Abstract
Pre-mRNA splicing is among the last known nuclear events before export of mature mRNA to the cytoplasm. At present, it is not known whether splicing and mRNA export are biochemically coupled processes. In this study, we have injected pre-mRNAs containing a single intron or the same mRNAs lacking an intron (Δi-mRNAs) into Xenopus oocyte nuclei. We find that the spliced mRNAs are exported much more rapidly and efficiently than the identical Δi-mRNAs. Moreover, competition studies using excess Δi-mRNA indicate that different factor(s) are involved in the inefficient export of Δi-mRNA vs. the efficient export of spliced mRNA. Consistent with this conclusion, spliced mRNA and Δi-mRNA, though identical in sequence, are assembled into different messenger ribonucleoprotein particles (mRNP) in vitro. Strikingly, the mRNA in the spliced mRNP, but not in the Δi-mRNP, is exported rapidly and efficiently. We conclude that splicing generates a specific nucleoprotein complex that targets mRNA for export. Our results, revealing a link between splicing and efficient mRNA export, may explain the reports that an intron is required for efficient expression of many protein-coding genes in metazoans.
Expression of protein-coding genes in eukaryotes requires synthesis and processing of pre-mRNA in the nucleus followed by transport of the mature mRNA to the cytoplasm for translation. The steps in pre-mRNA processing include capping at the 5′ end, removal of introns by splicing, and cleavage and polyadenylation at the 3′ end. In contrast to the other events in gene expression, little is known about how mRNA is packaged and exported to the cytoplasm.
Cellular and viral mRNAs that naturally lack introns contain specific cis-acting elements that may promote export (refs. 1–3 and references therein). However, the vast majority of metazoan mRNAs, which are derived from genes containing introns, do not appear to contain specific export sequence elements. The presence of introns retains unspliced pre-mRNAs in the nucleus because of an association with spliceosomal components (4–6). This selective retention is thought to prevent the accumulation of potentially deleterious unspliced pre-mRNAs in the cytoplasm. An exception occurs with RNA viruses, in which intron-containing RNAs are exported during the viral life cycle. In complex retroviruses, such as HIV-1, the virally encoded protein Rev binds to a specific sequence element (RRE) and mediates export of the intron-containing RNA (7, 8). In simpler retroviruses, such as simian virus type D, a cellular factor (TAP) binds to a specific cis-acting element (CTE) in the viral RNA to mediate its export (9–13).
Although significant progress has been made in understanding viral mRNA export, much less is known about the export of cellular mRNAs, especially in metazoans. The prevailing model for this process envisions that shuttling heterogeneous nuclear ribonucleoprotein (hnRNP) proteins bind to mRNAs in the nucleus and mediate their export (for reviews, see refs. 14–17). However, given that an export complex containing mRNA has yet to be identified and characterized biochemically, it remains to be established whether hnRNP proteins are critical components of the export machinery.
Genetic studies in yeast have identified several proteins, including Mex67p, Gle1p, Gle2p, and an RNA helicase, Dbp5p, that are specifically required for mRNA export (12, 18–24). Significantly, TAP is the metazoan counterpart of Mex67p, consistent with studies indicating that TAP is a cellular mRNA export factor in metazoans (12, 13, 25). Gle1p and Gle2p also have metazoan counterparts involved in mRNA export (hGle1p and hRAE1, respectively) (26–28). Finally, specific nucleoporins that function in mRNA export have been identified in yeast as well as metazoans (21, 28–31).
Although splicing is among the last known steps that precedes mRNA export, the possibility that splicing is biochemically coupled to mRNA export has not been investigated. Indeed, mRNA export in metazoans is routinely studied by using RNA transcripts of cDNAs that are derived from genes that normally contain introns (9, 10, 32–37). Thus, the potential involvement of splicing in export was not addressed in these studies. Here, we provide direct evidence that splicing and mRNA export are functionally coupled. In addition, we show that the mechanistic basis for this coupling is the formation of a specific nucleoprotein complex that targets the spliced mRNA for rapid and efficient export.
Materials and Methods
Plasmids.
Plasmids encoding AdML (adenovirus major late) pre-mRNA, Ftz (Fushi tarazu) pre-mRNA, AdMLΔ5′, and AdMLΔ5′Δ3′ were described (38–41). Plasmids encoding the corresponding Δi-mRNAs were constructed by exact deletion of the intron for each pre-mRNA. pUC19-U6 plasmid was a gift from M. Moore (42).
In Vitro Splicing.
Pre-mRNAs or Δi-mRNAs (50 ng) were incubated for 1 hr in standard splicing reaction mixtures (25 μl) containing 30% HeLa cell nuclear extract (43) and 12% Xenopus nuclear extract (44). Data similar to that shown in Figs. 3 and 4 were obtained in HeLa nuclear extracts alone, except that the splicing is more rapid and efficient in the HeLa/Xenopus extract than in HeLa extract alone. For native gel analysis, samples were mixed with glycerol loading dye (without heparin) and fractionated on a 1.5% low-melting-point agarose gel in 0.5× TBE (90 mM Tris/90 mM boric acid/2.5 mM EDTA, pH 8.3; 6 V/cm at 4°C). Gel slices containing each complex were excised from a parallel lane, and total RNA was analyzed on a denaturing 15% polyacrylamide gel. Complexes were isolated by gel filtration as described (38).
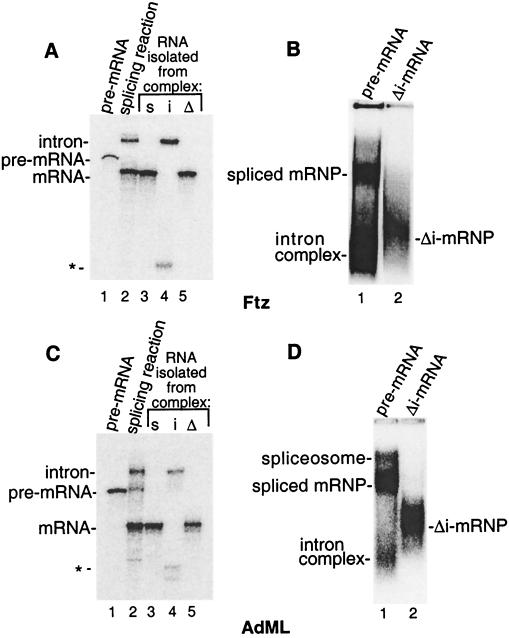
Figure 3.
Distinct complexes assemble on spliced mRNA vs. the identical Δi-mRNA. (A) Ftz pre-mRNA was incubated under splicing conditions for 1 hr, and then total RNA was fractionated on a 15% denaturing gel (lane 2). Pre-mRNA run as a marker is shown in lane 1. In lanes 3–5, total RNA was extracted from the spliced mRNP (s), intron complex (i), or Δi-mRNP (Δ) indicated in B and fractionated on the denaturing gel. The asterisk indicates a breakdown product of the lariat intron. (B) Ftz pre-mRNA (lane 1) or Δi-mRNA (lane 2) was incubated under splicing conditions as in A and fractionated on a native gel. (C and D) The same as A and B except using AdML pre-mRNA and Δi-mRNA. Note that the AdML intron complex is faint because the intron degrades. RNA analysis showed that the band above the spliced mRNP is the spliceosome (D, lane 1). Note that Ftz pre-mRNA is spliced so efficiently that the Ftz spliceosome is almost undetectable on the native gel (B, lane 1). As expected, the spliceosome is detected at higher levels when splicing reactions are incubated for shorter times (data not shown).
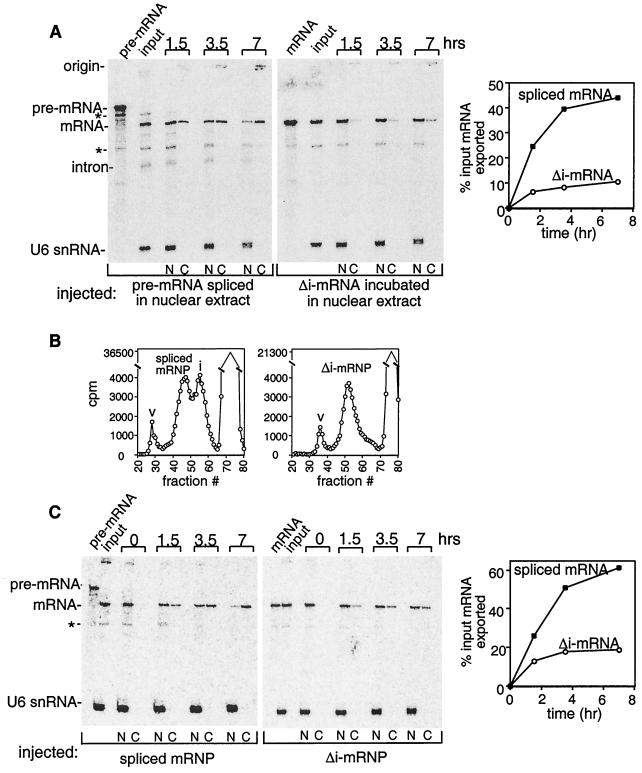
Figure 4.
The spliced mRNP assembled in vitro targets mRNA for export. (A) Ftz pre-mRNA or Δi-mRNA was incubated under splicing conditions, and aliquots of these reactions were mixed with U6 snRNA and then injected into oocyte nuclei. Oocytes were incubated at 18°C for the indicated times and dissected, and total RNA was prepared from the nucleus (N) or cytoplasm (C). RNAs were analyzed on an 8% denaturing polyacrylamide gel. Breakdown products of the pre-mRNA or mRNA are indicated by the asterisks. Bands were quantitated by phosphorimager analysis, and the percent input mRNA exported was plotted (mRNA in cytoplasm/mRNA in the input lane). (B) Ftz pre-mRNA or Δi-mRNA was incubated under splicing conditions. Each reaction mixture then was fractionated on a Sephacryl-S 500 column. The positions of the spliced mRNP, Δi-mRNP, intron complex (i), and void volume (v) are indicated. (C) Same as A, except that the injected complexes first were partially purified by gel filtration (see B).
Xenopus Oocyte Microinjection and RNA Analysis.
Injection of Xenopus laevis oocytes was performed as described (6). 32P-labeled RNAs, complexes, or aliquots of in vitro splicing reactions were mixed in a buffer containing 50 mM NaCl, 10 mM Tris⋅HCl (pH 7.5), 0.6 mM DTT, and 0.5 unit/μl RNAsin and microinjected into nuclei. Oocytes were incubated at 18°C. After dissection, the nucleocytoplasmic distributions of the RNAs were analyzed on denaturing 8% polyacrylamide gels. Blue dextran and U6 small nuclear RNA (snRNA), both of which cannot be exported (45, 46), were used as controls for the accuracy of injection and oocyte dissection.
Results
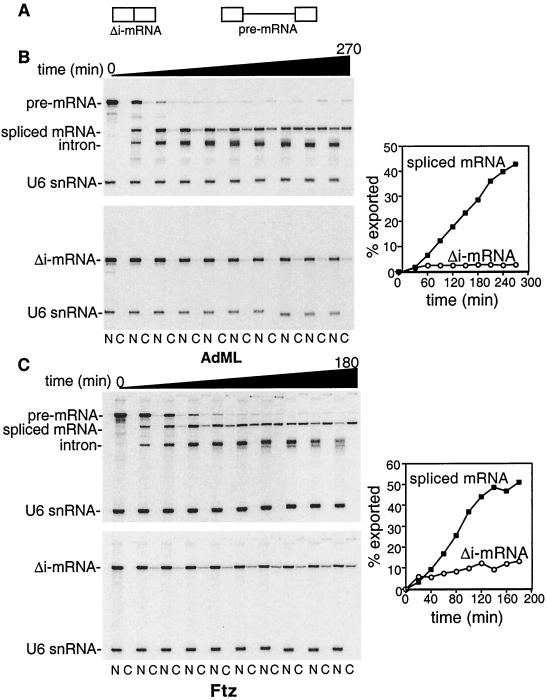
To determine whether splicing affects mRNA export, we synthesized 32P-labeled AdML pre-mRNA containing a single intron and the corresponding Δi-mRNA lacking this intron (see Fig. 1A). These RNAs were injected into Xenopus oocyte nuclei and incubated for the times indicated (Fig. 1B). The nucleocytoplasmic distributions of the Δi-mRNA and in vivo spliced mRNA were compared. As expected from previous studies, AdML pre-mRNA was mostly spliced by ≈60 min (6). Strikingly, the spliced mRNA was exported both much more rapidly and efficiently than the identical Δi-mRNA. Specifically, a low level (<5%) of Δi-mRNA was detected in the cytoplasm by 1 hr after injection, and the levels did not increase significantly over time. In contrast, the levels of spliced mRNA detected in the cytoplasm continued to increase linearly throughout the time course. Similar results were obtained comparing Ftz pre-mRNA and Δi-mRNA (Fig. 1C) as well as two other pre-mRNA/Δi-mRNA pairs (dihydrofolate reductase and β-globin, data not shown). On the basis of these observations, we conclude that splicing targets mRNA for rapid and efficient export. Moreover, the effect of splicing on export appears to be general because efficient export was observed with multiple pre-mRNAs.
Figure 1.
Splicing is required for efficient mRNA export. Equal molar amounts of the indicated pre-mRNAs or the corresponding Δi-mRNAs were mixed with U6 snRNA and injected into oocyte nuclei. After incubation at 18°C for the times indicated, oocytes were dissected, and the nucleocytoplasmic distribution of total RNA was analyzed. Data were quantitated by PhosphorImager. The percent mRNA exported was determined by calculating the fraction of mRNA in the cytoplasm relative to the total mRNA at the zero time point (for Δi-mRNAs) or to the total mRNA at the earliest time point when maximal spliced mRNA is present (the 90-min time point for AdML and 60-min time point for Ftz). Note that the graphs for both the Δi-mRNA and the spliced mRNA are plotted beginning with the actual zero time point shown on the gels. Thus, the kinetics of spliced mRNA export are actually faster than indicated in the graphs because the pre-mRNA is spliced before export.
Evidence That Export of Spliced mRNAs Requires Distinct Factors.
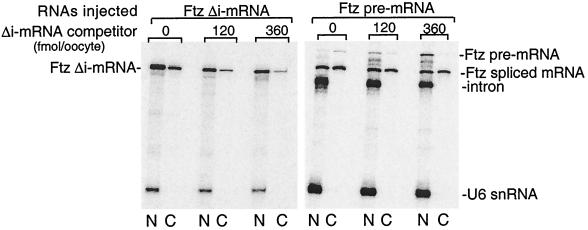
Given the observation that most metazoan mRNAs are generated by splicing and our observation that Δi-mRNAs are exported inefficiently relative to spliced mRNAs, the biological significance of Δi-mRNA export is not clear. Nevertheless, it is possible that the inefficient export of Δi-mRNAs involves the same factors and occurs by the same mechanism used for the export of spliced mRNAs. Several distinct pathways of RNA export have been defined by using different classes of RNAs as competitors, including tRNA, snRNAs, and Δi-mRNAs (9, 33, 35). To determine whether different factors are involved in the export of Δi-mRNAs vs. spliced mRNAs, we first carried out an export assay with excess unlabeled Ftz Δi-mRNA as a competitor. (We note that pre-mRNAs cannot be used as competitors because they inhibit splicing.) As shown in Fig. 2 and consistent with previous work (9, 33, 35), increasing levels of cold Ftz Δi-mRNA competitively inhibit export of the labeled Ftz Δi-mRNA in a dose-dependent manner. Thus, a factor(s) involved in export of Ftz Δi-mRNA is titrated by the competitor. In the case of Ftz pre-mRNA, increasing levels of the Ftz Δi-mRNA competitor decrease the efficiency of splicing in a dose-dependent manner, as indicated by the accumulation of pre-mRNA in the nucleus (Fig. 2). However, the spliced mRNA is exported about as efficiently as it is in the absence of competitor (Fig. 2). We conclude that Δi-mRNA competitively inhibits export of Δi-mRNA, but not export of the identical mRNA generated by splicing. Thus, export of the spliced mRNA requires distinct factors that cannot be titrated by Δi-mRNA. These factors may account for the rapid and efficient export of spliced mRNA that is not observed with Δi-mRNA.
Figure 2.
Δi-mRNA competitively inhibits Δi-mRNA but not spliced mRNA export. Ftz pre-mRNA or the corresponding Δi-mRNA was coinjected into oocyte nuclei with the indicated amounts of Ftz Δi-mRNA competitor. Oocytes were incubated at 18°C for 1 hr, and the nucleocytoplasmic distribution of the spliced vs. Δi-mRNA was analyzed.
Splicing Promotes Export by Generating a Specific Nucleoprotein Complex.
To further address whether Δi-mRNA and spliced mRNA export involve distinct factors, we analyzed complex assembly on these RNAs. To do this, Ftz pre-mRNA (Fig. 3A, lane 1) was incubated in nuclear extract to generate spliced mRNA and the lariat intron (Fig. 3A, lane 2), and an aliquot of this reaction mixture was fractionated on a native gel. As shown in Fig. 3B, two main complexes were detected (lane 1). The faster mobility complex contained the lariat intron, whereas the slower mobility complex contained the spliced mRNA (Fig. 3A, lanes 3 and 4). When Ftz Δi-mRNA was incubated in nuclear extract under identical conditions, a complex with very different mobility than the spliced mRNP was detected on the native gel (Fig. 3B, compare lanes 1 and 2). We conclude that mRNA generated by splicing in vitro is assembled into a complex distinct from that assembled on the identical Δi-mRNA. Similar results were obtained with three other pre-mRNA/Δi-mRNA pairs (e.g., AdML, Fig. 3 C and D, and data not shown), indicating that formation of these complexes is general. Note that both the spliced mRNA and the intron are released from the spliceosome that migrates just above the spliced mRNP (Fig. 3D, lane 1, and see legend).
We next asked whether the differences in the spliced mRNP and the Δi-mRNP affect the export behavior of the RNAs in these complexes. The pre-mRNA or the corresponding Δi-mRNA was incubated in nuclear extract, and then an aliquot of each reaction containing either the spliced mRNP or the Δi-mRNP was injected into Xenopus oocyte nuclei (Fig. 4A). Significantly, the mRNA in the spliced mRNP was exported more rapidly and efficiently than the identical mRNA in the Δi-mRNP.
To determine whether the complexes could be isolated from the nuclear extract and still retain their export behaviors, the complexes were partially purified by gel filtration (47). Consistent with the native gel pattern (Fig. 3), the spliced mRNP and the intron complex were detected in two discrete peaks, with the spliced mRNP eluting as a larger particle than the intron complex (Fig. 4B). The spliced mRNP eluted between the spliceosome and the hnRNP complex (H) (data not shown), consistent with the conclusion that the spliced mRNP is a specific nucleoprotein complex released from the spliceosome. As expected from the native gel analysis, the Δi-mRNP eluted as a single peak (Fig. 4B).
The gel filtration-isolated spliced mRNP and Δi-mRNP then were injected into oocyte nuclei, and the export efficiencies of the mRNA in these complexes were compared. Importantly, export from the partially purified spliced mRNP is still significantly more efficient than that from the isolated Δi-mRNP (Fig. 4C). Moreover, the spliced mRNP retains similar export efficiency before and after gel filtration (compare Fig. 4 A and C), indicating that the features of this complex critical for export remain intact. The spliced mRNP may contain specific factors that promote export, and/or splicing may prevent the binding of factors that retain the mRNA in the nucleus. Similarly, the Δi-mRNP may contain specific retention factors or lack factors that promote export. Export from the Δi-mRNP is slightly more efficient after, compared with before, partial purification (compare Fig. 4 A and C). It is possible that retention factors dissociate during isolation of this complex.
Together, our data indicate that splicing generates a specific, isolable complex that promotes rapid and efficient mRNA export. This complex is either an “exportosome” or a functional precursor to such an export complex. Because most mRNAs in metazoans are generated by splicing, and splicing promotes export, we conclude that the spliced mRNP, and not the Δi-mRNP, is the biologically relevant export substrate in metazoans.
Discussion
In this study, we provide direct evidence that splicing and mRNA export are biochemically coupled processes. Specifically, we have shown that mRNA generated by splicing in vivo is exported rapidly and efficiently compared with the corresponding mRNA lacking an intron. The effect of splicing on export is general as it was observed with multiple RNAs. Moreover, the effect is direct because the export substrates that we used contained a single intron, lacked a polyadenylation site, and were transcribed in vitro. Thus, the enhanced export is not due to indirect effects such as stimulation of splicing of other introns, stimulation of polyadenylation, or release of mRNA from the site of transcription.
Studies over the past 20 years have revealed numerous examples of recombinant proteins whose expression in mammalian cells requires the presence of an intron (e.g., refs. 48–56). Specifically, mRNAs that are transcribed from cDNAs are expressed poorly relative to the same mRNAs transcribed from a gene containing introns. Indeed, many commercially available expression vectors contain an intron for this reason. It is possible that our data, linking splicing to export, explains the intron requirement. Some recently developed expression vectors, however, do not contain an intron. For example, many cDNAs can be expressed from vectors containing the cytomegalovirus (CMV) promoter. However, this expression relies on the presence of a strong simian virus 40 poly(A) site (R. Davis, personal communication). Thus, it is possible that polyadenylation targets the Δi-mRNA for efficient export. Finally, even though not strictly required, insertion of an intron into CMV expression cassettes does significantly increase gene expression in the cases that have been examined (e.g., refs. 57–60).
Our in vitro studies revealed that the mechanistic basis for the link between splicing and export is the formation of a specific nucleoprotein complex that targets the mRNA for rapid and efficient export. This complex does not assemble on the identical Δi-mRNA, indicating that specific factors that potentiate export may be recruited to the mRNA during splicing. Among possible candidates are members of the SR family of splicing factors (reviewed in refs. 61 and 62). SR proteins are specifically bound to the pre-mRNA during splicing and remain bound to the mRNA after splicing is completed (reviewed in refs. 61–63). Significantly, some members of the SR family recently were shown to shuttle between the nucleus and cytoplasm (64). In addition to specific factors that may bind to the mRNA during splicing, it is also possible that proteins are selectively modified during the splicing reaction, and this modification may target the mRNA for export. Purification of the spliced mRNP and analysis of its protein components should lead to important insights into how the complex promotes export.
Acknowledgments
We are indebted to J. Newport and J. Walter for useful discussions and for suggesting and providing technical assistance with the Xenopus extracts. We also thank J. Biggers, M. Mercola, and M. Kirschner for the equipment used in this study and D. Melton and M. Whitman for assistance in setting up the microinjection assay. We are grateful to M. Rosbash, K. Chua, Z. Zhou, K. N. Clouse, and B. Graveley for useful discussions and comments on the manuscript.
Abbreviations
- AdML
adenovirus major late
- Ftz
Fushi tarazu
- Δi-mRNA
mRNA lacking an intron
- snRNA
small nuclear RNA
- hnRNP
heterogeneous nuclear ribonucleoprotein
- mRNP
messenger ribonucleoprotein particle
References
- 1.Huang Y, Carmichael G G. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Z M, Yen T S. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Mertz J E. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 4.Chang D D, Sharp P A. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 5.Legrain P, Rosbash M. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 6.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 7.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saavedra C, Felber B, Izaurralde E. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 11.Kang Y, Cullen B R. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 14.Izaurralde E, Adam S. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 15.Nakielny S, Dreyfuss G. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 16.Lee M S, Silver P A. Curr Opin Genet Dev. 1997;7:212–219. doi: 10.1016/s0959-437x(97)80131-1. [DOI] [PubMed] [Google Scholar]
- 17.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 18.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy R, Watkins J L, Wente S R. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy R, Wente S R. Nature (London) 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 22.Snay-Hodge C A, Colot H V, Goldstein A L, Cole C N. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng S S, Weaver P L, Liu Y, Hitomi M, Tartakoff A M, Chang T H. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues J P, Boscheron C R G, Wilm M, Seraphin B, Carmo-Fonseca M, Izaurralde E. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung J U, Hurt E. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watkins J L, Murphy R, Emtage J L, Wente S R. Proc Natl Acad Sci USA. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraemer D, Blobel G. Proc Natl Acad Sci USA. 1997;94:9119–9124. doi: 10.1073/pnas.94.17.9119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard C E, Fornerod M, Kasper L H, van Deursen J M. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailer S M, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. EMBO J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastos R, Lin A, Enarson M, Burke B. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ullman K S, Shah S, Powers M A, Forbes D J. Mol Biol Cell. 1999;10:649–664. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 33.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izaurralde E, Kutay U, von Kobbe C, Mattaj I W, Gorlich D. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasquinelli A E, Powers M A, Lund E, Forbes D, Dahlberg J E. Proc Natl Acad Sci USA. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett M, Michaud S, Kingston J, Reed R. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 39.Reed R, Maniatis T. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 40.Michaud S, Reed R. Genes Dev. 1993;7:1008–1020. doi: 10.1101/gad.7.6.1008. [DOI] [PubMed] [Google Scholar]
- 41.Das R, Reed R. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolff T, Bindereif A. Genes Dev. 1993;7:1377–1389. doi: 10.1101/gad.7.7b.1377. [DOI] [PubMed] [Google Scholar]
- 43.Krainer A R, Maniatis T, Ruskin B, Green M R. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 44.Walter J, Sun L, Newport J. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- 45.Hamm J, Mattaj I W. EMBO J. 1989;8:4179–4187. doi: 10.1002/j.1460-2075.1989.tb08603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terns M P, Dahlberg J E, Lund E. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- 47.Reed R. Proc Natl Acad Sci USA. 1990;87:8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchman A R, Berg P. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chung S, Perry R P. Mol Cell Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng T L, Li Y, Johnson L F. Nucleic Acids Res. 1989;17:645–658. doi: 10.1093/nar/17.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gasser C S, Simonsen C C, Schilling J W, Schimke R T. Proc Natl Acad Sci USA. 1982;79:6522–6526. doi: 10.1073/pnas.79.21.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hamer D H, Leder P. Cell. 1979;18:1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- 53.Jonsson J J, Foresman M D, Wilson N, McIvor R S. Nucleic Acids Res. 1992;20:3191–3198. doi: 10.1093/nar/20.12.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuberger M S, Williams G T. Nucleic Acids Res. 1988;16:6713–6724. doi: 10.1093/nar/16.14.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rafiq M, Suen C K, Choudhury N, Joannou C L, White K N, Evans R W. FEBS Lett. 1997;407:132–136. doi: 10.1016/s0014-5793(97)00325-6. [DOI] [PubMed] [Google Scholar]
- 56.Ryu W S, Mertz J E. J Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee A, Suh Y S, Sung J H, Yang S H, Sung Y C. Mol Cells. 1997;7:495–501. [PubMed] [Google Scholar]
- 58.Simari R, Yang Z Y, Ling X, Stephan D, Perkins N D, Nabel G J, Nabel E G. Mol Med. 1998;4:700–706. [PMC free article] [PubMed] [Google Scholar]
- 59.DeYoung M B, Zamarron C, Lin A P, Qiu C, Driscoll R M, Dichek D A. Hum Gene Ther. 1999;10:1469–1478. doi: 10.1089/10430349950017806. [DOI] [PubMed] [Google Scholar]
- 60.Yew N, Wysokenski D, Wang K, Zeiger R, Marshall J, McNeilly D, Cherry M, Osburn W, Cheng S. Hum Gene Ther. 1997;8:575–584. doi: 10.1089/hum.1997.8.5-575. [DOI] [PubMed] [Google Scholar]
- 61.Burge C B, Tuschl T H, Sharp P A. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 525–560. [Google Scholar]
- 62.Fu X D. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 63.Reed R. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 64.Caceres J F, Screaton G R, Krainer A R. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]