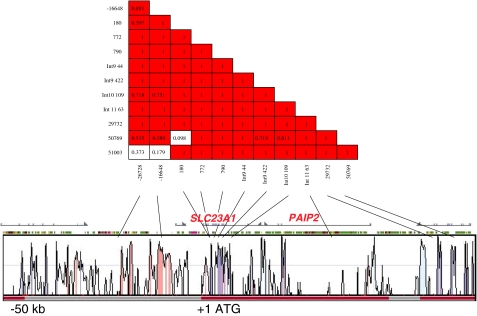
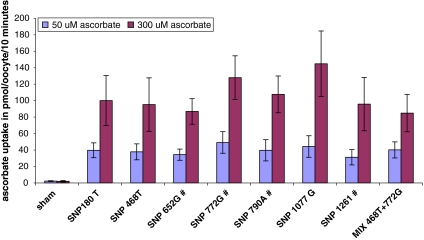
Twenty-two SNPs, including four nonsynonymous were detected in SLC23A1. Nearly all of the SNP in SLC23A1 were population specific in either African Americans or Caucasians, including the four nonsynonymous SNPs (Table 1). All nonsynonymous SNPs but the Exon 8 V264M conversion, which had a very low frequency in the Caucasian population, were only observed in the African American population. Haplotype analysis indicated a single haplotype block for the SLC23A1 gene as demonstrated for African American in Fig. 1. Therefore the African American population would be mainly affected by functional consequences of an variation. The current recommendations for Vitamin C are based in part on pharmacokinetic studies in healthy Caucasian volunteers. Vitamin C plasma concentration is under tight control, mediated by bioavailability, tissue transport, and renal excretion. Therefore, variations in SVCT1 could change vitamin C pharmacokinetics if transporter functions are altered. Functional studies in the Xenopus laevis oocyte expression system did not show significant differences in transport function between the proteins reflecting the variant genotypes of SVCT1 identified to date (Fig. 2). Our model represents a fundamental step toward the application of genomics to refine nutrient recommendations and may serve as a paradigm for micronutrients.
Table 1.
Common genetic variants in SLC23A1
| DbSNP a | Location | Variantb | Estimated allele frequencyc | |
|---|---|---|---|---|
| AA | CA | |||
| rs6596475 | 5′UTR | a-716g | 0.37 | 0.71 |
| 5′UTR | a-584g | 0.70 | 0.32 | |
| Ex 3 (I60) | C180T | 0.09 | 0.01 | |
| Ex 7 (I218V) | A652G | 0.03 | – | |
| In 7 | c106t | 0.08 | – | |
| Ex 8 (M258V) | A772G | 0.17 | – | |
| Ex 8 (V264M) | G790A | 0.10 | 0.02 | |
| In 9 | a44c | 0.17 | – | |
| In 9 | c422t | 0.14 | 0.67 | |
| In 10 | t109c | 0.84 | 0.29 | |
| In 11 | a63g | 0.17 | – | |
| rs6596473 | In 13 | c2515g | 0.54 | 0.31 |
| rs6596471 | In 14 | t2088c | 0.54 | 0.28 |
Only variants observed in two or more subjects are included in the table. Nine singletons were observed at the following positions (either in AA or CA): 5′UTR g-219a (AA); Exon 6 (V156) C468T (CA); Intron 7 g59a (CA); Intron 9 g188t (CA); Exon 10 (G359) C1077G (CA); Intron10 c92g (AA); Exon 11 (A421S) G1261T (AA); Intron 12 t49c (CA); Intron 12 c389t (CA)
adb-SNP numbers are pending for new SNPs
bFor exonic SNPs, the numerical designation refers to the nucleotide position in the published cDNA sequence relative to the first nucleotide of the initiation codon, ATG. For intronic SNPs, the numerical designation refers to the base sequence of the specific intron only
cAllele frequencies are based on the sequence analysis of both sets of control subjects (n = 55 for CA and n = 48 for AA). In some cases, the minor allele in CA is the major allele in AA and the reciprocal has also been observed
Fig. 1.
Linkage disequilibrium for SNPs in the membrane transport gene SLC23A1
Fig. 2.
Uptake of ascorbic acid (50 and 300 μM) in Xenopus laevis oocytes 3 days after injection of 16 ng cRNA encoding different SVCT1 genotypes. No significant differences were observed
Acknowledgments
Nutrigenomics New Zealand is a collaboration between AgResearch Limited, Crop and Food Research, HortResearch and the University of Auckland and is funded by the Foundation for Research, Science and Technology.
Footnotes
Peter Eck and Hans Christian Erichsen shared first authorship, Stephen J. Chanock and Mark Levine shared senior authorship.
References
- 1.Wang H, Dutta B, Huang W et al (1999) Human Na(+)-dependent vitamin C transporter 1 (hSVCT1): primary structure, functional characteristics and evidence for a non-functional splice variant. Biochim Biophys Acta 1461:1–9 [DOI] [PubMed]
- 2.Stratakis CA, Taymans SE, Daruwala R, Song J, Levine M (2003). Mapping of the human genes (SLC23A2 and SLC23A1) coding for vitamin C transporters 1 and 2 (SVCT1 and SVCT2) to 5q23 and 20p12, respectively [in process citation]. J Med Genet 37:E20 [DOI] [PMC free article] [PubMed]
- 3.Eck P, Erichsen HC, Taylor JG et al (2004) Comparison of the genomic structure and variation in the two human sodium-dependent vitamin C transporters, SLC23A1 and SLC23A2. Hum Genet 115(4):285–294 [DOI] [PubMed]
- 4.Tsukaguchi H, Tokui T, Mackenzie B et al (1999) A family of mammalian Na+-dependent L-ascorbic acid transporters. Nature 399:70–75 [DOI] [PubMed]